
Evaluation of Resistance Improvement of Soybean (Glycine Max (L)
Merr.) against Salinity using Mass Selection and Gene Expression of
Salinity Tolerant
Nini Rahmawati
1
, Rosmayati
1
, Delvian
2
, Mohammad Basyuni
2
and Hirosuke Oku
3
1
Faculty of Agriculture, Universitas Sumatera Utara, Medan 20155, Sumatera Utara, Indonesia
2
Faculty of Forestry, Universitas Sumatera Utara, Medan20155, Sumatera Utara, Indonesia
3
Centers of Molecular Biosciences, University of the Ryukyus, Okinawa, Japan
okuhiros@comb.u-ryukyu.ac.jp
Keywords: Soybean, Saline Tolerant Gene, Mass Selection.
Abstract: Saline soils are land that has not been utilized widely for the cultivation of soybean due to the toxic effects
that interfere with the growth of the plant. This research aims to get the soybean genotypes that have salinity
resistant through mass selection methods and gene expression testing of third generation of saline tolerant of
the soybean genotypes that salinity resistant (F3). The research of mass selection conducted on research’s
land with salinity of land 5-6 mmhos/cm and molecular analysis carried out at the Center of Molecular
Biosciences University of the Ryukyus, Japan. Molecular analysis of saline tolerant gene at the root of
soybean selection results F3 and soybean grobogan varieties showed mRNA expression gene DREB5,
GPRP3, P5CS, bZIP, ERF and NHX1 higher in selected soybean that salinity resistant F3 that was treated by
salinity compared to the controls, while the level of gene expression GmCLC1 and PAP3 lower than the
control. Comparison of gene expression levels in soybeans that given salinity stress show there has been an
increased of expression genes that associated with the ability of adaptation of plants to salinity stress.
1 INTRODUCTION
Salinity is one of the important abiotic factors that
limiting the production of soybean in the
world. Reclamation of soil is not an economical
option to increase soybean production that
experiencing in salinity stress. Therefore, genetic
improvement for salt tolerance is a more cost
effective option. Conventional breeding has
contributed significantly to enhancement of soybean
production in the last 50 years. Through conventional
breeding, it is easy to manipulate the inheritance of
qualitative properties that are less sensitive to
environment changes, but quantitative properties like
yield or tolerance to abiotic stress were significantly
influenced by the environment (Pathan et al., 2007).
Some plants develop mechanisms to cope with
these stresses, in addition there are also being
adapted. The majority of the cultivation of plants are
vulnerable and could not survive in high salinity
condition, or even survive, but with yields reduced. A
study of the response plant to salinity is important in
efforts to achieve effective plant filtering technique.
Soybean varieties showed broad spectrum in its
ability to tolerate salt. Filtering of soybean genotypes
have been conducted to identify genetic properties
that show a high tolerance to salt stress. Currently, the
breeding is the main strategy to improve salt tolerance
in soybean (Phang et al., 2009).
Plant breeding in the future will be further lead to
the use of techniques and methodologies of molecular
breeding using genetic markers. The use of
"molecular breeding" has been promising simplicity
of the constraints and challenges in plant breeding
complexity. Selection indirectly using molecular
markers that are bound to the desired properties has
allowed individual studies on the related to the
selection of the double properties and inaccuracy of
measurement due to the expression of properties that
caused by external factors of double genetic locus
(Sudarmi, 2013).
Genes that induced by abiotic stresses such as
high salinity have been found and provide an
important opportunity to improve the tolerant
30
Rahmawati, N., Rosmayati, ., Delvian, ., Basyuni, M. and Oku, H.
Evaluation of Resistance Improvement of Soybean (Glycine Max (L) Merr.) against Salinity using Mass Selection and Gene Expression of Salinity Tolerant.
DOI: 10.5220/0008387500300037
In Proceedings of the International Conference on Natural Resources and Technology (ICONART 2019), pages 30-37
ISBN: 978-989-758-404-6
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

properties to high salinity through genetic
engineering approach. Target genes include genes
that encode the enzymes that necessary for the
biosynthesis of various osmoprotectant, enzymes that
eliminated reactive oxygen species, end
embryogenesis proteins (LEA), enzymes to detoxify
and transcription factors (Santoso et al., 2012).
Molecular approach through screening is done at
the plant to determine the genes that are play a role in
the resistance mechanism (Turan et al., 2012;
Ermawati, 2011).
The main mechanism for salt tolerance is to
minimize salt that was taken up by the roots, split
them at the level of cells and tissues so as do not reach
concentrations that contain of toxic on the cytosol in
the leaves. Candidate genes, among others for ion
transport, osmoprotectant, and make plants grow
faster in saline soil. A study of gene expression in the
roots and leaves has been widely reviewed and
various suggestions have been submitted to increase
plant resistance in saline soils (Munns, 2005). Some
researchers have reported several genes that
associated with tolerance soybean against salinity
stress, among others P5CS, GmCLC1, GmSOS1,
GmNHX1, GmbZIP, GmDREB, GPRP3 (Phang et
al., 2008; Peng et al., 2012; Lan et al., 2011; Celik
and Atak, 2011; Gao et al., 2011; Zhang et al., 2009;
Li et al., 2006; Liao et al., 2003; Sun et al., 2013).
2 MATERIALS AND METHODS
2.1 Study Area
Mass selection starts from elders to the fourth
generation (F4) was conducted in the Paluh Merbau
village, Percut Sei Tuan, Deli Serdang district with a
height of ± 1.5 m above sea level and salinity levels
5-6 mmhos/cm that was conducted in May 2011 to
the May 2013.
2.2 Procedures
Molecular analysis carried out in the greenhouse and
laboratory of Center of Molecular Biosciences
(COMB) University of the Ryukyus, Okinawa, Japan,
from October to December 2012. The limit of
selection that used was 10% of the plant
population. And the heritability was analyzed using
the following formula:
Criteria of heritability:
h
2
> 0.5 : High
h
2
0,2 - 0,5 : Moderate
h
2
< 0.2 : Low
(Stansfield, 1991).
Soybeans that will be used for molecular analysis
planted in greenhouse in a tub of plastic for 14 days
and were given the control treatment (without salinity
stress) and the treatment of salinity stress given for 72
hours after 14 HST by using a commercial salt
powder (Red Sea Salt, Houston, TX, USA) by DHL
5-6 mmhos/cm. Media salinity level is done every day
by using a salinity refractometer S/Mill-E (Atago Co.
Ltd., Tokyo, Japan). RNA isolation is done by using
RNA easy Plant Mini Kit (Qiagen). Total RNA
quantitative test using nanodrop, while the qualitative
test using electrophoresis and agarose. The next stage
is the synthesis of cDNA with Hexamer Random
Method using High Capacity RNA-to-cDNA
(Applied Biosystems). Primary will be used for the
analysis of gene expression and housekeeping genes
(for normalization of target genes) were 8 primers
that designed using data from NCBI and software
Genetyx (Table 1).
Table 1: Primary that Designed and Used for Gene
Expressions Analysis.
Gene
Forward primer
sequence [5’-3’]
Reverse primer
sequence [5’-3’]
DREB5
GTGAGCGATGAC
CAGGTTCATG
CATCCAAATC
ATCCCACATGGG
GPRP3
CTGCTGCTGCTT
ATGGTGCTCA
ATGCTTTCCA
TGCTTGCCAAAC
P5CS
GGAAGTGCACAT
ACTGATTCCG
TGGACCCCGA
GCATGAATCCTG
BZIP
CACCAGTTGGTG
ACTGTTCAGA
CCTTTACCAG
CTTTCCCAGTTG
ERF3
GCTCCTGAGATC
TCATCCATGC
GTCACCAGAT
AGCAAGGATTCC
CLC1
TTACATGGGTGG
AGCTGGCTGA
GCGAAGTCCT
ACTTGTCTGAAG
NHX1
GCATCACGATAA
CCACAGATCC
CATCAAACTT
ACGCCACAAGCG
PAP3
GTCGGAGATGGT
GGAAATCAAG
GCCATCATCA
TTGCGATTCCAG
ACTI I
ATTTTGACTGAG
CGTGGTTATTCC
GCTGTCTCCA
GTTCTTGCTCGT
2.3 Data Analysis
Analysis of genes expression were using Real Timer-
PCR that using Fast SYBR®GreenMaster Mix. RT-
PCR result data were analyzed to determine the level
of expression of each variety and treatment.
eg
g
p
g
h
22
2
2
2
2
Evaluation of Resistance Improvement of Soybean (Glycine Max (L) Merr.) against Salinity using Mass Selection and Gene Expression of
Salinity Tolerant
31

3 RESULT AND DISCUSSIONS
3.1 Heritability, Selection Limits and
Selection Advancement of Salinity
Resistant Soybean Selection
Selection of soybeans to obtain plants that have the
potential to be developed as saline tolerant varieties
showed the progress of selection is not yet stable, but
observation variable of production character has high
heritability values. Soybean production is determined
by genetic factors, besides that, it also strongly
influenced by environmental conditions, especially
changes in soil salinity levels. Selection of salinity
resistant soybean ranging from elders to the fourth
generation showed the average of each observation
variable of growth and production showed diverse
results (Table 2 and Table 3). Growth and production
of best soybean is reached on the second generation
of selection. Soybean production per plant in the
second generation reached 12.00 g. While on the third
generation of plant production (weight of seeds per
plant) reached its low point of 1.6 g per plant. The
growth and production of soybean increased again in
the fourth generation of selection, where production
per plant was 10.55 g.
The highest soybean production was obtained in
the second generation, namely 12.0 g/plant (Table 2).
This is supported by the environmental conditions
with the level of soil salinity is relatively
stable. While in the third generation, there was a
decrease of the production average due to changes in
soil salinity is very high, reaching 10.36 mmhos/cm.
Ghassemi-Golezani and Taifeh-Noori (2011)
reported the production of soybeans that grown in soil
with DHL 9 dS/m decreased by 354.55% compared
to the DHL 0 dS/m.
Heritability value of the first generation to fourth
generation showed a change (Table 4 and Table
5). Observation variables of production character
such as harvest age, number of pods, number of
containing pods, the number of empty pods, and the
weight of seeds/plants that are tend to have high
heritability values in each generation. Heritability is
one of the most important considerations in plants
evaluating, selection method and crossbreeding
system. More specifically, heritability is part of the
total variation in properties that caused by genetic
differences among observed plants. Heritability is the
ratio between the genetic variance to the phenotypic
variance. Phenotypic variance is influenced by
genetic and environmental factors. Heritability value
of production characters at every stage of selection is
likely to be stable and included in high or moderate
criteria. This shows that genetic factors are more
dominant to controlling the characters. Roy (2000)
stated that the success of selection is determined by
the existing of diversity that controlled by genetic
factors, while Ceccarelli et al. (2007) suggested that
selection on the stress environment conducted in the
target environment so as to maximize the expression
of genes that control the yield capability and plant
adaptability. Marquez-Ortiz et al., (1999) stated a low
heritability value means that environmental factors
have greater influence than genetic factor.
Table 2: The Average of Agronomic Observation Variable
from Elders to Second Generation.
Variable Observation
Generation
Elders
First
Second
Plant Height (cm)
20.30
20.86
34.60
Number of
Branches(branch)
1.60
1.80
5.40
Flowering Age (day)
29.80
29.12
30.50
Harvest Age (day)
68.00
86.53
84.90
Number of Pods (pod)
10.10
3.83
44.40
Containing Pod (pod)
9.80
3.64
42.00
Empty Pod (pod)
0.30
0.30
2.40
Seed/Plant Weight (g)
2.90
0.66
12.00
Table 3: The Average of Agronomic Observation Variable
from Third to Fourth Generation and Optimal Condition.
Variable Observation
Generation
Optimal
Condition
Third
Fourth
Plant Height (cm)
29.20
34.10
61.24
Number of
Branches (branch)
3.90
4.00
2.90
Flowering Age (day)
29.00
29.20
35.00
Harvest Age (day)
72.90
73.15
76.12
Number of Pods (pod)
9.80
32.15
60.50
Containing Pod (pod)
8.60
27.7
53.15
Empty Pod (pod)
1.30
4.45
6.35
Seed/Plant Weight (g)
1.60
10.55
14.04
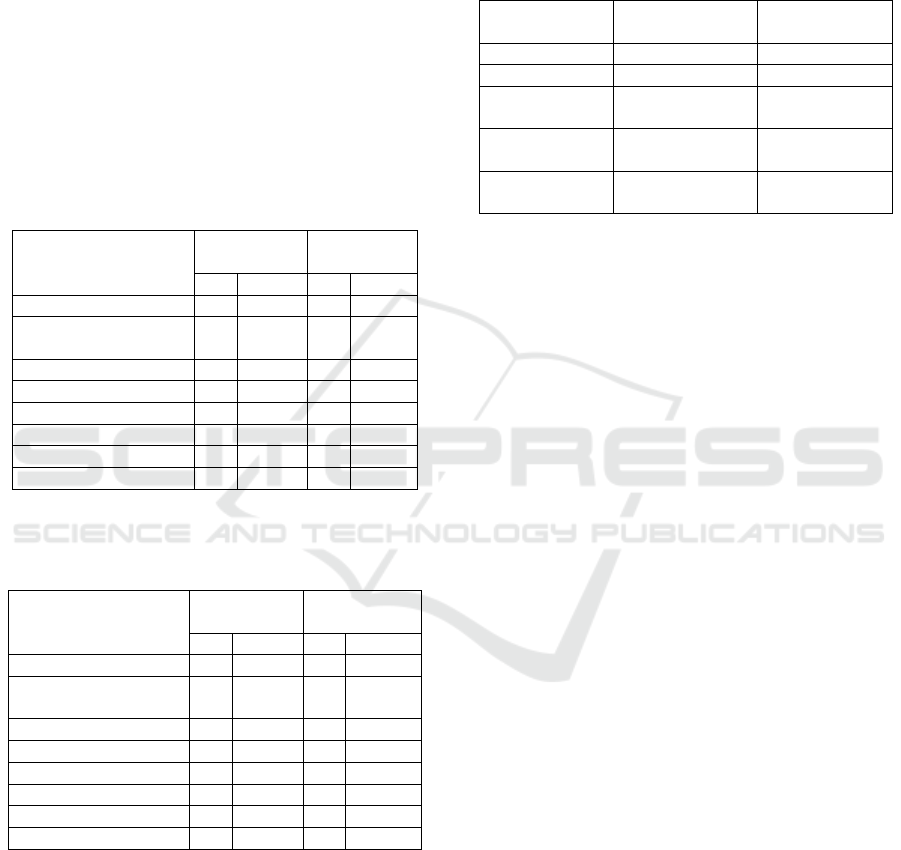
Selection limit and selection advancement of each
stage of selection can be seen in Table 6. The highest
selection limit was on the second and fourth
generation, namely 10.16 g/plant, while the lowest
selection limit was in the third generation, namely
0.21 g/plant. Selection progress also showed diverse
results. Selection regress occurred from the second
generation to the third generation, there was 2.38.
Selection progress rebound in the second and fourth-
generation, namely 3.57 and 3.93.
Changes in saline environments also causes the
selection boundary to the character of production is
not yet stable. The highest selection limit is achieved
by the second generation, namely 10.16 and
decreased to 0.21 in the third generation due to
ICONART 2019 - International Conference on Natural Resources and Technology
32
increased soil salinity. Selection boundary on the
fourth generation increased again to 10.16 and 1.69
for the selection progress. Selection progress in this
research showed an increase in the third and fourth
generation. Selection progress is a value which is a
parameter of the success of the selection that we
done. Sketchily, the value of the selection progress is
the difference of the initial population and the further
population that has experiencing selection (Idris et al.,
2011). The high value of selection progress is a
manifestation of the value of diversity additive in a
population. The diversity of additive itself is a
necessary component for recurrent selection (Sutoro,
2006).
Table 4: Heritability Value of the First Generation and
Second Generation.
Variable Observation
First
Generation
Second
Generation
H
Criteria
h
Criteria
Plant Height (cm)
0.89
H
0.45
M
Number of Branches
(branch)
0.00
L
0.00
L
Flowering Age (day)
0.00
L
0.05
L
Harvest Age (day)
0.39
M
0.00
L
Number of Pods (pod)
0.98
H
0.50
H
Containing Pod (pod)
0.97
H
0.50
H
Empty Pod (pod)
0.88
H
0.41
M
Seed/Plant Weight (g)
0.98
H
0.50
H
H = High, M = Medium, L = Low
Table 5: Heritability Value of the Third Generation and
Fourth Generation.
Variable Observation
Third
Generation
Fourth
Generation
h
Criteria
h
Criteria
Plant Height (cm)
0.76
H
0.95
H
Number of Branches
(branch)
0.00
L
0.20
L
Flowering Age (day)
0.26
M
0.13
L
Harvest Age (day)
0.81
H
0.52
H
Number of Pods (pod)
0.67
H
0.91
H
Containing Pod (pod)
0.68
H
0.93
H
Empty Pod (pod)
0.00
L
0.83
H
Seed/Plant Weight (g)
0.75
H
0.82
H
H = High, M = Medium, L = Low
Uncontrolled environmental conditions are one of
the obstacles to assemble soybean that salinity
resistant that has high yield capability. Ashraf (2004)
stated that the direct selection in the field about
quantitative properties which is tolerant to high
salinity still difficult because of uncontrolled
environmental factors. One approach to improve the
efficiency of the breeding program is to adopt a new
selection criterion based on the knowledge of
physiological processes, which is the delimiter from
production plants at the time of exposure of high
salinity.
Table 6: Selection Limit and Selection Progress every
Generation.
Generation
Selection limit Of
Products/Plants
Selection Progress
Elders
2.37
-
First Generation
0.48
4.68
Second
Generation
10.16
2.38
Third
Generation
0.21
3.57
Fourth
Generation
10.16
3.93
3.2 Molecular Analysis of Saline
Tolerant Genes in Soybean Roots
Resistant to Salinity
Strong interactions between agronomic properties
which are morphological markers with environmental
factors encourage the use of molecular breeding
methodology using genetic markers to support the
selection of soybean that salinity resistant. Salinity
stress caused some changes in physiological
processes, metabolism, and the expression of several
genes that allegedly played an important role in the
adaption response of plants to salinity stress. Santoso
et al., (2012) describes the genes that was induced by
abiotic stresses such as high salinity have been found
and provide an important opportunity to improve the
properties of tolerant to high salinity through genetic
engineering approach. Target genes include genes
that encode the enzymes that necessary for the
biosynthesis of various osmoprotectant, enzymes that
eliminated the reactive oxygen species, late
embryogenesis proteins (LEA), enzyme for
detoxification and transcription factors. This research
examined the expression of several genes that are
responsive to salinity stress, namelyDehydration
Responsive Element Binding Protein 5 (DREB5),
Glycine and Proline Rich Proteins 3 (GPRP3), Δ1-
Pyrroline-5-carboxylate synthetase (P5CS), bZIP
Transcription Factor (ZIP), EREBP/AP2
Transcription Factor (ERF), Gm Chloride Channel 1
(GmCLC1), Gm putative Na+/ H+ antiporter
(NHX1), and Purple Acid Phosphatases 3 (PAP3).
Molecular test of saline tolerant gene at the root
of soybean selection result F3, soybean grobongan
varieties and Burangrang varieties can be seen in
Table 7, Figure 1 and Figure 2. In Table 7 and Figure
Evaluation of Resistance Improvement of Soybean (Glycine Max (L) Merr.) against Salinity using Mass Selection and Gene Expression of
Salinity Tolerant
33
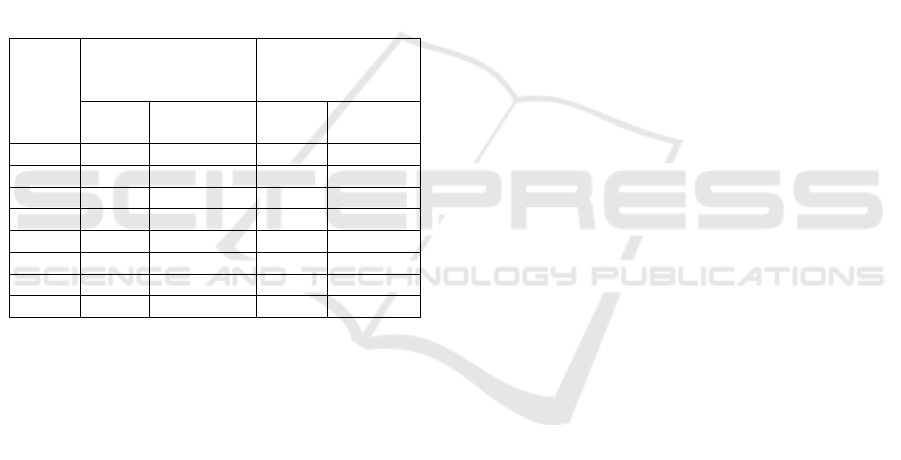
1, it can be seen that the mRNA expression of
Dehydration Responsive Element Binding Protein 5
( DREB5) genes, Glycine and Proline Rich Proteins 3
(GPRP3), Δ1-Pyrroline-5-carboxylate synthetase
(P5CS), bZIP Transcription Factor (ZIP),
EREBP/AP2 Transcription Factor (ERF) and Gm
putative Na+/H+ Antiporter (NHX1 ) higher (up-
regulated) in selected soybean that salinity resistant
F3that was given salinity treatment compared with
the control treatment, while the level of gene
expression of Gm Chloride Channel 1 (GmCLC1)
and Purple Acid phosphatases 3 (PAP3) lower (down-
regulated) than control treatment. P5CS genes
showed the highest level of expression enhancement
that is 1.75 fold in soybean that treated salinity.
Table 7: Level of mRNA Expression of Salinity Tolerant
Genes (fold) on soybean genotypes that salinity resistant
and Grobogan varieties on Control Treatment and Salinity
Stress 5-6 mmhos/cm.
Gene
Soybean Result of
Selection Salinity
Resistant F3
Grobogan Varieties
Soybean
Control
Salinity Stress
Control
Salinity
Stress
DREB5
0.61
0.94
0.54
0.71
GPRP3
0.40
0.56
0.46
0.49
P5CS
0.76
1.75
1.09
1.30
BZIP
0.35
0.51
0.35
0.25
ERF3
0.59
0.92
0.60
0.68
CLC1
0.63
0.28
0.60
0.38
NHX1
0.44
0.54
0.60
0.38
PAP3
0.43
0.31
0.50
0.34
Shinozaki and Yamaguchi-Shinozaki (1997) also
mentions that some of the genes that responsive to
drought stress, high salinity, and cold temperatures at
the level of transcription (mRNA) have been widely
reported. The amount of mRNA of the gene that is
responsive to salinity decreases if the stress on the
plant is stopped. This is an evidence that these genes
induced by salinity on plant growth environment.
Gene expression that induced by environmental
stress will produce proteins that function as a signal
conductor from the surface of plant cells into the cell,
enzymes that involved in the biosynthesis of
molecules that influence the defense mechanisms
(such as proline, some types of carbohydrates and
polyamine), or a transcription factor that activates the
expression of genes that play a role in plant defense
mechanisms against the stress. Cis and elements trans
involved in gene expression that induced by stress has
been widely analyzed in detail and carefully to ravel
the mechanisms of plants in defend themselves
against environmental stress. Dehydration-responsive
element (DRE) is cis elements that contained in a
promoter and co-regulate the expression of genes in
times of drought stress, high salinity and cold
temperatures. This element has a motive sequences
A/GCCGAC that detected by DRE-binding protein
(DREB) transcription factor (Hardiarto, 2010). One
of the DREB gene subfamily is GmDREB5. This
gene plays an important role in the soybean plant
resistance to drought stress by recognizing the
response of dehydration (Lan et al., 2011). The results
of research showed the level of gene expression
DREB 5 and GPRP3 gene (Glycine and Proline Rich
Proteins 3) in third generation of selected soybean
salinity increases with salinity stress (Figure
1). Penget al. (2012) also reported an increase in gene
expression, particularly in soybean roots that induced
by salinity stress. DREB5 and GPRP3 allegedly play
an important role in mediating independent pathways
of ABA (abscisic acid) of the salinity stress (Phang et
al., 2008; Peng et al., 2012). Decrease in leaf water
potential to stimulate the synthesis of ABA. ABA
concentration in the crown would affect the
expression of genes that determine the synthesis of
proteins (including protein function and protein
regulator). Functional protein that referred is among
LEA proteins, proteinase, detoxifying enzymes, and
synthesis regulator and osmotic controller enzymes
osmotic, namelyproline, betaine and sugar. It appears
that the rapid response of plants is closing of
stomata. While the density of stomata allegedly
changed after the plant experienced continual stress
in a relatively long time (weekly to monthly) other
changes in the crown, leaf senescence, changes in
root growth, vernalization changes, when flowering
and seed filling. There appears to be the link between
a decrease of relative water content of leaves that
followed by an increase in ABA with synthesis of
proline as osmotic control compound (Shinozaki and
Yamaguchi-Shinozaki, 1997; Passiora, 1996;
Swasono, 2012).
MRNA expression of several genes in unselected
soybean Grobongan varieties that treated salinity also
showed an enhancement that is DREB5, P5CS and
ERF3 compared with controls (Table 5 and Figure
2). P5CS genes showed the highest level of
expression of 1.30 fold. While the level of gene
expression GPRP3, ZIP, CLC1, NHX1 and PAP3 in
soybean Grobongan varieties that are subjected lower
salinity treatment than the control.
ICONART 2019 - International Conference on Natural Resources and Technology
34

Figure 1: Level of Gene Expression in Selection Soybean
that Salinity Resistant F3, Grobogan varieties (**t <0.001,
*t> 0.05 Compared Control Using t test).
Figure 2: Level of Gene Expression in Soybean Grobogan
Varieties (** t <0.001, * t> 0.05 Compared Control Using t
test).
Proline is the main compounds that can protect the
cells through stabilization of proteins and cell
membranes. On a variety of plant species, found
accumulation of proline in salinity stress. Proline
accumulation is regulated by a balance between the
synthesis and catabolism. Δ1-Pyrroline-5-
Carboxylate Synthetase (P5CS) is a key enzyme in
the mechanism of proline. , P5CS is primarily
responsible genes in the biosynthesis of proline and
proline decomposition by the proline dehydrogenase
enzymes that is an enzyme in mitochondria and the
other mechanisms that can increase the concentration
of proline. The level of gene expression is very
important to understand the mechanism of proline
accumulation (Ashraf and Foolad 2007; Lutts et al.,
1999). The results of research showed the expression
of gene P5CS in selected soybean salinity F3
increases with salinity stress, as well as soybean
Grobogan varieties despite an increase in that gene
expression was lower than selected soybean salinity
(Figure 1 and Figure 2). Celik and Atak’s research
(2011) also showed an increase in gene expression
P5CS by increasing the salt concentration in the
growing media, especially in soybeans that salinity
tolerant.
BZIP gene expression Transcription Factor (ZIP)
in selected soybean that salinity resistant also showed
an increase in the presence of salinity stress, while in
soybean Grobongan varieties and Burangrang
decreased due to salinity stress. Gao et al. (2011)
explained that over-expression of GmbZIP1increases
the response of transgenic plant to forming of an
independent ABA and triggering stomatal closure
under stress conditions, thereby potentially increasing
the tolerance to multiple abiotic stresses including
high salt stress. So far, the relationship between salt
stress and stomata is still largely unknown. In this
research, it is known that GmbZIP protein play a
positive role in stomatal closure as effect of salt
induction. The results showed that GmbZIP can act as
a positive regulator of the salinity response by
controlling stomatal closure. Thus, under salinity
stress, excess GmbZIP in transgenic plants may be
able to prevent the entry of Na+ and Cl-, stomatal
closure, and reduce membrane cell damage that
caused by ions and so that increases tolerance to salt
stress.
The results also showed gene expression
EREBP/AP2 Transcription Factor 3 (ERF3) increases
in selected soybean that salinity resistant that
experiencing salinity, in soybean Grobogan varieties
increased expression of the gene is not as high in
selected soybean (Figure 1 and Figure 2). The same
result has also been reported to Zhang et al., (2009) in
the treatment of salt stress, mRNA GmERF3
accumulates after 5 hours of initiation and reached a
peak after 10 hours of initiation. GmERF3 transgenic
tobacco than given 200 mM NaCl stress is able to
maintain the green leaves and roots that can grow
well. Allegedly biotic and abiotic stresses, including
salinity stress can regulate the expression of GmERF3
and abundance of transcriptional activator GmERF3
and most likely regulate transcription, up-regulate
several genes that induced by stress, and their protein
products contribute to increase resistance to stress
conditions. In addition, the accumulation of soluble
sugars and proline as osmolite, plays an important
role in plants that exposed to stress.
In addition to increased osmolite, maintenance of
ion homeostasis is an important strategy in plants to
survive in salinity stress. This mechanism will
prevent the toxicity effect that repercussions of ion
poisoning which causes cytoplasmic organelle
membrane damage. GmNHX1 and GmCLC1 are
genes that regulate ion homeostasis in soybean (Li et
al., 2006). The results of research showed that the
salinity stress also increases the gene expression of
Evaluation of Resistance Improvement of Soybean (Glycine Max (L) Merr.) against Salinity using Mass Selection and Gene Expression of
Salinity Tolerant
35

Gm Putative Na+/H+ antiporter (NHX1) in selected
soybean that salinity resistant F3, while in soybean
Grobogan varieties and Burangrang, gene expression
is decreased in the presence of salt stress. It shows
gene expression of NHX1 in selected soybean that
salinity resistant F3 more resistant to salinity stress
than soybean Grobogan varieties. Staal et al. research
results (1991) also showed the activities of NHX
tonoplast higher on Plantogomaritima NaCl tolerant
compared to Plantogomaritima NaCl
sensitive. Research Li et al., (2006) showed the
transgenic cell vacuole of GmNHX1-YFPcontained
Na+ accumulation, whereas in control YFD vacuoles
is no accumulation of Na+. Allegedly ion transporter
that located in the plasma membrane and tonoplast
can help the release of ion from cells and ions
compartment within the cell, thereby reducing the
effects of ion poisoning.
GmCLC1 gene encodes a protein of Cl- to the
vacuole, namely by transferring ions from the
cytoplasm into the vacuole to reduce the toxic effects
of salts (Li et al., 2006). GmCLC1 genes also play an
important role to increase plant tolerance to salinity,
reducing damage to the structure of the membrane,
increasing osmotic adjustment and regulation of
antioxidant enzymes in salinity stress conditions (Sun
et al., 2013). Gene expression Gm Chloride Channel
1 (GmCLC1) in selected soybean that salinity
resistant F3, soybean Grobogan varieties on NaCl
stress conditions lower than those in the control
treatment. Instead, the research of Sun et al. (2006)
showed increased expression of GmCLC1 which
increases the tolerance of transgenic plants
Populusdeltoides × P. euramericana 'Nanlin 895'
against salinity stress. This difference is expected
because Cl compartment not only increased because
GmCLC1 expression and the presence of other
mechanism of Cl released that reduce toxic effects on
plants.
Salinity stress not only induces the accumulation
of proline, sugars and other osmolite, but it can also
lead to increased production of reactive oxygen
species (ROS) that can cause damage to lipid
membranes, proteins and nucleic acids that can cause
cell death. Plants develop enzymatic protection
mechanisms that can be scavenged ROS and prevent
the damaging effects of free radicals. Liao et
al. (2003) stated that the GmPAP3 genes alleged
related to soybean adaptation to NaCl stress through
its involvement in the depuration of reactive oxygen
species (ROS). The results showed GmPAP3
expression in selected soybean that salinity resistant
lower than the treatment without salt stress, so it is
necessary to put other efforts to help plants cope with
oxidative stress that caused by ROS. Exogenous
antioxidants applications such as ascorbic acid,
salicylic acid, glutathione, tocoferol, ubiquinone,
ubiquinol, and cysteine are expected to help soybean
plants to overcome the problems of the free
radicals. Research of Sitinjak et al. (2012)
demonstrated the application of ascorbic acid at a
dose of 500 ppm on soybean Grobogan varieties in
saline soil with DHL 5-6 mmhos/cm that produce the
highest production. Based on these results, in the
third stage studies of ascorbic acid is applied to
increase soybean resistance to salinity stress.
4 CONCLUSIONS
Salinity-resistant soybean selection shows success
which is indicated by high and moderate heritability
in production characters and increased selection
progress. Molecular test of the third generation
salinity tolerant gene shows the expression of
DREB5, GPRP3, P5CS genes, bZIP Transcription
Factor (ZIP), EREBP/AP2 Transcription Factor
(ERF3) and Gm Putative Na
+
/H
+
Antiporter (NHX1)
were higher in selected F3 salinity-resistant soybeans
that received salinity stress compared to control
treatment, whereas GmCLC1 and PAP3 genes is
lower than the control treatment. Increasing the
heritability and expression of several salinity tolerant
genes in salinity-resistant soybean related to their role
in plant defense mechanisms against salinity stress
shows the potential for selected soybeans to be
developed as a salinity tolerant variety.
ACKNOWLEDGEMENTS
The authors wish to express sincere thankfulness to
Directorate General of Higher Education, Ministry of
Research and Higher Education, Indonesia for
financial support on Sandwichlike Program and Prof.
Hirosuke Oku (Centers of Molecular Biosciences,
University of the Ryukyus, Okinawa, Japan) as
supervisor and facility support in this study.
REFERENCES
Ashraf, M. 2004. Some important physiological selection
criteria for salt tolerance in plants. Flora, 199: 361-376.
Ashraf, M., Foolad, M. R. 2007. Roles of glycine betaine
and proline in improving plant abiotic stress resistance.
Environmental and Experimental Botany, 59: 207-216.
ICONART 2019 - International Conference on Natural Resources and Technology
36

Ceccarelli, S., Erskine, W., Humblin, J., Brando, S. 2007.
Genotype by environment interaction and international
breeding program. http://www.icrisat.com.
Celik, O., Atak, C. 2011. Evaluation of the proline
accumulation and Δ'-pyrroline-5-carboxylate
synthetase (P5CS) gene expression during salinity
stress in two soybean (Glycine max L. Merr.) Varieties.
Plant Molecular Biology Reporter, 30: 566-577.
Ermawati, N. 2011. Ermawati, N. 2011. Isolation and
characterization of intrinsic membrane protein-coding
genes of plants halofit tonoplas Salicornia herbacea.
Journal of Basic Science, 12 (1): 23-29.
Gao, S. Q, Chen, M., Xu, Z. S., Zhao, C. P., Li, L., Xu, H.,
Tang, Y., Zhao, X., Ma, Y.Z. 2011. The soybean
GmbZIP1 transcription factor enhances multiple abiotic
stress tolerances in transgenic plants. Plant Molecular
Biology, 75: 537-553.
Ghassemi-Golezani, K., Taifeh-Noori, M. 2011. Soybean
performance under salinity stress. soybean. In
Biochemistry, Chemistry and Physiology. Intechopen,
631-642.
Hardiarto, T. 2010. Transcription factors of OsDREB1A
and OsDREB1B in rice. Research and Development
Center for Biotechnology and Genetic Resources
Agriculture. Institution of Agricultural Research and
Development, Ministry of Agriculture. http://biogen.
litbang.pertanian.go.id/index.php/2010/11/faktor-
transkripsi-osdreb1a-dan-osdreb1b-pada-padi/
Idris, U. M., Yakop, Farida, N. 2011. Progress in mass
selection of local corn cultivar (kebo) intercropping
with peanuts after one selection cycle. Kemajuan
seleksi massa pada jagung kultivar lokal kebo setelah
satu siklus seleksi dalam pertanaman tumpangsari
dengan kacang tanah. Crop Agronomy, 4 (2): 37-42.
Lan, C. H., Nguyen, T. A., Nguyen, V. T. T., Nguyen, H.
H., Mau, C. H. 2011. The characterization of the
GmDREB5 gene isolated from soybean cultivar
XanhTiendai, Vietnam. In Proceedings of International
Conference on Biology, Environment and Chemistry 1:
354-358.
Li, W. F., Wong, F. L., Tsai, S, N., Phang, T. H., Shao, G.,
Lam, H. M. 2006. Tonoplast-located GmCLC1 and
GmNHX1 from soybean Enhance NaCl tolerance in
transgenic bright yellow (BY) -2 cells.Plant, Cell and
Environment, 29: 1122-1137.
Liao, H., Wong, T., Phang, T. H., Cheung, M. Y., Li, W. F.,
Shao, G., Yan X., Lam, H. M. 2003. GmPAP3, a novel-
like purple acid phosphatase gene in soybean induced
by NaCl stress but not phosphorus deficiency. Gene,
318: 103- 111.
Marquez-Ortiz, J. J., Lamb, J. F. S., Johnson, L.D., Barnes,
D.K., and Stucker, R.E. 1999. Heritability of crown
properties in alfalfa. Crops Science, 39: 38 -43.
Munns, R. 2005. Genes and salt tolerance: bringing them
together. New Phytologist, 167 (3): 645-663.
Passiora, J. B., 1996. Drought and drought tolerance. Plant
Growth Regulation, 20: 79-83.
Pathan, M. S., Lee, J. D., Shannon, J. G., Nguyen, H. T.
2007. Recent advances in breeding for drought and salt
stress tolerance in soybean. In Advances in molecular
breeding toward drought and salt tolerant crops
Springer, Dordrecht. 739-773.
Peng, H., Feng, Y., Zhang, H., Wei, X., Liang, S. 2012.
Molecular cloning and characterization of genes coding
for Glycine- and Proline-Rich Proteins (GPRPs) in
Soybean. Plant Molecular Biology Reporter, 30: 566-
577.
Phang, T. H., Shao, G., Lam, H. M. 2008. Salt tolerance in
soybean. Journal of Integrative Plant Biology, 50(10),
1196-1212.
Roy, D., 2000. Plant breeding: Analysis and exploitation of
variation. Narosa Publishing House Calcutta.
Santoso, T. J., Abdullah, B., Carsono, N., Apriana, A.,
Sisharmini, A., Trijatmiko, K. R. 2012. Synergy and
stability expression of the Os F1 and OsDREB1A genes
in the transgenic Ciheran x Nipponbare crossing
program for high salinity tolerance. In SiNas
Proceedings 2012.
Shinozaki, K., Yamaguchi-Shinozaki, K. 1997. Gene
expression and signal transduction in water stress
response. Plant Physiology, 115: 327-334.
Sudarmi. 2013. The role of molecular biology in plant
breeding. Magistra, 84 (XXV): 75-80.
Sun, W., Deng, D., Yang, L., Zheng, X., Yu, J., Pan, H.,
Zhuge, Q. 2013. Overexpression of the chloride
channel gene (GmCLC1) from soybean increases salt
tolerance in transgenic Populus deltoides × P.
euramericana 'Nanlin895'. Omics Plant Journal, 6 (5):
347-354
Sutoro, A., Bari, Subandi, Yahya, S. 2006. The genetic
parameters of Bisma corn population on different
fertilization. Variety of additives-dominant corn seed
weight. AgroBiogen Journal, 2 (2): 60-67.
Swasono, F. D. H., 2012. The role of ABA and proline in
the mechanism of adaptation of shallots to drought
stress in coastal sand soil. AgriSains Journal, 4 (5): 71-
78.
Turan, S., Cornish, K., Kumar, S. 2012. Salinity tolerance
in plants: Breeding and genetic engineering. Australian
Journal of Crop Science, 6 (9): 1337-1348
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., Ma,
Y. 2009. The soybean GmERF3 overexpression of the
gene, an AP2 / ERF type transcription factor for
Increased tolerances to salt, Drought, and diseases in
transgenic tobacco. Journal of Experimental Botany, 60
(13): 3781-3796.
Evaluation of Resistance Improvement of Soybean (Glycine Max (L) Merr.) against Salinity using Mass Selection and Gene Expression of
Salinity Tolerant
37
