
Semantic Search for Biomedical Texts using Predicate-Argument
Structure
Mohammed Alliheedi
1
and Robert E. Mercer
2
1
Department of Computer Science, Al Baha University, Prince Mohammad Bin Saud, Al Bahah 65527, Saudi Arabia
2
Department of Computer Science, The University of Western Ontario, 1151 Richmond St., London, Ontario, Canada
Keywords:
Biomedical Ontology, Semantic Search, Semantic Roles, Knowledge Representation, Formal Ontology.
Abstract:
In this position paper we argue that using semantic roles in addition to using biologically-oriented ontologies
and databases (or knowledge bases) will further enhance the generation of RDF triples that can be collected
from biomedical text. RDF triples have been used to enhance semantic search beyond the simple use of
linguistically oriented additions such as synonyms. We wish to focus on drug-virus interactions.
1 INTRODUCTION
In this position paper we argue that using seman-
tic roles in addition to using linguistically-oriented
and biologically-oriented ontologies and databases
(or knowledge bases) will further enhance the gen-
eration of RDF triples that can be collected from
biomedical text. RDF triples can then be used to
enhance semantic search beyond the simple use of
linguistically oriented additions such as synonyms
(Kohlschein et al., 2018). Linguistically-oriented on-
tologies such as WordNet (Miller, 1995) can provide
synonyms, hyponyms, hypernyms, and meronyms.
VerbNet (Kipper-Schuler, 2005; Kipper et al., 2008)
and BioFrameNet (Dolbey et al., 2006), an im-
portant biologically-oriented extension to FrameNet
(FrameNet, 2020), provides means to connect verbs
with semantic roles. BioFrameNet also provides con-
nections to biological ontologies, such as, GO (Ash-
burner et al., 2000) and Entrez Gene (Entrez Gene,
2020). Recently, interest to extend VerbNet (Kipper-
Schuler, 2005; Kipper et al., 2008) into the biolog-
ical domain has reemerged with BioVerbNet (Chiu
et al., 2019), which follows on the earlier work of
Lippincott et al. (Lippincott et al., 2013), and a sepa-
rate nascent investigation of biochemistry experimen-
tal method procedure verbs (Alliheedi et al., 2019b;
Alliheedi and Mercer, 2019; Alliheedi et al., 2019a;
Alliheedi et al., 2019c).
We wish to focus on drug-virus interactions and
are particularly interested in searching the COVID-19
Open Research Dataset (Wang et al., 2020) for drug-
virus interactions and how the drugs interact with the
viruses. While these just mentioned linguistic ontolo-
gies provide important information about the verbs in
the form of semantic roles that the verbs take as lin-
guistic arguments, what is missing are the relations
among biological entities. These relationships can be
captured by machine accessible RDF triples, subject-
verb-object triples that use the standard nomenclature
provided by various terminology ontologies. Triples
in the biology domain are collected by tools such as
those found in Bio2RDF.org (Bio2RDF, 2020). This
type of machine-accessible knowledge is strongly ar-
gued for in the FAIR guidelines (Wilkinson et al.,
2016). Our immediate objective is to use the seman-
tic role information found in the linguistic ontologies
to generate the RDF triples found in scientific pub-
lications and the text sections found in many of the
biological knowledge bases. Our long term goal is
to use these RDF triples in a semantic search engine
targeted on the COVID-19 Open Research Dataset.
2 ONTOLOGIES AND
KNOWLEDGE SOURCES
In this section, we give an overview about exist-
ing biologically oriented ontologies in the litera-
ture. Table 1 shows some of well known ontolo-
gies that have been developed over the last decades.
These ontologies include the Gene Ontology (GO)
(Ashburner et al., 2000), the Foundational Model of
Anatomy (FMA) (Rosse and Mejino Jr, 2003), the
Relation Ontology (RO) (Smith et al., 2005), the
Alliheedi, M. and Mercer, R.
Semantic Search for Biomedical Texts using Predicate-Argument Structure.
DOI: 10.5220/0010150702990306
In Proceedings of the 12th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2020) - Volume 2: KEOD, pages 299-306
ISBN: 978-989-758-474-9
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
299

Table 1: List of developed ontologies in the literature.
Ontology name Domain Developed by
The Gene Ontology (GO)
describes knowledge of the gene
domain.
(Ashburner et al., 2000;
Gene Ontology Consortium, 2011)
The Foundational Model of
Anatomy Ontology (FMA)
is an ontology about human anatomy (Rosse and Mejino Jr, 2003)
The Relation Ontology (RO)
consists of relations intended to be
used across various ontologies in OBO
Foundry
(Smith et al., 2005)
The Ontology of Scientific
Experiments (EXPO)
defines the general knowledge about
scientific experimental aspects (e.g.,
methodology and design)
(Soldatova and King, 2006)
The Ontology for Chemical
Entities of Biological
Interest (ChEBI)
a database of molecular entities
focusing on small molecules
(Degtyarenko et al., 2007)
The Molecular Methods
Database (MolMeth)
a resource consists of scientific
protocol ontologies
(Klingstr
¨
om et al., 2013)
The Ontology of Scientific
Experiments (EXACT)
describes scientific protocols and
experiments
(Soldatova et al., 2013)
Semanticscience Integrated
Ontology
describes scientific protocols and
experiments
(Dumontier et al., 2014)
The Ontology for
Biomedical Investigations
(OBI)
a resource for annotating biomedical
investigations
(Bandrowski et al., 2016)
ontology of Scientific Experiments (EXPO), the on-
tology for Biomedical Investigations (OBI) (Solda-
tova and King, 2006), the ontology for Chemical En-
tities of Biological Interest (ChEBI), the ontology
of scientific experiments (EXACT) (Soldatova et al.,
2013), and the Molecular Methods Database (Mol-
Meth) (Klingstr
¨
om et al., 2013). These ontologies are
discussed briefly in this section. Most of these on-
tologies describe a set of concepts and categories in
the biological domain that shows their properties and
the relations between them. The goal of these ontolo-
gies is to provide definitive controlled terminologies
that describe entities in the biomedical genre. At-
tention towards designing and building ontologies has
become increasingly central in the biomedical domain
(Rosse and Mejino Jr, 2003). The main aspect of GO
is to provide information that describes gene prod-
ucts using a precisely defined vocabulary (Ashburner
et al., 2000). GO was built using various resources
such as those in (FlyBase Consortium, 2003; Blake
et al., 2000; Ringwald et al., 2000; Ball et al., 2000).
Similarly to GO, ChEBI (Degtyarenko et al., 2007)
was created using data from several resources such
as IntEnz (Fleischmann et al., 2004), KEGG COM-
POUND (Kanehisa et al., 2006), and the Chemical
Ontology. While Go and ChEBI focus on Gene and
molecular entities, OBI
1
(Bandrowski et al., 2016), on
1
http://purl.obolibrary.org/obo/obi
the other hand, focuses on the annotations of biomed-
ical investigations and provides standard tools to rep-
resent study design, protocols and instrumentation
used, the data generated, and the types of analysis
performed on the data. Several ontologies (Courtot
et al., 2008; Brinkman et al., 2010; Zheng et al., 2013;
Soldatova et al., 2013; Dumontier et al., 2014) were
developed and based on the OBI ontology.
EXPO provides detailed descriptions of various
aspects of scientific experiments and their relation-
ships (Soldatova and King, 2006) .
Descriptions of experimental processes are pro-
vided by OBI, and three real-world applications are
discussed in (Brinkman et al., 2010). Some of the
relations in these applications (e.g., inputs, outputs)
add to the ontologies that describe biological entities.
The beta cell genomics application ontology (BCGO)
(Zheng et al., 2013) also uses OBI, but it tends to be a
more descriptive ontology than some of the others that
use OBI, but some of the relations in RO, the relation
ontology (Smith et al., 2005), that are used (e.g., pro-
duces, translate to) do have an ordering sense.
EXACT (Soldatova et al., 2013) and the Se-
manticscience Integrated Ontology (Dumontier et al.,
2014) describe scientific protocols and experiments.
MolMeth is a database which contains scientific
protocol ontologies that conform to a set of laboratory
protocol standards (Klingstr
¨
om et al., 2013).
Other ontologies describe general concepts that
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
300

Table 2: List of Semantic Roles developed in the work of (Alliheedi and Mercer, 2019).
Semantic role Definition
Agent Generally a human or an animate subject.
Patient Participants that have undergone a process.
Predicate A word that initiates the frame.
Theme Participants in a location or undergoing a change of location.
Goal Identifies a thing toward which an action is directed or a place to which something
moves.
Factitive A referent that results from the action or state identified by a verb.
Location The physical place where the experiments took place.
Instrument an object or force that comes in contact with an object and causes some change in them
are useful to a biochemistry procedure-oriented on-
tology include: Ontologies consist of process such as
(Lenat et al., 1985) and (Schlenoff et al., 2000), on-
tology for units of measure (Rijgersberg et al., 2013),
classification of scenarios and plans (CLASP) (De-
vanbu and Litman, 1996), and materials ontology
(Ashino, 2010). Foundational theories such as pro-
cess calculus and regular grammar are essential for
the formalization of procedure-oriented ontologies.
We consulted three knowledge sources in our dis-
cussion in Section 3. MedChemExpress (MCE)
2
, a
company, offers a wide range of high quality research
chemicals and biochemicals. Its website provided
synonym information for some biological elements.
GeneCards
3
is a database of information on all known
and predicted human genes. UniProt
4
is a database of
protein sequence and functional information.
3 SEMANTIC ROLES
Semantic roles are defined as “the underlying re-
lationships that participants have with a verb in a
clause”
5
. Minsky defined frame as “a data-structure
representing a stereotyped situation” (Minsky, 1974),
with frames having a header, slots and slot fillers. Fill-
more (Fillmore, 1976) introduced the notion of frame
semantics as a theory of meaning. A semantic frame
is defined by Fillmore as “any coherent individuatable
perception, memory, experience, action or object”. In
other words, these are coherently world events or ex-
periences. In this work, we are interested in develop-
ing frame semantics at the verb level so that our head-
ers are verbs and our slots are semantic roles, filled
by the words which represent these roles. For exam-
ple, to understand the word “buy”, one would access
the knowledge contained in the commercial transac-
2
https://www.medchemexpress.com
3
https://www.genecards.org
4
https://www.uniprot.org
5
https://glossary.sil.org/term/semantic-role
tion frame which includes words such as the person
who buys the goods (buyer), the goods that are being
sold (goods), the person who sells the goods (seller),
and the currency that the buyer and seller agree on
(money). Motivated by Fillmore’s theory of frame
semantics, FrameNet (Baker et al., 1998) was devel-
oped to create an online lexical resource for English.
This framework includes more than 170,000 manu-
ally annotated sentences and 10,000 words. The com-
putational linguistic community has been attracted to
the concept of frame semantics and developed com-
putational resources using this concept, such as Verb-
Net (Kipper-Schuler, 2005), an on-line verb lexicon
for English and PropBank (Palmer et al., 2005), an
annotated corpus with basic semantic propositions.
Since we are focusing on drug-virus interaction
verbs with the associated semantic roles, we are par-
ticularly interested in analyzing the COVID-19 Open
Research Dataset (Wang et al., 2020). Verbs evoke se-
mantic roles in writing. Semantic roles provide salient
pieces of information about experimental steps. Se-
mantic roles are crucial aspects of identifying relevant
information in the biomedical texts. This relevant in-
formation is essential to generate the RDF triples (i.e.,
subject, predicate, and object) which have been used
in the literature (Hu et al., 2017). We can use the
work by (Alliheedi and Mercer, 2019) to label each
sentence with the appropriate semantic roles (see Ta-
ble 2). We have used an annotation scheme for iden-
tifying the structured representation of knowledge in
a set of sentences of biochemistry articles (Alliheedi
and Mercer, 2019). The list of semantic roles include:
Theme, Patient, Agent, Location, Goal. These seman-
tic roles identify the arguments of both verbs (e.g.,
administered) and nominalised verbs (e.g., inhibitor
is a nominalized form of the verb inhibit). Further-
more, the use of ontology that describes drug-virus
interactions is salient because it provides the relations
among various molecules.
Semantic Search for Biomedical Texts using Predicate-Argument Structure
301
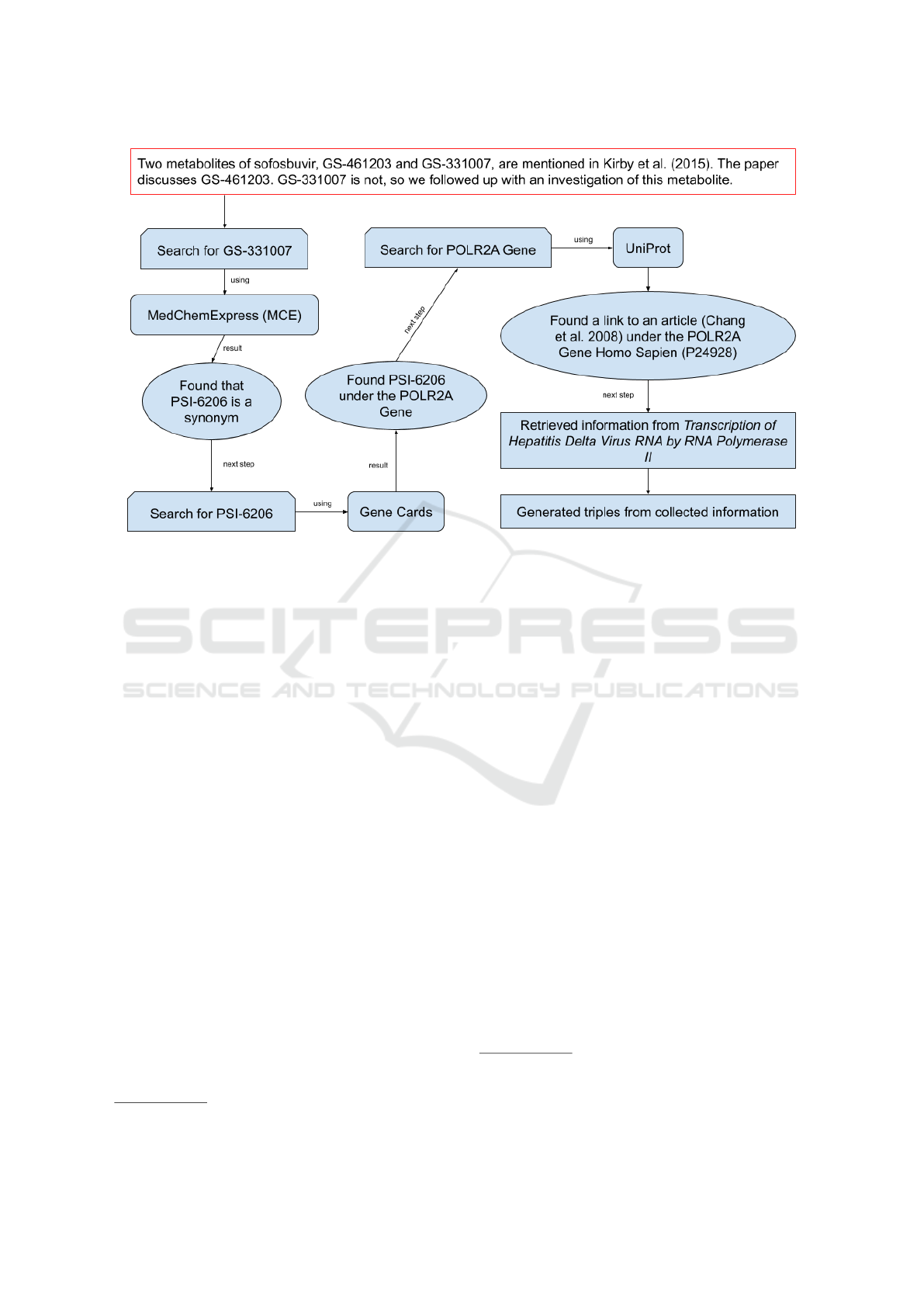
Figure 1: Steps of the example to find information about GS-331007, a metabolite of sofosbuvir.
4 EXAMPLE
We use as an example, a paper that discusses the anti-
viral drug sofosbuvir (Kirby et al., 2015). The title
and abstract of this paper states that sofosbuvir is an
inhibitor of the virus. This produces a very high-level
RDF triple. We would like to have triples that de-
scribe this inhibition in more detail. What is the bio-
chemistry that causes this inhibition? Reading further,
we encounter some information about how this inhibi-
tion takes place. We first find mention of two metabo-
lites of sofosbuvir, GS-461203 and GS-331007. But
the abstract is not that informative regarding why
these metabolites are mentioned. In the full text of
the paper we find “GS-461203, the pharmacologically
active nucleoside analog triphosphate metabolite of
sofosbuvir, is incorporated into HCV RNA by NS5B
polymerase, where it acts as a chain terminator.” So,
this provides us with the information that we need for
the GS-461203 metabolite. We remain interested in
following up with GS-331007. We will need to look
at other information sources. Figure 1 provides a vi-
sual presentation of the search that we now describe.
We cannot find mention of GS-331007 in some of
the knowledge bases that we search. However, we
find in MedChemExpress (MCE) that PSI-6206 and
GS-331007 are synonyms
6
. Now searching for PSI-
6206, it is found in GeneCards under the POLR2A
6
https://www.medchemexpress.com/PSI-6206.html
Gene
7
. We note information about PSI-6206 and its
importance as an inhibitor of the Hepatitis virus. But
no other information is forthcoming. So, we then
turned to UniProt
8
. While looking in UniProt under
POLR2A gene under Homo Sapien
9
we find “(Micro-
bial infection) Acts as an RNA-dependent RNA poly-
merase when associated with small delta antigen of
Hepatitis delta virus, acting both as a replicate and
transcriptase for the viral RNA circular genome.” and
a link to a publication (Chang et al., 2008). In this
publication we find important information about how
the hepatitis delta virus uses host RNA polymerase
which is the protein encoded by the POLR2A gene.
“Previous studies have indicated that the replication
of the RNA genome of hepatitis delta virus (HDV)
involves redirection of RNA polymerase II (Pol II), a
host enzyme that normally uses DNA as a template.
. . . Taken together, we have demonstrated that with
a low concentration of amanitin that only inhibited
Pol II transcription and did not affect host Pol I or
Pol III transcription there was a significant inhibition
for HDV genomic and antigenomic RNAs. Thus, we
believe that Pol II is required for the transcription of
both genomic and antigenomic HDV RNAs.” (Chang
et al., 2008).
7
https://www.genecards.org/Search/Keyword?
queryString=PSI-6206
8
https://www.uniprot.org
9
https://www.uniprot.org/uniprot/P24928
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
302

We now have a number of sources that have pro-
vided factual information and textual information.
The factual information (e.g., sofosbuvir is a drug)
can be readily made into an RDF triple. Knowledge
about drugs (drugs ending in ir are anti-viral drugs)
can be used to further refine the triple. The textual
information can be analyzed using the semantic roles
that we have discussed in Section 3. The semantic
roles can be used to form RDF triples (agent is the
subject, predicate is the verb, and theme and instru-
ment are the objects). As an example, we can la-
bel “GS-461203, the pharmacologically active nucle-
oside analog triphosphate metabolite of sofosbuvir, is
incorporated into HCV RNA by NS5B polymerase...”
using the semantic roles in Table 2 as follows:
GS-461203/Patient; is incorporated/Predicate; into
HCV RNA/Theme; by NS5B polymerase/Agent.
This example produces a number of RDF triples:
Triples
– sofosbuvir is-a drug
– sofosbuvir is-a anti-viral drug
– hepatitis delta is-a virus
– sofosbuvir inhibits Hepatitis
– GS-461203 is-a metabolite of sofosbuvir
– hepatitis C virus is-a-synonym-of HCV
– HCV NS5B polymerase incorporates GS-461203
into HCV RNA
– GS-461203 acts-as chain terminator of HCV RNA
synthesis
– GS-331007 is-a metabolite of sofosbuvir
– hepatitis delta virus is-a-synonym-of HDV
– POLR2A gene encodes RNA polymerase
– RNA polymerase is-essential-for cell function
– RNA polymerase acts-as an RNA-dependent RNA
polymerase
– RNA polymerase associates-with small delta anti-
gen of Hepatitis delta virus
– RNA polymerase acts-as replicate for the viral RNA
circular genome
– RNA polymerase acts-as transcriptase for the viral
RNA circular genome
– RNA polymerase II is-a RNA polymerase
– RNA polymerase II is-required-for transcription of
genomic HDV RNA
– RNA polymerase II is-required-for transcription of
antigenomic HDV RNA
5 SEMANTIC SEARCH
Standard information retrieval has relied on the vec-
tor space model (Salton et al., 1975). The vec-
tor space model for representing documents in high-
dimensional vector space has been validated by
decades of research and development. This model
represents documents and queries as vectors (based
in various ways, such as the almost universally used
TF-IDF (Salton et al., 1975), on the words occurring
in the document and the query) and then using some
measure (typically, the cosine difference between the
query and each document vector) to retrieve relevant
documents. Although very popular these types of rep-
resentation of document semantics based solely on
first order document-term statistics, such as TF-IDF,
are limited in their expressiveness and search recall.
Semantic search (or semantic information re-
trieval) started in the early days of Artificial Intelli-
gence (Raphael, 1964). There have been two main
directions to improve the vector space model: 1)
improve the vector representation of the query and
the document in order to improve the relationship
between the query and the relevant document vec-
tors, and 2) provide meta-knowledge, typically in the
form of synonyms, hyponyms, and hypernyms, but
sometimes with the more distantly related meronyms,
homonyms, or semantic fields, in general. Some of
the methods that have been developed are directly ap-
plicable to semantic search.
The typical techniques used are to include syn-
onyms, hyponyms, and hypernyms from the WordNet
ontology (Fern
´
andez et al., 2011). Although we will
investigate this knowledge source, we will also use
the biomedical ontologies (Bandrowski et al., 2016;
Courtot et al., 2008; Brinkman et al., 2010; Zheng
et al., 2013; Soldatova et al., 2013; Dumontier et al.,
2014; Soldatova and King, 2006; Zheng et al., 2013)
to build our own ontologies (Liang et al., 2017; Seit-
ner et al., 2016) because the specialized biomedical
domains have specialized meanings for many com-
mon words and many technical terminologies that re-
quire specialized ontologies. In addition to these lin-
guistically motivated semantic search extensions, we
will also use RDF triples that have been derived from
biomedical texts and information sources using our
suggested application of semantic roles.
As an example of how a query with additional se-
mantic information is used in a semantic search:
Query: How does sofosbuvir help infected-patients
with hepatitis C virus?
A search request is processed using the following pro-
cedure: The user search query is analyzed with lin-
guistic tools and is matched against the word ontolo-
gies and the RDF data source, retrieving synonyms
and RDF descriptions that semantically match each
entity in the query with semantic information encoded
in the RDF triples. A full text search query of various
databases, based on the data returned by the previ-
Semantic Search for Biomedical Texts using Predicate-Argument Structure
303

ous step, is generated. All resulting information is
returned to the user.
For this example query, the search would return
papers and other information that talks about how so-
fosbuvir works to inhibit hepatitis viruses, such as the
drug acting to prevent hepatitis C RNA synthesis.
6 CONCLUSION AND FUTURE
WORK
The long-term goal that has motivated this position
paper is to provide a semantic search system to query
the COVID-19 Open Research Dataset (Wang et al.,
2020). We have argued that RDF triples can assist this
semantic search. Moving toward this goal, our imme-
diate objective is to use the previously presented se-
mantic roles to assist the automatic generation of the
RDF triples found in scientific publications and the
text sections found in many of the biological knowl-
edge bases. This step together with information de-
rived from ontologies and knowledge bases are nec-
essary for building a semantic search system that is
capable of extracting the relevant information from
biomedical texts that goes beyond simple keyword
searches.
Our proposed idea is the first step towards devel-
oping an automated semantic search. We aim to refine
the methods discussed in this paper and to develop a
semantic search system. We intend to focus on the
drug-virus interactions (e.g., how different antiviral
drugs interact with COVID-19). A few drug interac-
tion ontologies exist
10
. A knowledge base with virus-
drug interactions exists
11
. Having the RDF triples de-
rived from text and this knowledge base would allow
us to move toward a drug-virus ontology.
REFERENCES
Alliheedi, M. and Mercer, R. E. (2019). Semantic roles:
Towards rhetorical moves in writing about experimen-
tal procedures. In Proceedings of the 32nd Canadian
Conference on Artificial Intelligence, pages 518–524.
Alliheedi, M., Mercer, R. E., and Cohen, R. (2019a). Anno-
tation of rhetorical moves in biochemistry articles. In
Proceedings of the 6th Workshop on Argument Min-
ing, pages 113–123.
Alliheedi, M., Mercer, R. E., and Haas-Neill, S. (2019b).
Ontological knowledge for rhetorical move analysis.
Computaci
´
on y Sistemas, 23(3):633–647.
10
https://bioportal.bioontology.org
11
https://go.drugbank.com
Alliheedi, M., Wang, Y., and Mercer, R. E. (2019c). Bio-
chemistry procedure-oriented ontology: A case study.
In Proceedings of the 11th International Conference
on Knowledge Engineering and Ontology Develop-
ment (KEOD 2019) – Volume 2 of Proceedings of the
11th International Conference on Knowledge Discov-
ery, Knowledge Engineering and Knowledge Manage-
ment (IC3K 2019), pages 164–173.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., et al. (2000). Gene ontol-
ogy: tool for the unification of biology. Nature Genet-
ics, 25(1):25.
Ashino, T. (2010). Materials ontology: An infrastructure
for exchanging materials information and knowledge.
Data Science Journal, 9:54–61.
Baker, C. F., Fillmore, C. J., and Lowe, J. B. (1998). The
Berkeley FrameNnet project. In Proceedings of the
17th International Conference on Computational Lin-
guistics - Volume 1, pages 86–90.
Ball, C. A., Dolinski, K., Dwight, S. S., Harris, M. A., Issel-
Tarver, L., Kasarskis, A., Scafe, C. R., Sherlock, G.,
Binkley, G., Jin, H., et al. (2000). Integrating func-
tional genomic information into the saccharomyces
genome database. Nucleic Acids Research, 28(1):77–
80.
Bandrowski, A., Brinkman, R., Brochhausen, M., Brush,
M. H., Bill Bug and, M. C. C., Clancy, K., Cour-
tot, M., Derom, D., Dumontier, M., Fan, L., Fos-
tel, J., Fragoso, G., Gibson, F., Gonzalez-Beltran, A.,
Haendel, M. A., He, Y., Heiskanen, M., Hernandez-
Boussard, T., Jensen, M., Lin, Y., Lister, A. L., Lord,
P., Malone, J., Manduchi, E., Monnie McGee and,
N. M., Overton, J. A., Parkinson, H., Peters, B.,
Rocca-Serra, P., Ruttenberg, A., Sansone, S.-A.,
Scheuermann, R. H., Schober, D., Smith, B., Solda-
tova, L. N., Christian J. Stoeckert, J., Taylor, C. F.,
Torniai, C., Turner, J. A., Vita, R., Whetzel, P. L., and
Zheng, J. (2016). The ontology for biomedical inves-
tigations. PLoS ONE, 11(4):e0154556.
Bio2RDF (2020). https://bio2rdf.org.
Blake, J. A., Eppig, J. T., Richardson, J. E., Davisson, M. T.,
Group, M. G. D., et al. (2000). The mouse genome
database (mgd): expanding genetic and genomic re-
sources for the laboratory mouse. Nucleic Acids Re-
search, 28(1):108–111.
Brinkman, R. R., Courtot, M., Derom, D., Fostel, J. M.,
He, Y., Lord, P., Malone, J., Parkinson, H., Peters,
B., Rocca-Serra, P., Ruttenberg, A., Sansone, S.-A.,
Soldatova, L. N., Jr., C. J. S., Turner, J. A., Zheng, J.,
and the OBI consortium (2010). Modeling biomedical
experimental processes with OBI. Journal of Biomed-
ical Semantics, 1 (Suppl 1):S7.
Chang, J., Nie, X., Chang, H. E., Han, Z., and Taylor, J.
(2008). Transcription of Hepatitis delta virus RNA by
RNA polymerase II. Journal of Virology, 82(3):1118–
1127.
Chiu, B., Majewska, O., Pyysalo, S., Wey, L., Stenius,
U., Korhonen, A., and Palmer, M. (2019). A neu-
ral classification method for supporting the creation
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
304

of BioVerbNet. Journal of Biomedical Semantics,
10(2):12.
Courtot, M., Bug, W., Gibson, F., Lister, A. L., Malone,
J., Schober, D., Brinkman, R. R., and Ruttenberg, A.
(2008). The OWL of biomedical investigations. In
Proceedings of the Fifth OWLED Workshop on OWL:
Experiences and Directions, page 12pp.
Degtyarenko, K., De Matos, P., Ennis, M., Hastings, J.,
Zbinden, M., McNaught, A., Alc
´
antara, R., Dar-
sow, M., Guedj, M., and Ashburner, M. (2007).
ChEBI: a database and ontology for chemical enti-
ties of biological interest. Nucleic Acids Research,
36(suppl 1):D344–D350.
Devanbu, P. T. and Litman, D. J. (1996). Taxonomic plan
reasoning. Artificial Intelligence, 84(1-2):1–35.
Dolbey, A., Ellsworth, M., and Scheffczyk, J. (2006).
BioFrameNet: A domain-specific FrameNet exten-
sion with links to biomedical ontologies. In Second In-
ternational Workshop on Formal Biomedical Knowl-
edge Representation (KR-MED 2006), CEUR Work-
shop Proceedings Vol. 222, pages 87–94.
Dumontier, M., Baker, C. J., Baran, J., Callahan, A., Che-
pelev, L., Cruz-Toledo, J., Rio, N. R. D., Duck, G.,
Furlong, L. I., Keath, N., Klassen, D., McCusker,
J. P., Queralt-Rosinach, N., Samwald, M., Villanueva-
Rosales, N., Wilkinson, M. D., and Hoehndorf, R.
(2014). The semanticscience integrated ontology (sio)
for biomedical research and knowledge discovery.
Journal of Biomedical Semantics, 5(1):14.
Entrez Gene (2020). https://www.ncbi.nlm.nih.gov.
Fern
´
andez, M., Cantador, I., L
´
opez, V., Vallet, D., Castells,
P., and Motta, E. (2011). Semantically enhanced
Information Retrieval: An ontology-based approach.
Journal of Web Semantics, 9(4):434–452.
Fillmore, C. J. (1976). Frame semantics and the nature of
language. Annals of the New York Academy of Sci-
ences, 280(1):20–32.
Fleischmann, A., Darsow, M., Degtyarenko, K., Fleis-
chmann, W., Boyce, S., Axelsen, K. B., Bairoch,
A., Schomburg, D., Tipton, K. F., and Apweiler,
R. (2004). Intenz, the integrated relational enzyme
database. Nucleic Acids Research, 32(suppl 1):D434–
D437.
FlyBase Consortium (2003). The flybase database of the
drosophila genome projects and community literature.
Nucleic Acids Research, 31(1):172–175.
FrameNet (2020). https://framenet.icsi.berkeley.edu.
Gene Ontology Consortium (2011). The gene ontology:
enhancements for 2011. Nucleic Acids Research,
40(D1):D559–D564.
Hu, W., Qiu, H., Huang, J., and Dumontier, M. (2017).
BioSearch: a semantic search engine for Bio2RDF.
Database, 2017.
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F.,
Itoh, M., Kawashima, S., Katayama, T., Araki, M.,
and Hirakawa, M. (2006). From genomics to chemical
genomics: new developments in kegg. Nucleic Acids
Research, 34(suppl 1):D354–D357.
Kipper, K., Korhonen, A., Ryant, N., and Palmer, M.
(2008). A large-scale classification of English verbs.
Language Resources and Evaluation, 42(1):21–40.
Kipper-Schuler, K. (2005). VerbNet: A broad-coverage,
comprehensive verb lexicon. PhD thesis, University
of Pennsylvania.
Kirby, B. J., Symonds, W. T., Kearney, B. P., and Mathias,
A. A. (2015). Pharmacokinetic, pharmacodynamic,
and drug-interaction profile of the hepatitis C virus
NS5B polymerase inhibitor sofosbuvir. Clinical Phar-
macokinetics, 54(7):677–690.
Klingstr
¨
om, T., Soldatova, L., Stevens, R., Roos, T. E.,
Swertz, M. A., M
¨
uller, K. M., Kala
ˇ
s, M., Lambrix,
P., Taussig, M. J., Litton, J.-E., Landegren, U., and
Bongcam-Rudlof, E. (2013). Workshop on labortory
protocol standards for the molecular methods data-
base. New Biotechnology, 30(2):109–113.
Kohlschein, C., Klischies, D., Paulus, A., Burgdorf, A.,
Meisen, T., and Kipp, M. (2018). An extensible se-
mantic search engine for biomedical publications. In
2018 IEEE 20th International Conference on e-Health
Networking, Applications and Services (Healthcom),
pages 1–6.
Lenat, D. B., Prakash, M., and Shepherd, M. (1985). Cyc:
Using common sense knowledge to overcome brittle-
ness and knowledge acquisition bottlenecks. AI mag-
azine, 6(4):65–65.
Liang, J., Zhang, Y., Xiao, Y., Wang, H., Wang, W., and
Zhu, P. (2017). On the transitivity of hypernym-
hyponym relations in data-driven lexical taxonomies.
In Proceedings of the Thirty-First AAAI Conference
on Artificial Intelligence, AAAI’17, page 1185–1191.
AAAI Press.
Lippincott, T., Rimell, L., Verspoor, K., and Korhonen,
A. (2013). Approaches to verb subcategorization
for biomedicine. Journal of Biomedical Informatics,
46(2):212–227.
Miller, G. A. (1995). Wordnet: A lexical database for en-
glish. Communications of the ACM, 38(11):39–41.
Minsky, M. (1974). A framework for representing knowl-
edge. Artificial Intelligence Memo No. 306, Mas-
sachusetts Institute of Technology A.I. Laboratory.
Palmer, M., Gildea, D., and Kingsbury, P. (2005). The
Proposition Bank: An annotated corpus of semantic
roles. Computational Linguistics, 31(1):71–106.
Raphael, B. (1964). SIR: A computer program for seman-
tic information retrieval. PhD thesis, Massachusetts
Institute of Technology, Cambridge, USA.
Rijgersberg, H., Van Assem, M., and Top, J. (2013). Ontol-
ogy of units of measure and related concepts. Seman-
tic Web, 4(1):3–13.
Ringwald, M., Eppig, J. T., Kadin, J. A., and Richardson,
J. E. (2000). Gxd: a gene expression database for the
laboratory mouse: current status and recent enhance-
ments. Nucleic Acids Research, 28(1):115–119.
Rosse, C. and Mejino Jr, J. L. (2003). A reference on-
tology for biomedical informatics: the foundational
model of anatomy. Journal of Biomedical Informat-
ics, 36(6):478–500.
Salton, G., Wong, A., and Yang, C. S. (1975). A vector
Semantic Search for Biomedical Texts using Predicate-Argument Structure
305

space model for automatic indexing. Commun. ACM,
18(11):613–620.
Schlenoff, C., Schlenoff, C., Tissot, F., Valois, J., and Lee,
J. (2000). The Process Specification Language (PSL)
overview and version 1.0 specification. Internal Re-
port (NISTIR) - 6459, NIST Interagency.
Seitner, J., Bizer, C., Eckert, K., Faralli, S., Meusel, R.,
Paulheim, H., and Ponzetto, S. P. (2016). A large
DataBase of hypernymy relations extracted from the
Web. In Proceedings of the Tenth International
Conference on Language Resources and Evaluation
(LREC’16), pages 360–367.
Smith, B., Ceusters, W., Klagges, B., K
¨
ohler, J., Kumar, A.,
Lomax, J., Mungall, C., Neuhaus, F., Rector, A. L., ,
and Rosse, C. (2005). Relations in biomedical ontolo-
gies. Genome Biology, 6(5):R46.
Soldatova, L., King, R., Basu, P., Haddi, E., and Saunders,
N. (2013). The representation of biomedical proto-
cols. EMBnet.journal, 19(B).
Soldatova, L. N. and King, R. D. (2006). An ontology of
scientific experiments. Journal of the Royal Society
Interface, 3(11).
Wang, L. L., Lo, K., Chandrasekhar, Y., Reas, R., Yang, J.,
Eide, D., Funk, K., Kinney, R. M., Liu, Z., Merrill,
W., Mooney, P., Murdick, D. A., Rishi, D., Sheehan,
J., Shen, Z., Stilson, B., Wade, A. D., Wang, K., Wil-
helm, C., Xie, B., Raymond, D. M., Weld, D. S., Et-
zioni, O., and Kohlmeier, S. (2020). CORD-19: The
covid-19 open research dataset. ArXiv.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Apple-
ton, G., Axton, M., Baak, A., Blomberg, N., Boiten,
J.-W., da Silva Santos, L. B., Bourne, P. E., Bouw-
man, J., Brookes, A. J., Clark, T., Crosas, M., Dillo,
I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R.,
Gonzalez-Beltran, A., Gray, A. J., Groth, P., Goble,
C., Grethe, J. S., Heringa, J., ’t Hoen, P. A., Hooft,
R., Kuhn, T., Kok, R., Kok, J., Lusher, S. J., Martone,
M. E., Mons, A., Packer, A. L., Persson, B., Rocca-
Serra, Philippeand Roos, M., van Schaik, R., Sansone,
S.-A., Schultes, E., Sengstag, T., Slater, T., Strawn,
G., Swertz, M. A., Thompson, M., van der Lei, J.,
van Mulligen, E., Velterop, J., Waagmeester, A., Wit-
tenburg, P., Wolstencroft, K., Zhao, J., and Mons, B.
(2016). The FAIR Guiding Principles for scientific
data management and stewardship. Scientific Data,
3(1):9.
Zheng, J., Manduchi, E., and Stoeckert Jr, C. J. (2013). De-
velopment of an application ontology for beta cell ge-
nomics based on the ontology for biomedical investi-
gations. In 4th International Conference on Biomedi-
cal Ontology, pages 62–67.
KEOD 2020 - 12th International Conference on Knowledge Engineering and Ontology Development
306
