
Information on Polyprenol Reductase Enzyme in the NCBI Online
Mohammad Basyuni
1,2
, Yuntha Bimantara
1
, Rahmah Hayati
1
, Jayusman
3
, Rizka Amelia
1
, Irma Deni
1
,
Arif Nuryawan
1,2
, Sumaiyah
2,4
and Etti Sartina Siregar
2,5
1
Department of Forestry, Faculty of Forestry, Universitas Sumatera Utara, Jl. Tri Dharma Ujung No. 1 Medan, North
Sumatera 20155, Indonesia
2
Center of Excellence for Mangrove, Universitas Sumatera Utara, Medan, North Sumatera 20155, Indonesia
3
Balai Besar Penelitian Dan Pengembangan Bioteknologi dan Pemuliaan Tanaman Hutan, Yogyakarta 55582, Indonesia
4
Faculty of Pharmacy, Universitas Sumatera Utara, Medan, North Sumatra 20155, Indonesia
5
Faculty of Mathematics and Natural Sciences, Universitas Sumatera Utara, Medan, North Sumatra 20155, Indonesia
ameliarizka0@gmail.com, irma.deni31@gmail.com, arif5@usu.ac.id, sumaiyah7777@gmail.com, ettisartina@yahoo.com
Keywords: Gene, Plasma Membrane, Polyprenol Reductase.
Abstract: Polyprenol reductase an enzyme is necessary to convert polyprenol to dolichol in the last phase of dolichol
biosynthesis. The present work reports search result of National Center for Biotechnology Information
(NCBI) databases on polyprenol reductase. To produce some useful data, the search for NCBI databases
(https:/www.ncbi.nlm.nih.gov/) was used. Results detected in 16 databases for polyprenol reductase. The
databases of the polyprenol reductase consist of literature, health, genomes, genes, protein, and chemical
features. The literature contained 16 bookshelves, 1 MeSH (Medical Subject Headings), 16 PubMed, and 58
PubMed Central. Health comprised 3532 Clincar documents, 163 dbGap, 899 GTR, 257 MedGen, 3 OMIM
(Online Mendelian Inheritance in Man database). Gene involves of 681 Genes, 303 GEO profiles, 1
HomoloGene, and 20 UniGenes. Proteins properties contained 933 Identical Protein Groups, 1,208 Proteins,
and 4 Protein Clusters. Genomes included 37,135 nucleotides, which are derived from 34,821 bacteria,
1,151 animals, 603 plants, 340 archaea, 141 fungi, and 7 viruses. The chemicals property represented 3648
BioSystems and five bioactivity screening studies. The present data provides indispensable information
about biotechnology of polyprenol reductase enzyme.
1 INTRODUCTION
Polyprenol reductase is an enzyme that stimulates
the reduction of polyprenols into dolichols in the
dolichol biosynthesis (Rosenwald et al., 1993;
Quellhorst Jr et al. 1997; Sagami et al. 2018). The
occurrence of polyprenol reductase has been
described as various organisms. In the plant
kingdom, for example, has been described in
Arabidopsis thaliana (Jozwiak et al., 2015),
Kandelia obovata leaves (Basyuni et al., 2018a,b,c),
spinach leaves (Sakaihara et al., 2000). Polyprenol
reductase has been shown in Haloferax volcani, a
halophilic archeon (Naparstek et al., 2012),
mammalian (Dsouzaschorey et al., 1994), yeasts
(Tateyama and Sagami, 2001; Szkapinska et al.,
2006), hamster (Chaves et al., 2015), and human
(Cantegral et al., 2010).
The biological and pharmacological activity of
polyprenol reductase has been described, such as
prostate cancer prevention (Nacusi and Tindall,
2011; Schmidt and Tindall, 2011). Polyprenol
reductase has been shown to play a role in
congenital disorders of glycosylation (CDG)
(Gründahl et al., 2012) and genetic defects of
dolichol metabolism in dolichol-related CDGs for
clinical and biochemical phenotypes (Buczkowska et
al., 2015).
In this context, it is essential to understand
further the polyprenol reductase enzyme relating
biotechnology from all databases available. Here we
report a brief review via a search engine to gather
useful data in biotechnology-related science studies.
Therefore, the present study aimed to report the use
of the databases of the National Center for
Biotechnology Information (NCBI) search to obtain
more insight on updated biotechnology related to
polyprenol reductase enzyme.
104
Basyuni, M., Bimantara, Y., Hayati, R., Jayusman, ., Amelia, R., Deni, I., Nuryawan, A., Sumaiyah, . and Siregar, E.
Information on Polyprenol Reductase Enzyme in the NCBI Online.
DOI: 10.5220/0008525901040107
In Proceedings of the International Conference on Natural Resources and Technology (ICONART 2019), pages 104-107
ISBN: 978-989-758-404-6
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
2 MATERIALS AND METHOD
The material of this work applied databases of
polyprenol reductase that were used to produce some
valuable information biotechnology can be
conducted using NCBI databases search engine
online (https://www.ncbi.nlm.nih.gov/). The
material was derived from typing on all database
search as mentioned earlier on January 22, 2019.
Information features include the Bookshelf, MeSH
(Medical Subject Headings), NLM (National Library
of Medicine) Catalog, PubMed, PubMed Central,
Clinvar, a database of Genotypes and Phenotypes
(dbGap), Genetic Testing Registry (GTR). MedGen,
Online Mendelian Inheritance in Man (OMIM),
BioSample, Clone, database of human genomic
structural variation (dbVar), Genome, GSS (Genome
Survey Sequences), Nucleotide, Probe, EST, Gene,
Gene Expression Omnibus (GEO) Profiles,
HomoloGene, UniGene, Identical Protein Groups,
Protein, Protein Clusters, Biosystems, and PubChem
BioAssay.
3 RESULT AND DISCUSSION
The exploration for the polyprenol reductase enzyme
showed sixteen databases in the NCBI database.
Table 1 depicts the literature available in the NCBI
concerning polyprenol reductase. Four biographies,
namely bookshelf, MeSH, PubMed, and PubMed
Central with a number of deposited documents. The
online NCBI literature provides online libraries and
free access to bookshelf data (sixteen books and
reports). MeSH contained one ontology used for
Pubmed indexing. PubMed covered the publication
year 1990-2018.
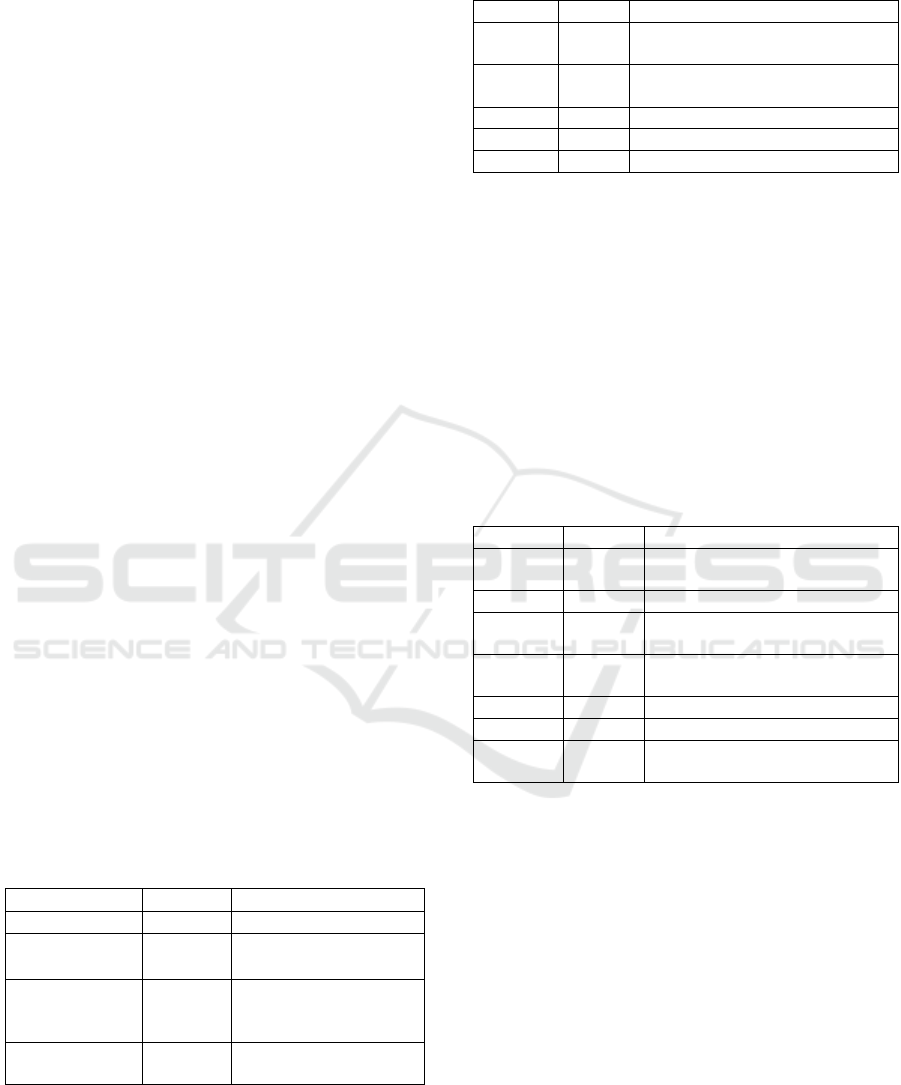
Table 1: Literature source NCBI database for polyprenol
Reductase.
Literature
Sum
Information
Bookshelf
16
Books and reports
MeSH
1
The ontology used to
index Pubmed
PubMed
16
Abstracts/quotations
from science and
medicine
PubMed Central
58
Full-text journal
articles
Table 2: Health source NCBI database for polyprenol
reductase.
Health
Total
Definition
Clinvar
3532
variations in human clinical
importance
dbGap
163
genotype/phenotype interaction
issues
GTR
899
genetic testing registry
MedGen
257
Medical genetics literature and links
OMIM
3
online mendelian inheritance in man
Information on health sources is displayed in
Table 2. This data is including 3,532 ClinVar
relating to individual variations of clinical
significance, 163 dbGap, 899 GTR, 257 MedGen. It
is important to mention that this research (Table 2)
identified the OMIM (Online Mendelian Inheritance
in Man). The OMIM consisted of Steroid 5-alpha-
reductase 3, SRD5A3, Steroid 5-alpha-reductase 1,
SRD5A1, and Congenital disorder of glycosylation
(CDG), CDG1Q as previously was reported
((Gründahl et al., 2012; Buczkowska et al., 2015).
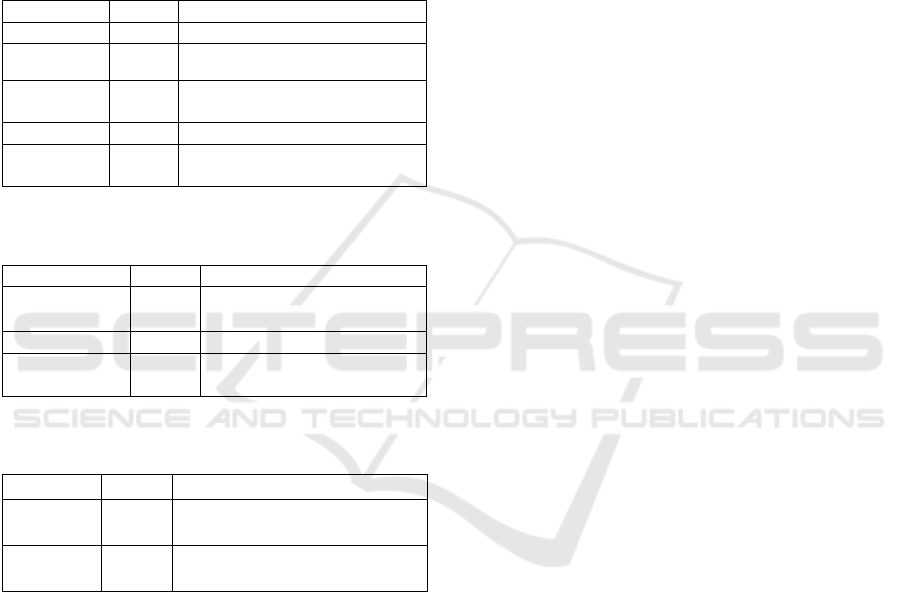
Table 3: Genomes source NCBI database for polyprenol
reductase.
Genomes
Number
Explanation
BioSample
302
descriptions of materials from
biological sources
Clone
12
genomic and cDNA clones
dbVar
21,375
studies of structural genome
variation
Genome
4
organizational genome
sequencing projects
GSS
1,514
genome project sequences
Nucleotide
37,135
DNA and RNA sequences
Probe
18
samples and primers based on
sequences
Furthermore, genome source for polyprenol
reductase enzyme namely 302 BioSample, 12 clones
of genomic and cDNA, 21,375 trials of structural
genome variability, 4 projects of genome sequencing,
1514 GSS, 37,135 DNA and RNA sequences, and
18 Probes. It is intersting that from 37,135
nucleotides in the databases comprised of 1,151
animals, 603 plants, 141 fungi, 78 protists, 34,821
bacteria, 340 archea, and 7 viruses while the
molecular type contained 36,192 DNA/RNA and
851 mRNA.
The Genes collection consisted of 1 Expressed
sequence tag (EST), 681 information about gene
loci, 303 gene expressions, one homologene, and 20
Unigenes as depicted in Table 4. Table 5 shows the
protein source for 933 identical protein groups,
1,208 protein, and 4 protein clusters. Four protein
Information on Polyprenol Reductase Enzyme in the NCBI Online
105
clusters are 3-oxo-5-alpha-steroid 4-dehydrogenase,
C-terminal domain containing Embryophyte protein
(Accession: PLN03164), 3-oxo-5-alpha-steroid 4-
dehydrogenase family protein conserved in
Arabidopsis thaliana (Accession: CLSN2912717),
probable polyprenol reductase 1-like conserved in
BOP clade, and probable polyprenol reductase 2-like
conserved in Glycine max (Accession:
CLSN2960920).
Table 4: Genes source NCBI database for polyprenol
Reductase.
Genes
Number
Definition
EST
1
expressed sequence tag sequences
Gene
681
collected gene loci data
GEO Profiles
303
gene expression and profiles of
molecular abundance
HomoloGene
1
homologous gene
UniGene
20
sets of expressed transcripts for
chosen organisms
Table 5: Proteins source NCBI database for polyprenol
reductase.
Proteins
Number
Explanation
Identical Protein
Groups
933
identity grouped protein
sequences
Protein
1,208
protein sequences
Protein Clusters
4
Sequence of protein clusters
based on similarity
Table 6: Chemicals source NCBI database for polyprenol
reductase.
Chemicals
Amount
Information
BioSystems
3,648
gene, protein and chemicals
linked molecular pathways
PubChem
BioAssay
5
screening of bioactivity
The biological activities have been well
documented (Nacusi and Tindall, 2011; Schmidt and
Tindall, 2011Buczkowska et al., 2015; Tao et al.,
2016). Polyprenol reductase has been reported to
play an essential role in CDG (Gründahl et al., 2012)
and the clinical and biochemical phenotypes in
dolichol-concerning CDG (Buczkowska et al., 2015).
Probable polyprenol reductases from K. obovata
had been reported (Basyuni et al., 2018a,b,c) to
different previous data on polyprenol reductase from
A. thaliana (Jozwiak et al., 2015). The position of
predicted polyprenol reductase from K. obovata sit
together with Ricinus communis and Iopoea nil
(Basyuni et al., 2018b,c; Basyuni and Wati, 2018).
Variation of chemical features of the polyprenol
reductase as was displayed in Table 6, in this report
only BioSystems and PubChem BioAssay were
detected. Biosystems contained two types, conserved
biosystems (206) and organism-specific biosystems
(3442). According to a record type, 3,608 pathways,
28 structural complexes, 12 functional sets. Relating
to source name consisting of BioCyc (2), GO (196)
and KEGG (3,337). Whereas the source of databases
derived from INSDC/GenBank was 24,798 and
RefSeq was 12,331. The PubChem BioAssays
comprised 5 bioactivities screening studies (Table
6). These five Assays are Upregulation of NADH-
cytochrome b5 reductase expression in Candida
albicans SC5314 at 200 uM after 24 hrs by MALDI-
TOF MS, Upregulation of Glutathione reductase
expression in Candida albicans SC5314 at 200 uM
after 24 hrs by MALDI-TOF MS, Upregulation of
Mitochondrial 2-enoyl thioester reductase
expression in Candida albicans SC5314 at 200 uM
after 24 hrs by MALDI-TOF MS, Upregulation of
Aldose reductase expression in Candida albicans
SC5314 at 200 uM after 24 hrs by MALDI-TOF
MS, and Inhibition of 5alpha-reductase in human
epidermis assessed as inhibition of [14C]testosterone
to DHT after 24 hrs.
4 CONCLUSIONS
The NCBI online discusses different data about
polyprenol reductase enzyme in biology and
biotechnology. The current study also delivers
important data regarding biotechnology of
polyprenol reductase.
ACKNOWLEDGMENTS
This work was in part assisted by World Class
Research from Directorate for Research and
Community Service, Ministry of Research,
Technology and Higher Education, the Republic of
Indonesia.
REFERENCES
Basyuni, M., Sagami, H., Baba, S., and Oku, H. 2018.
Genome sequence analysis of predicted polyprenol
reductase gene from mangrove plant Kandelia
obovata. IOP Conference Series: Earth and
Environmental Science 130, 012039.
ICONART 2019 - International Conference on Natural Resources and Technology
106

Basyuni, M., Wati, R., Sagami, H., Oku, H., and Baba, S.
2018. Bioinformatics approach of three partial
polyprenol reductase genes in Kandelia obovata.
Journal of Physics: Conference Series 978, 012044.
Basyuni, M., Baba, S., Wati, R., Sumardi, Sulistiyono, N.,
Oku, H., and Sagami, H. 2018. Isolation and
phylogenetic analysis of new predicted polyprenol
reductase from mangrove plant (Kandelia obovata
Sheue, HY Liu & J. Yong). AIP Conference
Proceedings 2002, 020041.
Basyuni, M., and Wati, R. 2018. Bioinformatics analysis
of the predicted polyprenol reductase genes in higher
plants. Journal of Physics: Conference Series 978,
012050.
Buczkowska, A., Swiezewska, E., Lefeber, D. J. 2015.
Genetic defects in dolichol metabolism. Journal of
inherited metabolic disease, 38(1), 157-169.
Cantagrel, V., Lefeber, D. J., Ng, B. G., et al., 2010.
SRD5A3 is required for converting polyprenol to
dolichol and is mutated in a congenital glycosylation
disorder. Cell, 142(2), 203-217.
Chávez, B., Ramos, L., García-Becerra, R., and Vilchis, F.
2015. Hamster SRD5A3 lacks steroid 5α-reductase
activity in vitro. Steroids, 94, 41-50.
Dsouzaschorey, C., McLachlan, K. R., Krag, S. S., and
Elbein, A. D. 1994. Mammalian glycosyltransferases
prefer glycosyl phosphoryl dolichols rather than
glycosyl phosphoryl polyprenols as substrates for
oligosaccharyl synthesis. Archives of Biochemistry
and Biophysics, 308(2), 497-503.
Gründahl, J. E. H., Guan, Z., Rust, S., et al., 2012. Life
with too much polyprenol: polyprenol reductase
deficiency. Molecular Genetics and Metabolism,
105(4), 642-651.
Jozwiak, A., Gutkowska, M., Gawarecka, K., et al., 2015.
POLYPRENOL REDUCTASE2 deficiency is lethal in
Arabidopsis due to male sterility. The Plant Cell,
27(12), 3336-3353.
Naparstek, S., Guan, Z., and Eichler, J. 2012. A predicted
geranylgeranyl reductase reduces the ω-position
isoprene of dolichol phosphate in the halophilic
archaeon, Haloferax volcanii. Biochimica et
Biophysica Acta (BBA)-Molecular and Cell Biology of
Lipids, 1821(6), 923-933.
Nacusi, L. P., and Tindall, D. J. 2011. Targeting 5α-
reductase for prostate cancer prevention and treatment.
Nature Reviews Urology, 8(7), 378.
Quellhorst Jr, G. J., Hall, C. W., Robbins, A. R., and Krag,
S. S. 1997. Synthesis of dolichol in a polyprenol
reductase mutant is restored by elevation of cis-
prenyltransferase activity. Archives of Biochemistry
and Biophysics, 343(1), 19-26.
Rosenwald, A. G., Stanley, P., McLachlan, K. R., & Krag,
S. S. 1993. Mutants in dolichol synthesis: conversion
of polyprenol to dolichol appears to be a rate-limiting
step in dolichol synthesis. Glycobiology, 3(5), 481-
488.
Sagami, H., Swiezewska, E., & Shidoji, Y. 2018. The
history and recent advances in research of polyprenol
and its derivatives. Bioscience, Biotechnology, And
Biochemistry, 82(6), 947-955.
Sakaihara, T., Honda, A., Tateyama, S., and Sagami, H.
2000. Subcellular fractionation of polyprenyl
diphosphate synthase activities responsible for the
syntheses of polyprenols and dolichols in spinach
leaves. The Journal of Biochemistry, 128(6), 1073-
1078.
Schmidt, L. J., and Tindall, D. J. 2011. Steroid 5 α-
reductase inhibitors targeting BPH and prostate
cancer. The Journal of Steroid Biochemistry and
Molecular Biology, 125(1-2), 32-38.
Stiles, A. R., and Russell, D. W. 2010. SRD5A3: a
surprising role in glycosylation. Cell, 142(2), 196-198.
Szkopinska, A., Swiezewska, E., and Rytka, J. 2006.
Interplay between the cis-prenyltransferases and
polyprenol reductase in the yeast Saccharomyces
cerevisiae. Biochimie, 88(3-4), 271-276.
Tateyama, S., and Sagami, H. 2001. Study on the
biosynthesis of dolichol in yeast: Recognition of the
prenyl chain length in polyprenol reduction. The
Journal of Biochemistry, 129(2), 297-302.
Tao, R., Wang, C., Ye, J., Zhou, H., and Chen, H. 2016.
Polyprenols of Ginkgo biloba enhance antibacterial
activity of five classes of antibiotics. BioMed
Research International, 2016.
Information on Polyprenol Reductase Enzyme in the NCBI Online
107
