
Biochemistry Procedure-oriented Ontology: A Case Study
Mohammed Alliheedi
1,2
, Yetian Wang
2
and Robert E. Mercer
2,3
1
Department of Computer Science, Al Baha University, Prince Mohammad Bin Saud, Al Bahah 65527, Saudi Arabia
2
David Cheriton School of Computer Science, University of Waterloo, 200 University Ave., Waterloo, Ontario, Canada
3
Department of Computer Science, The University of Western Ontario, 1151 Richmond St., London, Ontario, Canada
Keywords:
Experimental Procedure, Procedural Steps, Sequence of Steps, Biomedical Ontology, Formal Ontology,
Knowledge Representation.
Abstract:
Ontologies must provide the entities, concepts, and relations required by the domain being represented. The
domain of interest in this paper is the biochemistry experimental procedure. The ontology language being
used is OWL-DL. OWL-DL was adopted due to its well-balanced flexibility among expressiveness (e.g., class
description, cardinality restriction, etc.), completeness, and decidability. These procedures are composed of
procedure steps which can be represented as sequences. Sequences are composed of totally ordered, partially
ordered, and alternative subsequences. Subsequences can be represented with two relations, directlyFollows
and directlyPrecedes that are used to represent sequences. Alternative subsequences can be generated by
composing a oneOf function in OWL-DL, referred to it as optionalStepOf in this work, which is a simple
generalization of exclusiveOR. Alkaline Agarose Gel Electrophoresis, a biochemistry procedure, is described
and examples of these subsequences are provided.
1 INTRODUCTION
Ontologies provide entities (known as individuals in
some ontological languages) and concepts, and rela-
tions among those entities and concepts. Ontologies
must provide relations that are required by the domain
being represented. Our interest is centered on the bio-
chemistry domain, the experimental methodology as-
pect, in particular.
A number of biologically oriented ontologies have
been created, one of the best known is the Gene On-
tology (GO) (Ashburner et al., 2000). Others have
been developed for a variety of other purposes. They
are discussed in detail in the next section. Most of
these ontologies describe a set of concepts and cate-
gories in the biological domain that shows their prop-
erties and the relations between them.
The type of domain that we are attempting to
represent consists of procedures, experimental pro-
cedures, in particular. Procedures are sequences of
procedure steps (simply, steps, henceforth). Some
ontologies provide descriptions of steps (Soldatova
et al., 2013). To the best of our knowledge no current
biologically oriented ontology represents sequences
of steps. An important aspect of the steps in a pro-
cedure is that they immediately follow one another.
‘Directly follows’ (and ‘directly precedes’) is an in-
transitive relation (i.e., if B directly follows A, and if
C directly follows B, then C does not directly follow
A). Transitive relations are the norm in the current bi-
ologically oriented ontologies (e.g., the omnipresent
‘subclass’ relation; ‘proper part of’, ‘precedes’ and ‘is
causally related to’ ((Dumontier et al., 2014), Figures
6 and 9)).
Procedures can contain sequences of steps that are
totally ordered (i.e., the steps must be done one after
the other in the sequence specified), steps that can be
partially ordered (i.e., subsequences of steps that can
be done in any order), and alternative subsequences
of steps (i.e., only one of the alternatives is done). In
addition to the intransitive relations ‘directly follows’
and ‘directly precedes’ our contribution also includes
these three types of sequence orderings.
Descriptions of experimental procedures exist in
scientific writing. The scientific domain of interest to
us is biochemistry. An important type of information
contained in the Method section of biochemistry ar-
ticles are references to standard biochemistry exper-
iment procedures. These protocols, which typically
involve several steps, are described in detail in man-
uals of standard biochemistry experiment procedures
(Boyer, 2012; Sambrook and Russell, 2001). In this
164
Alliheedi, M., Wang, Y. and Mercer, R.
Biochemistry Procedure-oriented Ontology: A Case Study.
DOI: 10.5220/0008167101640173
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 164-173
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

paper, we propose a biochemistry procedure-oriented
ontology that explicitly identifies all of the steps of an
experimental procedure and provides the relations be-
tween the steps of an experimental procedure. A case
study investigates one experimental procedure, Alka-
line Agarose Gel Electrophoresis, that exists in the
manual of standard biochemistry experimental proce-
dures.
2 RELATED WORK
Developing ontologies has become increasingly cru-
cial in the biomedical domain in general (Rosse and
Mejino Jr, 2003). Several ontologies have been de-
veloped in recent years such as the Gene Ontology
(Ashburner et al., 2000), the Ontology for Chem-
ical Entities of Biological Interest (ChEBI) (Degt-
yarenko et al., 2007), the Ontology for Biomedical In-
vestigations (OBI) (Bandrowski et al., 2016), and the
Foundational Model of Anatomy (FMA) (Rosse and
Mejino Jr, 2003). Mainly, the goal of these ontologies
is to provide definitive controlled terminologies that
describe entities in the biomedical genre.
The main aspect of Gene Ontology (GO) is to
provide information that describes gene products us-
ing precisely defined vocabulary (Ashburner et al.,
2000). GO intially used three model organism
databases including FlyBase (FlyBase Consortium,
2003), Mouse Genome Informatics (Blake et al.,
2000; Ringwald et al., 2000), and the saccharomyces
Genome Database (Ball et al., 2000). Recently, the
number of model organism databases has increased
dramatically (Gene Ontology Consortium, 2011).
The Chemical Entities of Biological Interest on-
tology (ChEBI) is a lexicon of molecular entities
concerned with small molecules (Degtyarenko et al.,
2007). To create ChEBI, data from several resources
(e.g., IntEnz (Fleischmann et al., 2004), KEGG
COMPOUND (Kanehisa et al., 2006), and the Chem-
ical Ontology) were used. ChEBI used various rela-
tions to describe the relationships between ontology
entities. These relations include relations required
by ChEBI (e.g., ‘is conjugate acid of’, and ‘is tau-
tomer of’) as well as relations which are defined by
the Relations Ontology
1
(e.g., ‘is a’ and ‘is part of’).
The Ontology for Biomedical Investigations (OBI),
http://purl.obolibrary.org/obo/obi, (Bandrowski et al.,
2016), a resource for annotating biomedical inves-
tigations, provides standard tools to represent study
design, protocols and instrumentation used, the data
generated and the types of analysis performed on
1
http://www.obofoundry.org/ontology/ro.html
the data. Several ontologies (Courtot et al., 2008),
(Brinkman et al., 2010), (Zheng et al., 2013), (Solda-
tova et al., 2013), (Dumontier et al., 2014) are based
on the OBI ontology. These ontologies are closest to
our interest in biochemistry procedures.
A work predating the above list, (Soldatova and
King, 2006), proposes EXPO, an ontology of scien-
tific experiments, in general. It remains a descriptive
ontology, providing a detailed description of various
aspects of scientific experiments and how they are re-
lated.
Descriptions of experimental processes are pro-
vided by OBI, and three real-world applications are
discussed in (Brinkman et al., 2010). Some of the
relations in these applications (e.g., inputs, outputs,
etc.) come very close to our purpose here. The beta
cell genomics application ontology (BCGO) (Zheng
et al., 2013) also uses OBI, but it tends to be a more
descriptive ontology than some of the others that use
OBI, but some of the relations in RO, the relation on-
tology (Smith et al., 2005), that are used (e.g., pro-
duces, translate to) do have an ordering sense.
The two ontologies that are most similar to the
work described below are EXACT (Soldatova et al.,
2013) and the Semanticscience Integrated Ontology
(Dumontier et al., 2014). Both are motivated by a
need to describe scientific protocols and experiments.
Where they differ from what we are proposing is that
they describe sets of actions in scientific protocols and
experiments, whereas we are proposing to represent
sequences of actions, or steps in a procedure, if you
like. Relations that describe orderings of actions (e.g.,
‘precedes’ (Dumontier et al., 2014)) are not applica-
ble to sequences since these relations are transitive.
The Molecular Methods Database (MolMeth) is a
database which contains scientific protocol ontologies
that conform to a set of laboratory protocol standards
(Klingstr
¨
om et al., 2013).
Other ontologies describe general concepts that
are useful to a biochemistry procedure-oriented on-
tology include: Ontologies consist of process such as
(Lenat et al., 1985) and (Schlenoff et al., 2000), on-
tology for units of measure (Rijgersberg et al., 2013),
classification of scenarios and plans (CLASP) (De-
vanbu and Litman, 1996), and materials ontology
(Ashino, 2010). Foundational theories such as pro-
cess calculus and regular grammar are essential for
the formalization of procedure-oriented ontologies.
Biochemistry Procedure-oriented Ontology: A Case Study
165

3 PROCEDURE-ORIENTED
ONTOLOGY
We propose a framework for procedure-oriented on-
tologies that explicitly identify all steps of an experi-
mental procedure and provide a set of relations to de-
scribe the relationships between the steps of an exper-
imental procedure. The novelty of this approach is to
allow creating a sequence of events (or steps in a pro-
cedure) using the ontological concept of “something
occurs before”. To accomplish this we need to have
an ontological concept of “sequence”. This is very
significant concept because one cannot simply call a
sequence of events “a sequence” unless these events
happen step by step in some sort of ordering.
This approach will be used to provide the neces-
sary information about the experimental procedures
for Knowledge Base systems with the required knowl-
edge about experimental processes. There are man-
uals of standard procedures in biochemistry (Boyer,
2012; Sambrook and Russell, 2001) which in turn will
help in building ontologies.
3.1 Classes and Properties
The proposed ontology framework consists of three
core classes: Step, State, and Action.
3.1.1 Step
The Step class (see Figure 1) represents each step
within a procedure. Orderings of each step can be de-
scribed by object properties such as ‘precedes’, ‘fol-
lows’, ‘parallel’, all being transitive. The properties
‘precedes’ and ‘follows’, inverses of each other, indi-
cate the chronological order of the steps. The prop-
erty ‘parallel’ is symmetrical which indicates steps
can happen simultaneously. Intransitive properties
‘directlyPrecedes’ and ‘directlyFollows’ are also used
to describe the ordering of steps. They are subprop-
erties of ‘precedes’ and ‘follows’ respectively. Sim-
ilar to ‘precedes’ and ‘follows’, they are also in-
verses of each other. Therefore, by stating step1.1
‘directlyPrecedes’ step1.2 and step1.2 ‘directlyPre-
cedes’ step1.3, a reasoner will automatically infer that
step1.1 ‘precedes’ step1.2 as well as step1.3. Also,
step1.3 ‘directlyFollows’ step1.2 but only ‘follows’
step1.1, both being inferable by a reasoner. For clean-
liness, we indicate only the ‘precedes’ relation in the
figures presented in this paper.
The structure of the procedure is outlined by
the properties ‘subStepOf’ and ‘optionalStepOf’ in
which both domain and range of the properties are
Step. ‘subStepOf’ indicates that the step(s) must be
Figure 1: Step class and example instances.
Figure 2: State and Action classes.
completed for the completion of the parent step, e.g.,
the triples (step1.1, subStepOf, step1) and (step1.2,
subStepOf, step1) state that step1.1 and step1.2 must
be completed in order to consider step1 to be com-
pleted. Conversely, ‘optionalStepOf’ indicates that
one of the steps (not both) must be completed in or-
der to complete the parent step, e.g., (step1.1a, op-
tionalStepOf, step1.1) and (step1.1b, optionalStepOf,
step1.1) state that one and only one of step1.1a or
step1.1b needs to be completed to complete step1.1.
Figure 1 illustrates a scenario in which all individ-
uals are Step instances. Also, step1 is parallel to step2
while step1.1 must complete before step1.2. Note,
there are no ordering relations between step1.1.1 and
step1.1.2 since they are optional steps of step1.1.
3.1.2 State and Action
The class Step with corresponding properties outlines
the structure of a procedure. The actual process in
each step is represented as states and their associ-
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
166
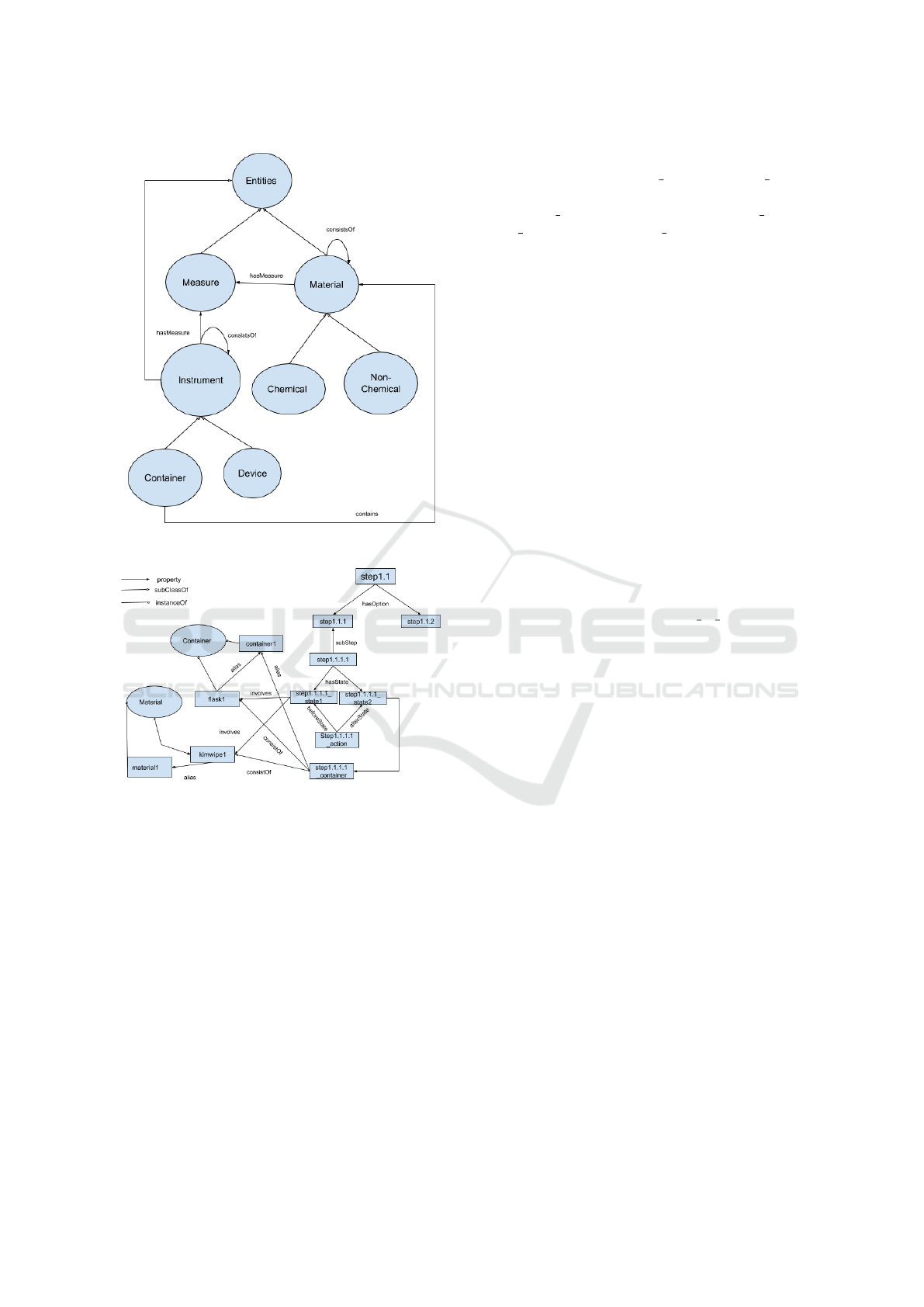
Figure 3: Demonstration of Entities class.
Figure 4: An example of alternative sub-sequences in steps
for preparing the Agarose solution.
ated actions. Each step involves a transition from
state to state via a single or a series of actions, rep-
resented by the classes State and Action (see Fig-
ure 2). State is connected to Step via the property
‘hasState’ and has three subclasses, InitialState, Mid-
State, and FinalState which are connected via proper-
ties such as ‘precedes’ and ‘follows’. InitialState can
only precede a state while FinalState can only follow
another state. Triples (StateX, precedes, StateY) im-
ply (StateY, follows, StateX), and vice versa, since
‘follows’ is an inverse property of ‘precedes’. Fig-
ures 1 and 2 omit ‘follows’ to keep the figures clean.
MidState can be connected to another state with both
‘precedes’ and ‘follows’ properties. Note that a step
has at most one instance of InitialState or FinalState
but may have multiple instances of MidState. For ex-
ample, an instance of Step, step1, may involve two
instances of State, i.e., step1 state1 and step1 state2,
represented by the following triples: (step1, has-
State, step1 state1), (step1, hasState, step1 state2),
(step1 state1, precedes, step1 state2).
3.1.3 Biochemistry Domain Knowledge
States are connected to the Action class via
‘beforeState’ and ‘afterState’, representing the states
before and after an action, respectively. The State
class is also connected to the Entities class (see Figure
3) via the property ‘involves’ which can be expanded
to describe instruments, materials, and devices in-
volved in a specific state. Thus, domain knowledge of
biochemistry can be described by extending the Enti-
ties class. For demonstration purposes, we have only
included selected general concepts related to experi-
mental procedures described in the Case Study. In-
strument includes Container and Device where Con-
tainer ‘contains’ Material which is a class for Chemi-
cal and Non-Chemical materials used in biochemistry
experiment procedures. Compound materials and as-
sembled instruments are represented using the prop-
erty ‘consistsOf’. Instrument and Material can be
connected to the class Measure which is a combina-
tion of numerical values and Unit of Measure, e.g.,
‘10m’ is a measure where the value is 10 with a unit
of measure of ‘meter’ (Rijgersberg et al., 2013). The
Measure class was extended with subclasses to repre-
sent absolute measures (e.g., 10m), range values (e.g.,
5m-10m), and ratio (e.g., 1/2).
3.2 Relations
We first need to examine the types of features that an
experimental procedure needs for its definition.
A procedure is a sequence of steps. These steps
can be totally ordered or partially ordered. Total
ordering needs a means to represent the concept that
one event precedes another event and this relation
needs to be transitive. Because a procedure is a
sequence of steps, there needs to be a means to rep-
resent the relation that one step immediately follows
another step and this relation needs to be intransi-
tive. These relations have been defined for OWL
(McGuinness et al., 2004) and are available from
http://www.ontologydesignpatterns.org/cp/owl/seque
nce.owl. Partial ordering is accomplished simply by
allowing more than one step to follow or to precede
another step.
Finally, we would like to be able to represent a
subsequence of steps and the choice of a subsequence
from one or more possible subsequences. This ‘op-
tionalStepOf’ relation would need to be crafted de-
Biochemistry Procedure-oriented Ontology: A Case Study
167
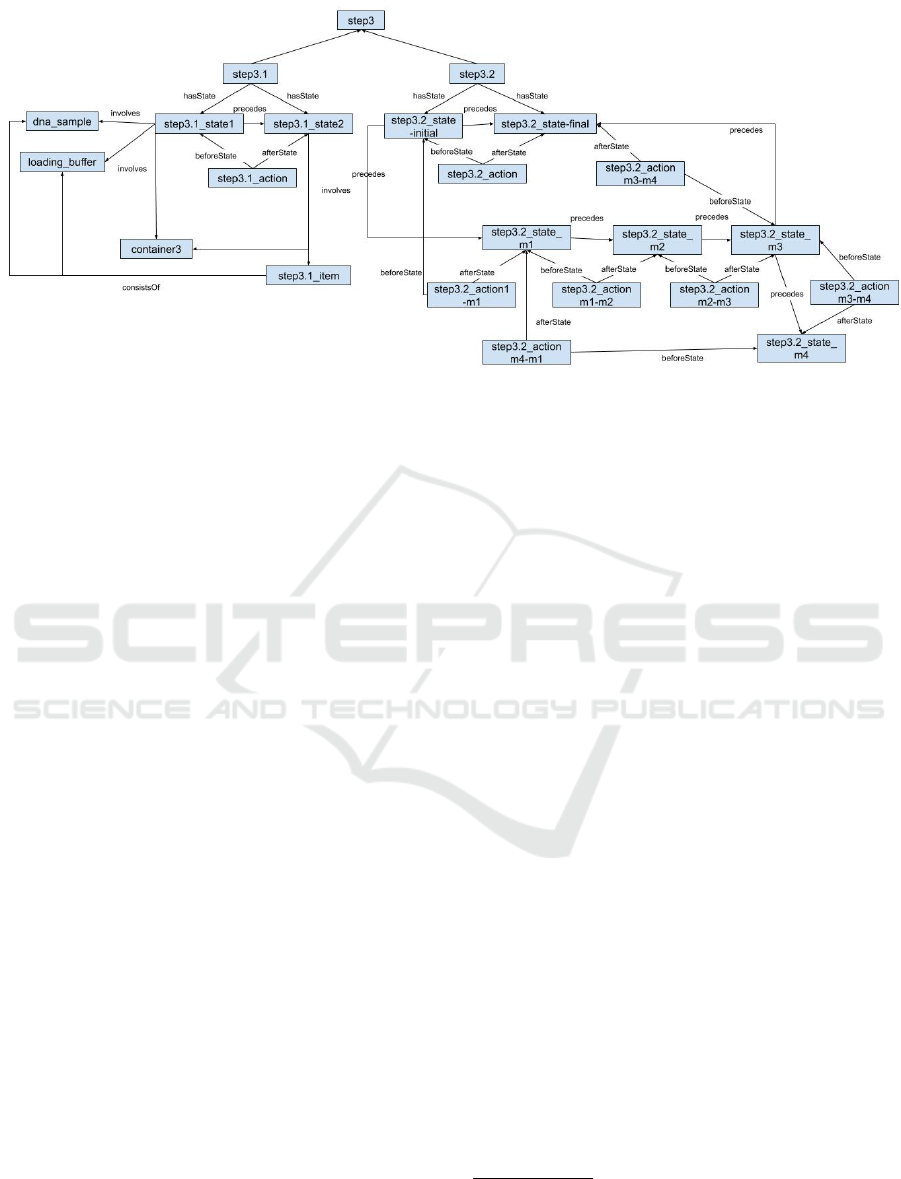
Figure 5: Instances related to Step3 which involves initiating the electrophoresis.
pending on how many choices are available. If two
choices, this relation is simply equivalent to exclu-
sive or otherwise it is simply a generalization of the
exclusive or. We have developed the concept of “pro-
cedure” based on these underlying relations.
4 CASE STUDY
We have designed a procedure-oriented ontology for
Alkaline Agarose Gel Electrophoresis (Sambrook and
Russell, 2001) using the set of relations described in
Section 3. Our motivation is analyzing the text in the
Method section of biochemistry articles. Since the
Method section in biochemistry articles is describing
experimental procedures, these procedures use some
steps that are not explicitly mentioned in the text be-
cause the article is intended for readers who have prior
knowledge of the field. Thus, without knowing this
implicit information, one cannot fully understand all
the steps of experimental procedures. For example, in
order to understand fully the sentence fragment, “the
resulting ca. 900 bp piece was gel purified and lig-
ated using T4 ligase into pUC19” (Carenbauer et al.,
2002), one needs to access the information involved
in gel purification and ligation. Thus, we have moved
to build an ontology that satisfies this requirement.
Figure 10 (see Appendix) shows the first steps
of Alkaline Agarose Gel Electrophoresis that are in-
volved in preparing both the agarose solution and
the DNA samples. Figure 4 describes step1.1, the
preparation of the agarose solution. Basically, step1.1
“adding the appropriate amount of powdered agarose
to a measured quantity of H2O” has two options ei-
ther: step1.1.1 “an Erlenmeyer flask” ‘exclusiveOR’
step1.1.2 “a glass bottle”. So we have a relation
that conveys the choice of using one container or an-
other. So, there is a choice of two sequences of steps:
If step1.1.1 “an Erlenmeyer flask” is selected then
‘directlyFollows’ step1.1.1.1 “loosely plug the neck
of the Erlenmeyer flask with Kimwipes” which in-
volves both initial and final states, action and con-
tainer as seen in Figure 4; else if step1.1.2 “a glass
bottle” is selected then ‘directlyFollows’ step1.1.2.1
“make sure that the cap is loose”
2
. In future steps of
the ontology, the instance Container1 appropriately
refers to the instances of either Erlenmeyer flask or
the glass bottle and material1 refers to the instances
of kimwipes or glass bottle cap. The two main steps
(step1, and step2) shown in Figure 1 are meant to be
partially ordered, that is, they can be performed in any
order (i.e., step1 then step2 or vice versa). In addition,
each one of these main steps consists of several steps
(mini-steps or sub-steps). Note that we only include
describe step 1.1.1 and step 3 in Figures 4 and 5 be-
cause these steps are representative of all other steps
in the procedure.
As one can see, Figure 4 shows a total ordered
sequence. Another example, shown in Figure 5, de-
scribes the instances of step3, step3.1 and step3.2
that are concerned with initiating the electrophore-
sis. Step3.1 is straightforward.
3
Since step3.2 in-
volves a condition to ensure the gel reaches a certain
length, this step requires several MidStates in addi-
tion to both the initial and finial states as is shown
in Table 1. All entities for step3.2 are described in
Table 1. Note that Step3.2 consists of a number of
MidStates which represents waiting until the desired
2
Due to the limited space of the paper, the option
step1.1.2 and its substeps are not included in Figure 4.
3
Due to the limited space of the paper, step3.1 and its
substeps are not described in Table 1.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
168

Table 1: Description of the entities involved in Step3.2.
Subject Property Object Description
step3.2 state initial rdf:type InitialState
involves electrophoresis
involves electrophoresis measure
precedes step3.2 state m1
step3.2 action initial m1 rdf:type TurnOn TurnOn is a subclass of Action
beforeState step3.2 state initial
afterState step3.2 state m1
step3.2 state m1 rdf:type MidState
involves electrophoresis
involves electrophoresis measure
involves bg migrate measure
n
measure for the migration of
bromocresol green
involves bromocresol green
involves gel
precedes step3.2 state m2
step3.2 action m1 m2 rdf:type DoNothing DoNothing is a subclass of Action
beforeState step3.2 state m1
afterState step3.2 state m2
step3.2 state m2 rdf:type MidState
involves bg migrate measure
a measure for the migration of
bromocresol green
involves bromocresol green
involves gel
involves gel length portion
a measure of current length of gel
that the bromocresol green has
migrated to
precedes step3.2 state m3
step3.2 action m2 m3 rdf:type TurnOff
beforeState step3.2 state m2
afterState step3.2 state m3
step3.2 state m3 rdf:type MidState
involves electrophoresis
involves electrophoresis measure
involves gel length portion
a measure of current length of gel
that the bromocresol green has
migrated to, less than 2/3
precedes step3.2 state m4
precedes step3.2 state final
step3.2 action m3 m4 rdf:type Action Put glass plate on gel
beforeState step3.2 state m3
afterState step3.2 state m4
step3.2 state m4 rdf:type MidState
involves gel
involves gel length portion
involves glass plate
step3.2 action m4 m1 rdf:type TurnOn
beforeState step3.2 state m4
afterState step3.2 state m1
step3.2 action m3 final rdf:type Action Put glass plate on gel
beforeState step3.2 state m3
afterState step3.2 state final
step3.2 state final rdf:type FinalState
involves electrophoresis
involves electrophoresis measure
involves gel length portion2
a measure of current length of gel
that the bromocresol green has
migrated to, equal to or more than
2/3
involves bromocresol green
involves gel
Biochemistry Procedure-oriented Ontology: A Case Study
169

amount of migration has been reached (i.e., 2/3 of gel
length). The instance step3.2 state initial and
step3.2 state final are instances of InitialState
and FinalState, respectively. The instances of Mid-
States are step3.2 state m1 to step3.2 state m4,
each representing a middle state described below:
• step3.2 state m1: Electrophoresis power is on
• step3.2 state m2: The state where bromocresol
green is migrating into gel
• step3.2 state m3: Bromocresol green has mi-
grated into gel approximately 0.5-1 cm, the power
of the electrophoresis has been turned off.
• step3.2 state m4: A glass plate has been
placed on top of the gel, bromocresol green has
migrated less than 2/3 of the gel length.
The process is a loop since step3.2 state m4
precedes step3.2 state m1. step3.2 state m4
differs with step3.2 state final in that the
bromocresol green has migrated to the targeted
amount in the latter state. step3.2 state m3
precedes both step3.2 state m4 and
step3.2 state final. An instance of Measure
could be used to track the amount that bromocresol
green has migrated.
4.1 Ontology Queries using SPARQL
We have used SPARQL to extract some domain
knowledge about the experimental procedure of Al-
kaline Agarose Gel Electrophoresis from our frame-
work. Figures 6, 8, 9, and 7 (see Appendix) show
the true power of knowledge representation by auto-
matically extracting the essential information that a
biochemist would use to perform experimental pro-
cedures in a lab. These figures show in a few ex-
amples how much information can be mined from
such a framework with only one experimental proce-
dure. What if all standard experimental procedures
in biochemistry (Boyer, 2012; Sambrook and Rus-
sell, 2001), for example, are modeled and built, one
simply cannot imagine how much time and effort will
be saved, knowing all essential information is just a
few clicks away. Figure 8 shows all of the instru-
ments involved in any state for all steps of the Alka-
line Agarose Gel Electrophoresis procedure whereas
Figure 7 shows a query that returned all materials in-
volved in the procedure. Figure 9 shows a query that
returned the states of step3 and its substeps which
are concerned with measuring the gel length and re-
turned their target values. The ontology was veri-
fied to be consistent using HermiT 1.3.8.3 reasoner
(Shearer et al., 2008).
5 CONCLUSIONS
We have proposed a framework that describes the re-
lations and steps of experimental procedures. This
framework will enrich the knowledge based systems
with necessary information about experimental proce-
dures that a scientist would automatically access such
as instruments (e.g., laboratory centrifuge) and mate-
rials (e.g., buffers). Most importantly, this approach
is an important step toward our ultimate goal to ana-
lyze biomedical articles. This work will be publicly
available for the research community to enhance and
expand upon. Such a work could be beneficial for
various genres that have similar procedure-oriented
characteristics. We also aim to expand our work by
incorporating existing ontologies that are essential to
this domain such as the ontology for units of measure
(Rijgersberg et al., 2013) and the materials ontology
(Ashino, 2010). Certain theoretical ontological mod-
elling of states and empirical observations in science
can be fruitfully incorporated into our ontology in the
future (Masolo et al., 2018).
REFERENCES
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., et al. (2000). Gene ontol-
ogy: tool for the unification of biology. Nature genet-
ics, 25(1):25.
Ashino, T. (2010). Materials ontology: An infrastructure
for exchanging materials information and knowledge.
Data Science Journal, 9:54–61.
Ball, C. A., Dolinski, K., Dwight, S. S., Harris, M. A., Issel-
Tarver, L., Kasarskis, A., Scafe, C. R., Sherlock, G.,
Binkley, G., Jin, H., et al. (2000). Integrating func-
tional genomic information into the saccharomyces
genome database. Nucleic acids research, 28(1):77–
80.
Bandrowski, A., Brinkman, R., Brochhausen, M., Brush,
M. H., Bill Bug and, M. C. C., Clancy, K., Cour-
tot, M., Derom, D., Dumontier, M., Fan, L., Fos-
tel, J., Fragoso, G., Gibson, F., Gonzalez-Beltran, A.,
Haendel, M. A., He, Y., Heiskanen, M., Hernandez-
Boussard, T., Jensen, M., Lin, Y., Lister, A. L., Lord,
P., Malone, J., Manduchi, E., Monnie McGee and,
N. M., Overton, J. A., Parkinson, H., Peters, B.,
Rocca-Serra, P., Ruttenberg, A., Sansone, S.-A.,
Scheuermann, R. H., Schober, D., Smith, B., Solda-
tova, L. N., Christian J. Stoeckert, J., Taylor, C. F.,
Torniai, C., Turner, J. A., Vita, R., Whetzel, P. L., and
Zheng, J. (2016). The ontology for biomedical inves-
tigations. PLoS ONE, 11(4):e0154556.
Blake, J. A., Eppig, J. T., Richardson, J. E., Davisson, M. T.,
Group, M. G. D., et al. (2000). The mouse genome
database (mgd): expanding genetic and genomic re-
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
170

sources for the laboratory mouse. Nucleic Acids Re-
search, 28(1):108–111.
Boyer, R. F. (2012). Biochemistry Laboratory: Modern
Theory and Techniques. Prentice Hall.
Brinkman, R. R., Courtot, M., Derom, D., Fostel, J. M.,
He, Y., Lord, P., Malone, J., Parkinson, H., Peters,
B., Rocca-Serra, P., Ruttenberg, A., Sansone, S.-A.,
Soldatova, L. N., Jr., C. J. S., Turner, J. A., Zheng, J.,
and the OBI consortium (2010). Modeling biomedical
experimental processes with obi. Journal of Biomedi-
cal Semantics, 1 (Suppl 1):S7.
Carenbauer, A. L., Garrity, J. D., Periyannan, G., Yates,
R. B., and Crowder, M. W. (2002). Probing
substrate binding to Metallo-β-Lactamase L1 from
Stenotrophomonas maltophilia by using site-directed
mutagenesis. BMC Biochemistry, 3(1):4.
Courtot, M., Bug, W., Gibson, F., Lister, A. L., Malone,
J., Schober, D., Brinkman, R. R., and Ruttenberg, A.
(2008). The owl of biomedical investigations. In Pro-
ceedings of the Fifth OWLED Workshop on OWL: Ex-
periences and Directions, page 12pp.
Degtyarenko, K., De Matos, P., Ennis, M., Hastings, J.,
Zbinden, M., McNaught, A., Alc
´
antara, R., Dar-
sow, M., Guedj, M., and Ashburner, M. (2007).
Chebi: a database and ontology for chemical enti-
ties of biological interest. Nucleic acids research,
36(suppl 1):D344–D350.
Devanbu, P. T. and Litman, D. J. (1996). Taxonomic plan
reasoning. Artificial Intelligence, 84(1-2):1–35.
Dumontier, M., Baker, C. J., Baran, J., Callahan, A., Che-
pelev, L., Cruz-Toledo, J., Rio, N. R. D., Duck, G.,
Furlong, L. I., Keath, N., Klassen, D., McCusker,
J. P., Queralt-Rosinach, N., Samwald, M., Villanueva-
Rosales, N., Wilkinson, M. D., and Hoehndorf, R.
(2014). The semanticscience integrated ontology (sio)
for biomedical research and knowledge discovery.
Journal of Biomedical Semantics, 5(1):14.
Fleischmann, A., Darsow, M., Degtyarenko, K., Fleis-
chmann, W., Boyce, S., Axelsen, K. B., Bairoch,
A., Schomburg, D., Tipton, K. F., and Apweiler,
R. (2004). Intenz, the integrated relational enzyme
database. Nucleic acids research, 32(suppl 1):D434–
D437.
FlyBase Consortium (2003). The flybase database of the
drosophila genome projects and community literature.
Nucleic acids research, 31(1):172–175.
Gene Ontology Consortium (2011). The gene ontology:
enhancements for 2011. Nucleic acids research,
40(D1):D559–D564.
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F.,
Itoh, M., Kawashima, S., Katayama, T., Araki, M.,
and Hirakawa, M. (2006). From genomics to chemical
genomics: new developments in kegg. Nucleic acids
research, 34(suppl 1):D354–D357.
Klingstr
¨
om, T., Soldatova, L., Stevens, R., Roos, T. E.,
Swertz, M. A., M
¨
uller, K. M., Kala
ˇ
s, M., Lambrix,
P., Taussig, M. J., Litton, J.-E., Landegren, U., and
Bongcam-Rudlof, E. (2013). Workshop on labortory
protocol standards for the molecular methods data-
base. New Biotechnology, 30(2):109–113.
Lenat, D. B., Prakash, M., and Shepherd, M. (1985). Cyc:
Using common sense knowledge to overcome brittle-
ness and knowledge acquisition bottlenecks. AI mag-
azine, 6(4):65–65.
Masolo, C., Botti Benevides, A., and Porello, D. (2018).
The interplay between models and observations. Ap-
plied Ontology, 13(1):41–71.
McGuinness, D. L., Van Harmelen, F., et al. (2004). Owl
web ontology language overview. W3C recommenda-
tion, 10(10):2004.
Rijgersberg, H., Van Assem, M., and Top, J. (2013). Ontol-
ogy of units of measure and related concepts. Seman-
tic Web, 4(1):3–13.
Ringwald, M., Eppig, J. T., Kadin, J. A., and Richardson,
J. E. (2000). Gxd: a gene expression database for the
laboratory mouse: current status and recent enhance-
ments. Nucleic acids research, 28(1):115–119.
Rosse, C. and Mejino Jr, J. L. (2003). A reference on-
tology for biomedical informatics: the foundational
model of anatomy. Journal of biomedical informat-
ics, 36(6):478–500.
Sambrook, J. and Russell, D. W. (2001). Molecular
Cloning: A Laboratory Manual. Cold Spring Harbor
Laboratory Press.
Schlenoff, C., Schlenoff, C., Tissot, F., Valois, J., and Lee,
J. (2000). The process specification language (PSL)
overview and version 1.0 specification. Citeseer.
Shearer, R., Motik, B., and Horrocks, I. (2008). Hermit: A
highly-efficient owl reasoner. In Owled, volume 432,
page 91.
Smith, B., Ceusters, W., Klagges, B., K
¨
ohler, J., Kumar, A.,
Lomax, J., Mungall, C., Neuhaus, F., Rector, A. L., ,
and Rosse, C. (2005). Relations in biomedical ontolo-
gies. Genome Biology, 6(5):R46.
Soldatova, L., King, R., Basu, P., Haddi, E., and Saunders,
N. (2013). The representation of biomedical proto-
cols. EMBnet.journal, 19(B).
Soldatova, L. N. and King, R. D. (2006). An ontology of
scientific experiments. Journal of the Royal Society
Interface, 3(11).
Zheng, J., Manduchi, E., and Jr, C. J. S. (2013). Develop-
ment of an application ontology for beta cell genomics
based on the ontology for biomedical investigations.
In 4th International Conference on Biomedical Ontol-
ogy, pages 62–67.
Appendix
SPARQL Queries
Query1. Return all devices involved in a state of all
steps (1.1, 1.2, 3)
SELECT ? s t e p ? s t a t e ? i te m
WHERE { ? s t e p r d f : t y p e : S t e p .
? s t e p : h a s S t a t e ? s t a t e .
? s t a t e : i n v o l v e s ? it em .
? i te m r d f : t y p e : D ev ic e }
Biochemistry Procedure-oriented Ontology: A Case Study
171

Query2. Return all materials involved in all steps
SELECT ? s t e p ? s t a t e ? i te m
WHERE { ? s t e p r d f : t y p e : S t e p .
? s t e p : h a s S t a t e ? s t a t e .
? s t a t e : i n v o l v e s ? it em .
? i te m r d f : t y p e / r d f s : s u b C la ss Of :
M a t e r i a l }
Query3. Return all instruments involved in all steps
SELECT ? s t e p ? s t a t e ? i te m
WHERE { ? s t e p r d f : t y p e : S t e p .
? s t e p : h a s S t a t e ? s t a t e .
? s t a t e : i n v o l v e s ? it em .
? i te m r d f : t y p e / r d f s : s u b C la ss Of :
I n s t r u m e n t }
Query4. Which states of step 3 and its substeps mea-
sure the gel length, and what is the target value?
SELECT ? s t e p ? s t a t e ? x
WHERE {
: s t e p 3 ˆ : s ub S t e p ? s t e p .
? s t e p : h a s S t a t e ? s t a t e .
? s t a t e : i n v o l v e s : g e l .
: g e l : h a sM ea su re / : hasNumValue ? x }
Figure 6: Result of Query1: extract all devices involved in
all steps of the Alkaline Agarose Gel Electrophoresis pro-
cedure.
Figure 7: Result of Query2: return all materials involved in
all steps of the Alkaline Agarose Gel Electrophoresis pro-
cedure.
Figure 8: Result of Query3: extract all instruments involved
in all steps of the Alkaline Agarose Gel Electrophoresis pro-
cedure.
KEOD 2019 - 11th International Conference on Knowledge Engineering and Ontology Development
172

Figure 9: Result of Query4: return which states of step3 and its substeps measure the gel length, and what is the target value.
1
Alkaline Agarose Gel Electrophoresis
1. Prepare the agarose solution
1.1 Adding the appropriate amount of powdered agarose to a measured
quantity of H2O in either:
1.1.1 An Erlenmeyer flask (Container 1)
1.1.1.1 Loosely plug the neck of the Erlenmeyer
flask with Kimwipes
1.1.2 OR a glass bottle (Container 1)
1.1.1.2 Make sure that the cap is loose
1.2 Heat the slurry (Item1) in (Conatiner1) for the minimum time
required to allow all of the grains of agarose to dissolve using
either:
1.2.1 A boiling-water bath
1.1.1.3 Check that the volume of the solution (Item
1) has not been decreased by evaporation
during boiling in (Container 1):
1.1.1.3.1 if yes: replenish with
H2O in (Container 1)
1.1.1.3.2 If no: do not add H2O in
(Container 1)
1.2.2 OR a microwave oven
2
1.2.2.1 Check that the volume of the solution (Item
1) has not been decreased by evaporation
during boiling in (Container 1):
1.2.2.1.1 if yes: replenish with
H2O in (Container 1)
1.2.2.1.2 If no: do not add H2O in
(Container 1)
1.3 Cool the clear solution (Item 1) to 55 C.
1.3.1 Add 0.1 volume of 10x alkaline agarose gel
electrophoresis buffer in (Container 1)
1.3.2 And immediately pour the gel (Item 1) into mold
(Container 2)
1.4 After the gel (Item 1) is completely set
1.4.1 Mount it (Item 1) in the electrophoresis tank (Container
3)
1.4.2 Add freshly made 1x alkaline electrophoresis buffer until
the gel (Item 1) is just covered.
2. Prepare DNA samples
2.1 Collect the DNA samples (Item 2) by standard precipitation with
ethanol
2.2 Dissolve the damp precipitates of DNA (Item 2) in 10-20 μl of 1x
gel buffer. (Item 3)
3
2.3 Add 0.2 volume of 6x alkaline gel-loading buffer.
2.3.1 It is important to chelate all Mg2+ with EDTA before
adjusting the electrophoresis samples to alkaline
conditions.
3. Initiate the electrophoresis
3.1 Load the DNA samples dissolved in 6x alkaline gel-loading buffer
into the wells of the gel (container 3)
3.2 Start the electrophoresis at <3.5 V/cm when the bromocresol green
has migrated into the gel approx. 0.5-1 cm; Turn off the power supply,
and place a glass plate on top of the gel in (Container 3) and then
continue electrophoresis until the bromocresol green has migrated
approximately two thirds of the length of the gel in (container 3).
4. Finalize the experiment
4.1 Process the gel according to one of the procedures either Southern
hybridization by:
4.1.1 Transfer the DNA either:
4.1.1.1 Directly (without soaking the gel) from the
alkaline agarose gel to a charged nylon
membrane. Please see Southern Blotting:
Capillary Transfer of DNA to Membranes
4.1.1.2 OR after soaking the gel in neutralizing
solution for 45 minutes at room
temperature to either:
4
4.1.1.2.1 An uncharged nitrocellulose as
described in Southern Blotting:
Capillary Transfer of DNA to
Membranes
4.1.1.2.2 OR nylon membrane as
described in Southern Blotting:
Capillary Transfer of DNA to
Membranes
4.1.2 Detect the target sequences in the immobilized DNA by
hybridization to an appropriate labeled probe. Please see Southern
Hybridization of Radiolabeled Probes to Nucleic Acids Immobilized
on Membranes
4.2 OR Staining
4.2.1 Soak the gel in neutralizing solution for 45 minutes at
room temperature.
4.2.1.1 Stain the neutralized gel with 0.5 μg/ml
ethidium bromide in 1x TAE or with SYBR
Gold.
4.2.1.1.1 A band of interest can be
sliced from the gel and
subsequently eluted by one of
the procedures described
Recovery of DNA from
Agarose Gels
Figure 10: The first steps of Alkaline Agarose Gel Electrophoresis.
Biochemistry Procedure-oriented Ontology: A Case Study
173
