
Search for Triterpene Synthase in the NCBI Database
Mohammad Basyuni
1,2
, Rahmah Hayati
1
, Yuntha Bimantara
1
, Arif Nuryawan
1
, Etti Sartina Siregar
3
and Sumaiyah
4
1
Department of Forestry, Faculty of Forestry, Universitas Sumatera Utara, Jl. Tri Dharma Ujung No. 1 Medan 20155,
North Sumatra, Indonesia
2
Mangrove and Bio-Resources Group, Center of Excellence for Natural Resources-Based Technology, Universitas
Sumatera Utara, Medan 20155, North Sumatra, Indonesia
3
Faculty of Mathematics and Natural Sciences, Universitas Sumatera Utara, Medan 20155, North Sumatra, Indonesia
4
Faculty of Pharmacy, Universitas Sumatera Utara, Medan 20155, North Sumatra, Indonesia
Keywords: -amyrin, lupeol, database, oxidosqualene, triterpene synthase
Abstract: Triterpenes are common chemical components of higher plants. The present work reports on triterpene
synthase through a search for National Center for Biotechnology Information (NCBI) databases
(https://www.ncbi.nlm.nih.gov/). To generate a amount of valued data, the main term triterpene synthase
was implemented. Results detected in 22 databases for triterpene synthase. All triterpene synthase databases
are composed of literature, genes, proteins, genomes and chemical properties. No data on genetics is
surprising. Bookshelf, MeSH (Medical Subject Headings), National Medicine Library (NML), PubMed, and
PubMed Central were discussed in the literature. Gene consisted of Gene, Gene Expression Omnibus (GEO)
DataSets, GEO Profiles, HomoloGene, and PopSet. Proteins included Conserved Domains, Identical Groups
of Proteins, Proteins, Clusters of Proteins, Sparcle, and Structure. Genomes associated in Nucleotide and
Probe. There were 98 nucleotides of plant triterpene synthases, which Arabidopsis thaliana were
predominant with 29 triterpene synthases. BioSystems, PubChem BioAssay, PubChem Compound and
PubChem Substance depicted the chemical property. The present study delivers crucial data regarding
biotechnology of triterpene synthases.
1 INTRODUCTION
Triterpenes are common chemical constituents of
higher plants. In the plant kingdom, triterpenes
along with phytosterols are biosynthesized by the
enzyme oxidosqualene cyclases (OCSs) from a
sustained precursor 2,3-oxidosqualene. 2,3-
Oxidosqualene occurs in triterpene or phytosterol
synthases at the branching point of the isoprenoid
pathway (Augustin et al., 2011). It is important to
note that more than hundreds of triterpenes are
engaged in the cyclicisation of 2,3-oxidosqualene to
generate mono- and multi-functional synthesis of
triterpene. (such as -amyrin synthesis, lupeol
synthesis or mix products of -amyrin, lupeol, and
-amyrin) (Basyuni et al. 2006, 2017a; Shibuya et
al., 2007 ).
The synthesis of triterpene was cloned and
abundantly described. Triterpene synthase for its
biological and physiological operations is also
extensively investigated (Basyuni et al., 2009,
2012a,b; Sheng and Sun, 2011; Lambert et al.,
2011). For instance, triterpene synthase's
physiological and molecular reactions to abiotic
stress such as salinity (Basyuni et al., 2009,
2012a,b). Salinity increased triterpene synthesis
genes against salt concentration in mangroves
(Basyuni et al., 2009, 2012a,b); the significance of
pentacyclic triterpenes as a multi-target technique
for metabolic and vascular disease prevention and
therapy (Sheng and Sun, 2011)
Although big numbers of research have been well
documented in triterpene synthases (Yu et al., 2009;
Agustin et al., 2011; Sawai and Saito, 2011; Moses
et al., 2014; Thimmappa et al., 2014; Isah et al.,
2016), limited work concentrated on biotechnology
data from all accessible databases in triterpene
syntheses. Here we report another technique to
collect useful data needed in latest biotechnology-
related science studies through a preferred search
engine. Therefore, the aim of this research was to
8
Basyuni, M., Hayati, R., Bimantara, Y., Nuryawan, A., Siregar, E. and Sumaiyah, .
Search for Triterpene Synthase in the NCBI Database.
DOI: 10.5220/0008386600080011
In Proceedings of the International Conference on Natural Resources and Technology (ICONART 2019), pages 8-11
ISBN: 978-989-758-404-6
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
explain the implementation of the National Center
for Biotechnology Information (NCBI) database
search to gain more insight into a lot of useful data
about updated triterpene synthesis biotechnology.
2 MATERIALS AND METHOD
The search engine for NCBI databases
(https:/www.ncbi.nlm.nih.gov/) was used to produce
many precious triterpene synthase data
biotechnologies. As stated previously on January 19,
2019, data bases were obtained by typing triterpene
synthase for all database searches. All databases
composed of triterpene synthase literature, genes,
protein, genomes and chemical properties were
performed using the Search button. Bookshelf,
MeSH (Medical Subject Headings), NLM (National
Medicine Library) catalogue, PubMed, PubMed
Central, EST, Gene, GEO datasets, PopSet, Identical
Protein Groups, Protein, Sparde, Structure,
Assembly, BioProject, BioSample, Genome, GSS,
Nucleotide, Probe, SRA, Taxonomy, Biosystems
and PubChem BioAssay were included in the data
variables.
3 RESULT AND DISCUSSION
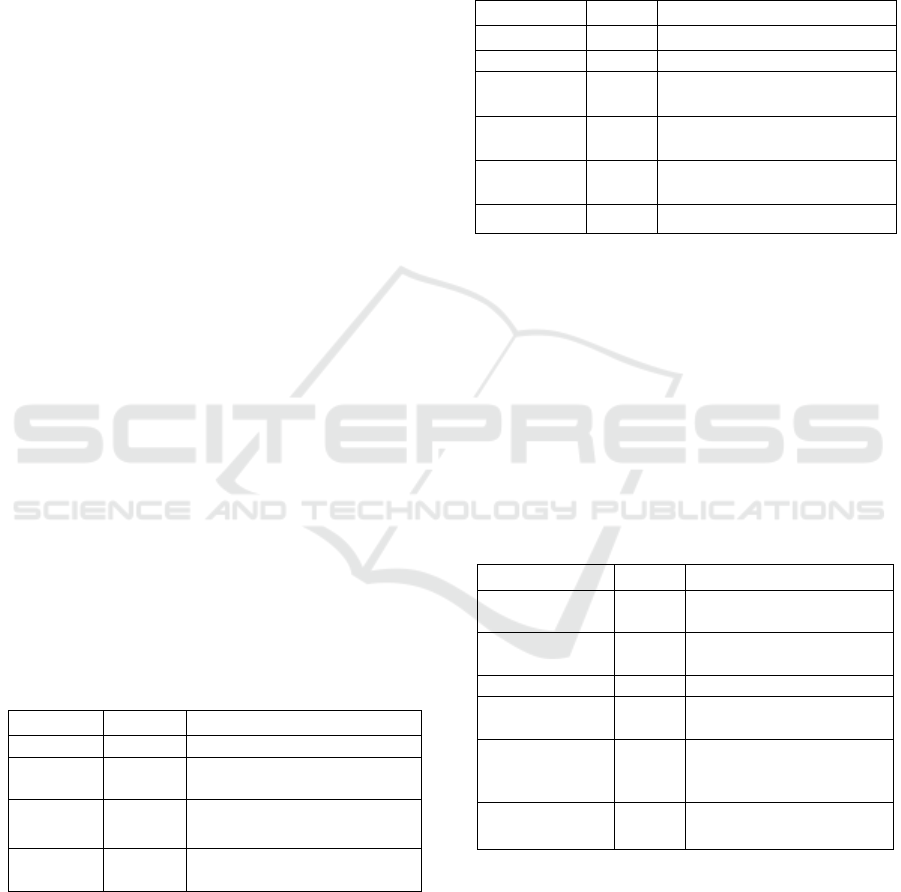
Table 1 illustrates the NCBI literature on triterpene
synthase. Four biographies stored in countless
numbers. The internet NCBI literature provides
internet libraries and free access to bookshelf data
(eight books and reports). The title of books was five
with a total of eight items.
Table 1: Literature source of NCBI database for plant
triterpene synthase
Literature
Total
Description
Bookshelf
8
Books and reports
NLM
1
Books, journals and more in
the NLM
Catalog
PubMed
1126
Collections abstract/ citations
of science and medicine
PubMed
Central
2857
Full-text journal articles
Furthermore, one NLM catalog reported as
known Plant isoprenoids: methods and protocols. In
the database, there were 1126 Pubmed, which within
five years consist of 2014 (99 documents), 2015
(114), 2016 (91), 2017 (88), and 2017 with 88
publications. Meanwhile, there was 2857 PubMed
Central documentation in the triterpene synthase,
with NIH grants of 474 documents (Table 1). The
PubMed database includes quotes from different
topics in triterpene synthase, many links to open
access papers from triterpene synthase reports.
Table 2: Genes source of NCBI database for plant
triterpene synthase
Gene
Sum
Definition
Gene
11
Collected gene loci data
Geo Data Sets
21
Functional genomic studies
Geo Profiles
728
Gene expression and profiles of
molecular abundance
HomoloGene
1
Homologous gene sets for the
organism chosen
PopSet
1
Sets of sequences from research
in phylogenetics and population
UniGene
7
Expressed transcript clusters
Facts on both gene sources and health are
presented in Table 2. This information includes
genes associated with Arabidopsis thaliana than
consisting of 9 genes, and they are three genes of
pentacyclic triterpene synthase (1,3, and 7), two
lupeol synthases (1 and 2), marneral synthase 1,
baruol synthase 1, camelliol C synthase 1, and
terpenoid cyclase family protein. It is important to
recognize that this research did not detect the OMIM
(Online Mendelian Inheritance in Man) (Table 2).
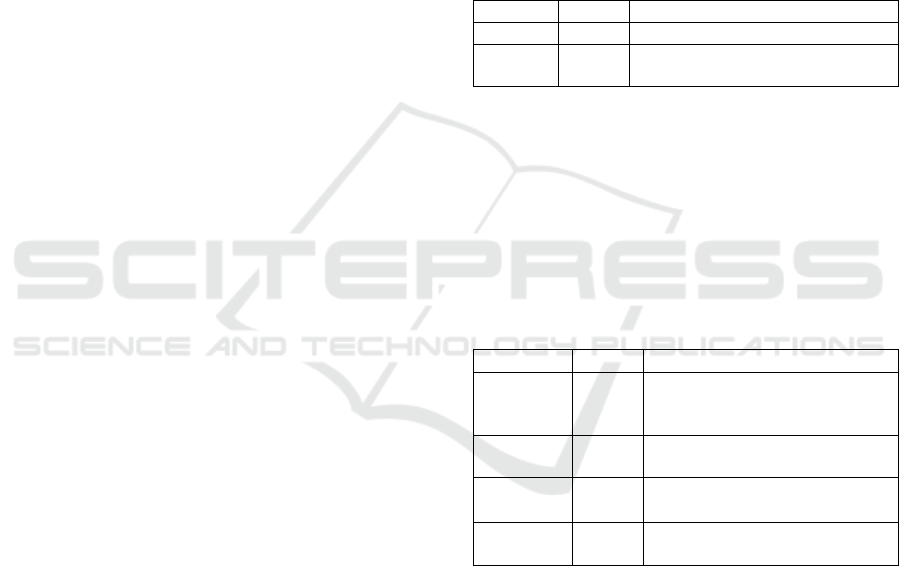
Table 3: Proteins source NCBI database for plant
triterpene synthase
Proteins
Number
Explanation
Conserved
Domains
2
Protein domains preserved
Identical Protein
Groups
17
Sequences of protein
grouped by identity
Protein
304
Protein sequences
Protein Clusters
1
Sequence-based clusters of
proteins
Sparcle
9
Functional protein
categorization by domain
architecture
Structure
16
Biomolecular structures
experimentally determined
Regarding with protein for triterpene synthase
observed in the database, two conserved protein
domains were found. Both are the enzymes of
Isoprenoid Biosynthesis, Class 1 and Trans IPPS
HH: Trans-Isoprenyl Diphosphate Synthases, head-
to-head as displayed in Table 3. There were 17
identical protein groups. These groups are
pentacyclic triterpene synthase 1 with 582 amino
Search for Triterpene Synthase in the NCBI Database
9
acid protein from A. thaliana (Mayer et al., 1999),
putative pentacyclic triterpene synthase 3 with 760
aa protein from A. thaliana (Tabata et al., 2000),
Putative pentacyclic triterpene synthase 7 with 706
aa protein from A. thaliana (Salanaubat et al., 2000),
thalianol synthase 1 with 758 aa protein from A.
thaliana (Tabata et al., 2000), pentacyclic triterpene
synthase 1 with 766 aa protein from A. thaliana
(Mayer et al., 1999), Putative pentacyclic triterpene
synthase 7 with 761 aa protein from A. thaliana.
Furthermore, taraxerol synthase (771 aa protein)
and germanicol synthase (759 aa protein), both are
from Rhizophora stylosa (Basyuni et al., 2007a),
low-quality protein putative pentacyclic triterpene
synthase 7 with 760 aa protein from A. lyrata subsp.
Lyrata (Hu et al., 2011), pentacyclic triterpene
synthase with 683 aa protein from A. thaliana (),
several multifunctional triterpene synthases from
Kandelia candel (Basyuni et al., 2006), R. stylosa
(Basyuni et al., 2007a), Costus speciosus (Kawano
et al., 2002), and Genlisea aurea (Leushkin et al.,
2013), pentacyclic triterpene synthase from A.
thaliana with 763 aa protein (Husselstein–Muller et
al., 2001), and three triterpene synthases from
Eugenia uniflora.
There were 304 proteins of triterpene synthase,
which 169 proteins belong plant. The top organisms
were A. thaliana (63), Panax quinquefolius (8),
Botryococcus braunii (8), P. ginseng (6), P.
notoginseng (6), Lycopodium clavatum (6), Lotus
japonicus (5), Ocimum basilicum (5), R. stylosa (4),
B. gymnorhiza (4), Pisum sativum (4), Malus
domestica (4). One protein cluster from Arabidopsis.
In this study, nine sparcles and sixteen structures
proteins were described (Table 3).
Recently, triterpene synthase protein modeling
has been characterized from mangrove trees
(Basyuni et al. 2018). In addition to protein from
NCBI database, another source from KEGG, with
search term: triterpene synthase found three entries:
ath: AT3G29255, putative pentacyclic triterpene
synthase 7, and ath:AT4G15340, K15823 arabdiol
synthase PEN1, pentacyclic triterpene synthase 1,
ath:At5G36150, K16205 tirucalldienol synthase,
PEN3; putative pentacyclic triterpene synthase 3.
In the enzyme database, search term of OSC
resulted of 5 hits, they were dammarenediol II
synthase (Tansakul et al., 2006), lanosterol synthase
(Suzuki et al., 2006), cycloartenol synthase such as
KcCAS and RsCAS (Basyuni et al., 2007b), -
amyrin synthase such as BgbAS (Basyuni et al.,
2007a), and -amyrin synthase such as mixed
amyrin synthase KcMS (Basyuni et at., 2007a).
These results implied a diversity of OSC genes in
the plant kingdom.
Nucleotide base sequences for genomes consisted
of 98 nucleotides and TaqMan probe (22) as shown
in Table 4. The top organisms for nucleotides were
A. thaliana (29), P. quinquefolius (8), P.
notoginseng (6), L. japonicus (5), M. domestica (5),
Ocimum basilicum (5), E. uniflora (3), Monteverdia
ilicifolia (3), P. ginseng (3), Lycopodium clavatum
(3), Medicago truncatula (2), P. sativum (2), R.
stylosa (2), B. gymnorhiza (2), Eleutherococcus
senticosus (2), Bupleurum kaoi (2), and Centella
asiatica (2).
Table 4: Genome source of triterpene synthase
Genomes
Total
Explanation
Nucleotide
98
RNA and DNA sequences
Probe
22
Probes and primers based on
sequences
Variation of the triterpene synthase chemical
features was shown in Table 5. There have been 120
molecular pathways linked to genes, proteins, and
chemicals. Online screening works are accessible for
one hundred and three bioactivities. One compound
from PubChem and one substance from PubChem in
the database.
Table 5: Chemicals source NCBI database for Plant and
triterpene synthase
Chemicals
Amount
Information
BioSystems
120
Molecular pathways associated
with genes, proteins and
chemicals
PubChem
BioAssay
103
Studies of bioactivity screening
PubChem
Compound
1
Chemical data with constructions,
understanding and connections
PubChem
Substance
1
Substance deposited and chemical
data
4 CONCLUSIONS
The online NCBI discusses numerous data on
triterpene synthase from biology and biotechnology.
The current research urged biotechnology scientists
to use the NCBI search engine to obtain more
advantages. The current research also provides
important information on triterpene synthase
biotechnology.
ICONART 2019 - International Conference on Natural Resources and Technology
10

ACKNOWLEDGMENTS
This research was in part funded by World Class
Research from Directorate for Research and
Community Service, Ministry of Research,
Technology and Higher Education, the Republic of
Indonesia.
REFERENCES
Augustin, J.M., Kuzina, V., Andersen, S.B., Bak, S., 2011
Molecular activities, biosynthesis and evolution of
triterpenoid saponins. Phytochemistry 72, 435-457.
Basyuni, M., Oku, H., Inafuku, M., Baba, S., Iwasaki, H.,
Oshiro, K., Okabe, T., Shibuya, M. and Ebizuka, Y.,
2006. Molecular cloning and functional expression of
a multifunctional triterpene synthase cDNA from a
mangrove species Kandelia candel (L.) Druce.
Phytochemistry, 67(23), 2517-2524.
Basyuni, M., Oku, H., Tsujimoto, E., Kinjo, K., Baba, S.
and Takara, K., 2007a. Triterpene synthases from the
Okinawan mangrove tribe, Rhizophoraceae. The FEBS
Journal, 274(19), 5028-5042.
Basyuni, M., Oku, H., Tsujimoto, E., and Baba, S. 2007b.
Cloning and functional expression of cycloartenol
synthases from mangrove species Rhizophora stylosa
Griff. and Kandelia candel (L.) Druce. Bioscience,
Biotechnology, and Biochemistry, 71(7), 1788-1792.
Basyuni, M., Baba, S., Inafuku, M., Iwasaki, H., Kinjo, K.
and Oku, H., 2009. Expression of terpenoid synthase
mRNA and terpenoid content in salt stressed
mangrove. Journal of Plant Physiology, 166(16),
1786-1800.
Basyuni, M., Baba, S., Kinjo, Y., Putri, L.A., Hakim, L.
and Oku, H., 2012a. Salt-dependent increase in
triterpenoids is reversible upon transfer to fresh water
in mangrove plants Kandelia candel and Bruguiera
gymnorrhiza. Journal of Plant Physiology, 169(18),
1903-1908.
Basyuni, M., Baba, S., Kinjo, Y., and Oku, H. 2012b.
Salinity increases the triterpenoid content of a salt
secretor and a non-salt secretor mangrove. Aquatic
Botany, 97(1), 17-23.
Basyuni, M., Wati, R., Sulistiyono, N., Hayati, R., Oku, H.,
Baba, S., & Sagami, H. 2018. Protein modelling of
triterpene synthase genes from mangrove plants using
Phyre2 and Swiss-model. Journal of Physics:
Conference Series, 978 (1), 012095. IOP Publishing.
Hu, T. T., Pattyn, P., Bakker, E. G., Cao, J., Cheng, J.,
Clark, R. M., et al. 2011. The Arabidopsis lyrata
genome sequence and the basis of rapid genome size
change. Nature Genetics, 43(5), 476.
Husselstein–Muller, T., Schaller, H., and Benveniste, P.
(2001). Molecular cloning and expression in yeast of 2,
3–oxidosqualene–triterpenoid cyclases from
Arabidopsis thaliana. Plant molecular biology, 45(1),
75-92.
Isah, M.B., Ibrahim, M.A., Mohammed, A., Aliyu, A.B.,
Masola, B. and Coetzer, T.H., 2016. A systematic
review of pentacyclic triterpenes and their derivatives
as chemotherapeutic agents against tropical parasitic
diseases. Parasitology, 143(10), 1219-1231.
Kawano, N., Ichinose, K., and Ebizuka, Y. (2002).
Molecular cloning and functional expression of
cDNAs encoding oxidosqualene cyclases from Costus
speciosus. Biological and Pharmaceutical Bulletin,
25(4), 477-482.
Leushkin, E. V., Sutormin, R. A., Nabieva, E. R., Penin, A.
A., Kondrashov, A. S., & Logacheva, M. D. (2013).
The miniature genome of a carnivorous plant Genlisea
aurea contains a low number of genes and short non-
coding sequences. BMC Genomics, 14(1), 476.
Mayer, K., Schüller, C., Wambutt, R., et al., 1999.
Sequence and analysis of chromosome 4 of the plant
Arabidopsis thaliana. Nature, 402(6763), 769.
Moses, T., Papadopoulou, K.K. and Osbourn, A., 2014.
Metabolic and functional diversity of saponins,
biosynthetic intermediates and semi-synthetic
derivatives. Critical Reviews in Biochemistry and
Molecular Biology, 49(6), 439-462.
Salanoubat, M., Lemcke, K., Rieger, M., et al., 2000.
Sequence and analysis of chromosome 3 of the plant
Arabidopsis thaliana. Nature, 408(6814), 820
Sawai, S. and Saito, K., 2011. Triterpenoid biosynthesis
and engineering in plants. Frontiers in Plant Science,
2, 25.
Sheng, H. and Sun, H., 2011. Synthesis, biology and
clinical significance of pentacyclic triterpenes: a
multi-target approach to prevention and treatment of
metabolic and vascular diseases. Natural Product
Reports, 28(3),543-593.
Shibuya, M., Xiang, T., Katsube, Y., Otsuka, M., Zhang,
H. and Ebizuka, Y., 2007. Origin of structural
diversity in natural triterpenes: direct synthesis of
seco-triterpene skeletons by oxidosqualene cyclase.
Journal of the American Chemical Society, 129(5),
1450-1455.
Suzuki, M., Xiang, T., Ohyama, K., et al., 2006.
Lanosterol synthase in dicotyledonous plants. Plant
and Cell Physiology, 47(5), 565-571.
Tabata, S., Kaneko, T., Nakamura, Y., et al., 2000.
Sequence and analysis of chromosome 5 of the plant
Arabidopsis thaliana. Nature, 408(6814), 823.
Tansakul, P., Shibuya, M., Kushiro, T., & Ebizuka, Y.
2006. Dammarenediol‐II synthase, the first dedicated
enzyme for ginsenoside biosynthesis, in Panax ginseng.
FEBS Letters, 580(22), 5143-5149.
Thimmappa, R., Geisler, K., Louveau, T., O'Maille, P. and
Osbourn, A., 2014. Triterpene biosynthesis in plants.
Annual Review of Plant Biology, 65, 225-257.
Yu, B. and Sun, J., 2009. Current synthesis of triterpene
saponins. Chemistry–An Asian Journal, 4(5), 642-654.
Search for Triterpene Synthase in the NCBI Database
11
