
Left Ventricle Computational Model based on Patients
Three-dimensional MRI
Maria Narciso
1 a
, Ana Isabel Sousa
1 b
, Fernando Crivellaro
1 c
, Rui Valente de Almeida
1 d
,
Ant
´
onio Ferreira
2 e
and Pedro Vieira
1 f
1
FCT Nova, Caparica, Portugal
2
Cardiology Department, Hospital de Santa Cruz, Centro Hospitalar Lisboa Ocidental, Lisbon, Portugal
Keywords:
Heart Computational Model, Parallel Simulation, Cardiac Electrophysiology, Arrhythmia, CHASTE,
MATLAB, Virtual Heart.
Abstract:
The propagation of electric signal in the heart is bonded to the cardiac muscle (myocardium) geometry and
condition. In this regard we aim to build a patient-specific computational model of the myocardium based on
noninvasive imaging, with the long-term goal of being used to run simulations in electrophysiology studies.
Three-dimensional (3D) Magnetic Resonance Images (MRI) were processed using MATLAB
R
to build a
volumetric mesh which embodies the Left Ventricle (LV) and can later be read by third party applications.
This feature was tested with the open source software CHASTE (Cancer, Heart and Soft Tissue Environment)
to solve and visualize the propagation of an excitation wave.
Furthermore, an algorithm was developed capable of defining the fibre orientation for the resulting mesh, based
on the geometry described in literature. This experiment substantiates the expectation that parallel computing
simulations of the heart maybe used, in the near future, as a monitoring and diagnostic tool for the assessment
of cardiac arrhythmias in clinical practice.
1 INTRODUCTION
The work here described is part of a project cur-
rently being developed in collaboration with Hospi-
tal de Santa Cruz (Carnaxide, Portugal), where Physi-
cians and Techs contribute with not only datasets but,
above all, guidance and expertise. The ultimate goal
is to create an arrhythmia risk-stratification tool using
noninvasive imaging as input. In order to accomplish
this, it is fundamental to develop a method capable of
assembling a 3D model of the heart, more specifically
of the LV, from the imaging datasets. This paper will
be focused on this very first step of the process.
Cardiac electric impulses begin at the sinoatrial
node (natural pacemaker) and spread through the my-
ocytes, cardiac cells with membranes selectively per-
meable to K, Na and Ca ions. The movement of these
a
https://orcid.org/0000-0001-5079-9381
b
https://orcid.org/0000-0003-2980-4742
c
https://orcid.org/0000-0002-7534-9149
d
https://orcid.org/0000-0002-2269-7094
e
https://orcid.org/0000-0002-1623-7382
f
https://orcid.org/0000-0002-3823-1184
ions in and out of the intracellular space causes the po-
tential difference responsible for driving the electrical
signals to the contractile myocytes (Tse, 2016)(Di Yu
et al., 2014).
The heart walls that enclose the four cardiac
chambers are formed by myocardium, the muscular
tissue responsible for the heart contraction. The in-
ner part of the myocardium is called endocardium,
while the outermost part is usually called epicardium.
Most of the ventricular arrhythmias are the result of
some form of structural heart disease that causes the
myocardial tissue to become heterogeneous in terms
of electric conduction. Since most of the clinically
significant ventricular arrhythmias arise from the LV,
and in order to simplify the model and reduce compu-
tational costs, we limited our analysis to this ventricle.
Cardiac magnetic resonance has started to re-
place ultrasound-based echocardiograms as the imag-
ing standard in clinical arrhythmology and electro-
physiology studies (De Maria et al., 2017). Beside
the advantage of being non-invasive while not us-
ing ionizing radiation. MRI provides excellent con-
trast between soft tissues, offering structural and func-
tional information. The images are computational re-
156
Narciso, M., Sousa, A., Crivellaro, F., Valente de Almeida, R., Ferreira, A. and Vieira, P.
Left Ventricle Computational Model based on Patients Three-dimensional MRI.
DOI: 10.5220/0008961601560163
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 2: BIOIMAGING, pages 156-163
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

constructions of the signal received from sensors that
measure the particles resonance when cut off from an
excitation pulse of radio waves. Although this tech-
nology alone is not capable of detecting fibres and
micro structures, the anatomical features extracted
from the resulting images provide the most advan-
tageous baseline to achieve in silico patient-specific
heart models. (Auricchio, Angelo, Singh, Jagmeet,
Rademakers, 2012). For this reason, this was the
imaging modality chosen as outset of our project.
Technical details of the method that has proven
to be able to generate three-dimensional models from
these images are described in the next chapter. Image
processing was done in MATLAB and the fibre ori-
entation estimation code was written in Python. The
diagram in figure 1 despicts the overall workflow.
3D MRI
Myocardium segmentation
Fibers
orientation
STL
Other formats
Simulations
Figure 1: Workflow of the three-dimensional model assem-
bling process.
2 DEVELOPMENT OF THE
VENTRICLE MODEL
This section reports the methods used for develop-
ing the 3D computational model of the LV, built from
magnetic resonance images of a specific patient. The
methods described were designed in MATLAB 2015
and trusted to be broad enough to be used with most
patients, aside from subjects with severe cardiac de-
formities, as these cases are not standardized and
would require specially designed tools. The result is a
Stereolithography (STL) file representing the cardiac
muscle, which may later be opened and modified by
several applications.
2.1 Dataset
Ant
´
onio Ferreira provided anonymized images in DI-
COM format from a contrast-enhanced 3D cardiac
MRI exam that was considered to be entirely normal.
Any areas of myocardial fibrosis would be visible in
the images as brighter areas amidst the darker cardiac
muscle. Therefore our process started from an ideal
case with homogeneous myocardial tissue.
The encoded images are made of voxels, the basic
element of a 3D image, in the same manner the pixel
is in 2D images. A voxel has three dimensions there-
fore it is referenced in space by three coordinates, its
intensity value can be in several different ranges as
the scales are not standardized.
In this acquisition, the whole slab is encoded re-
sulting in 3D image with high spatial resolution - size
of an individual voxel - and high signal-to-noise ratio.
The voxels are nearly isotropic, allowing reconstruc-
tion in any direction.
2.2 Segmentation of MR Data Set
From now on, the first dimension elements of any 3D
matrix will be referred to as rows (along yy), the sec-
ond columns (along xx) and the third slices (along zz).
Information as voxel size, number of rows and
columns of each image, was obtained from the DI-
COM file info section and stored with the purpose
of being accessed along the process. The images,
which represent axial slices of the torso, were read to
a three-dimensional matrix with the dimensions rows
x columns x number of images, meaning they were
turned into two dimensional matrices stacked along
the z axis. The intensity values of every image are di-
vided by the maximum value of intensity, to obtain a
range from 0(lightest) to 1 (darkest), therefore the rest
of the transformations can be applied in any set inde-
pendently of the scale used in the exam, as of today
there is still not a standard range.
Our aim was not to develop an algorithm capable
of automatically segment the myocardium but to de-
velop semi-automatic tools to do so, as in this context
it was required that a physician guided and supervised
the entire process.
The first required transformation is the rotation
of the matrix in order to attain the short-axis (SAX)
view. In this view, slices are perpendicular to the
long-axis (LAX) of the LV which allows us to exam-
ine cross-sections of both ventricles, where the cham-
bers walls are clearly distinguishable and the left ven-
tricle sections are usually annular.
Heart orientation and consequently the left ventri-
cle alignment, differs between individuals. On this
account and following the traditional cardiologist ap-
proach, we chose to navigate trough the different per-
spectives using cutting planes, illustrated by lines the
user defines.
As the licensed version of Matlab we had avail-
able did not support the imrotate3d function - which
allows the rotation of three dimensional objects in any
Left Ventricle Computational Model based on Patients Three-dimensional MRI
157
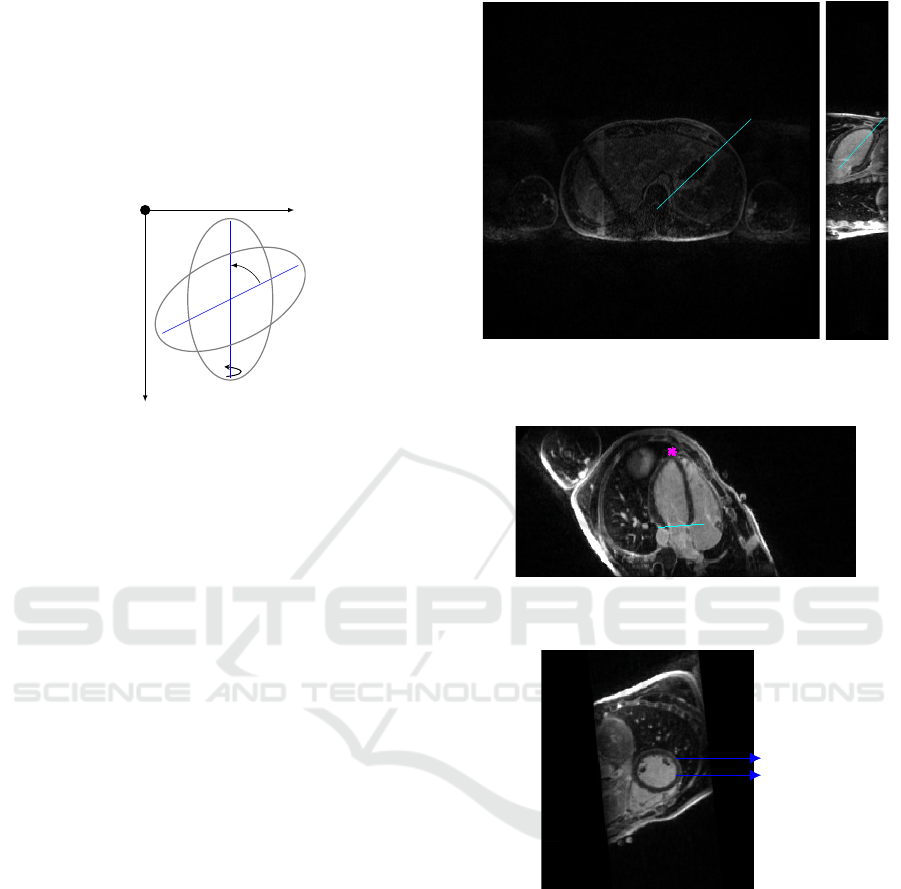
direction at any angle - we defined a function which
rotates each slice (two dimensional rotation) around
its centre, the same number of degrees the drawn line
makes with the vertical axis. Next the whole matrix is
rotated 90
o
clockwise around the y axis, as the func-
tion rot90 is available in this version of the software
and allows the use of angles multiples of 90
o
. The
logic of the cutting plan is exemplified in figure 2.
z
x
y
θ
90
o
Figure 2: Rotation of 2D images based on the drawn lines,
first in the amplitude of θ around the z axis and then 90
o
around the y axis.
For each transformation triggered by a line, two
new matrices are created, first a copy of the origi-
nal, with the same size but with each slice rotated and
cropped, and with the second transformation another
copy with the first and third dimensions permuted.
Starting from the axial view, the first line must be
parallel to the inter-ventricular septum, right picture
of figure 3, the rotation results in the vertical long axis
view shown in the left picture of 3. Then one should
draw a bisecting line from the LV apex to the mitral
valve, this leads to the horizontal long axis view. The
user is then asked to mark the LV ventricle apex with
a point and the base with a line, figure 4, finally we
are presented with the SAX view. This cut generates
a matrix with the dimensions number of rows of each
image x columns x number of slices from base to apex.
An example of a slice from this object is shown in
figure 5.
Inside the LV chamber, the pixels represent blood
therefore the intensity levels are lower than that at the
pixels representing tissue. A point in this area and an-
other one at the myocardium outside border are man-
ually marked.
The grayconnected function is applied to select
the contiguous regions with similar intensities for
both points. The output is a binary image where
the pixels that are part of the region have the value
one. The two resulting areas enclose every pixel
with values between seedvalue-tolerance and seed-
value+tolerance. For the area inside the chamber
seedvalue is the pixel intensity level at the selected
point and the tolerance is a quarter of the difference
between this value and the intensity value at the sec-
Figure 3: Right: Axial view of the torso with a line drawn
parallel to the septum. Left: LAX vertical view with the
bisetrix line.
Figure 4: Horizontal LAX view with the apex marked in
pink and base of LV in cyan.
Pericardium
Myocardium
RV
LV
Figure 5: SAX view where both ventricles are visible.
ond point, hence adding only pixels with values closer
to the blood intensity than to the muscle. The shape
centre of mass is calculated and stored. The second
region is the actual segmented myocardium, gener-
ated following the same logic. There is a thin pro-
tetive layer surrounding the whole heart named peri-
cardium , figure 5. Because it is almost white, its pix-
els are beyond the intensity threshold and will not be
aggregated, however this layer is so thin that some-
times is borders are not distinguishable, therefore the
distance between the centre of mass is set as the maxi-
mum distance the value of the pixels above this radius
are change to zero. The result is a black and white im-
BIOIMAGING 2020 - 7th International Conference on Bioimaging
158

Figure 6: Binary image of the LV myocardium segmented
from the SAX slice
Figure 7: SAX view with the myocardium borders con-
toured in white and the centre of mass marked with a red
cross.
age representing a slice of the myocardium, figure 6,
its contours are shown in figure 7 on top of the origi-
nal slice.
The process above is repeated for every slice, the
ones representing the apex myocardium is visible as
a small circle, instead of a annulus, therefore only the
second point is needed. Each resulting image is saved
into a matrix with the same size as the last one created,
stacking the cross sections of the ventricle along the
third dimension.
2.3 3D Model and Formatting
At this stage the object is represented in a regular grid
in 3D space, thus the location of each voxel is de-
scribed by discrete values of row, column and page, to
get the real spacial coordinates the find function was
called, as it returns the indexes of each voxel with a
value not zero and the respective coordinates where
saved in a matrix [r,c,p], then each element was mul-
tiplied by the voxel size for the respective dimension.
This transformation was applied at this stage and not
later for the simplicity of working with only point co-
ordinates. Regardless, this is a crucial step consider-
ing that, although STL structures are dimensionless
and the files contain no scale information, the output
must be ready to be handled by applications where
distances and dimensions matter.
It is possible to increase the resolution, multiply-
ing the number of points. Using linspace to generate
a vector with the size of each dimension, changing
the spacing from one to the desired integer number.
two meshgrids are generated one from the new vec-
tors and one from three vectors with the original spac-
ing. Then using bilinear interpolation with interpn, a
new matrix is created interpolating the original mesh-
grid with the new one. Higher resolution does not af-
fect the results, however, during the simulations, the
signal propagation can be observed with more detail,
as the mesh will be made of smaller blocks. As ex-
pected, this comes with a high computational price.
The function alphashape creates a surface around
all the defined points, using the variable alpha to de-
fine the tightness, the user defines the optimal alpha
value so that the chamber is open while the walls are
closed. Next, boundaryFacets is called to get the tri-
angles faces and each concerning vertex that form the
surfaces of the volume, this process is known as tes-
sellation, filling a surface with geometric shapes (in
this case triangles) without overlapping or forming
gaps). smoothpatch (Kroon, 2010) was applied to
polish the rough edges, this function takes each ver-
tex, checks the neighbours from the same facet and
moves them in a way that the distance between them
becomes more uniform, keeping the number of trian-
gles.
The last step in MATLAB is the exportation to a
STL file, supported by many software and applica-
tions.
The stlwrite (Holcombe, 2011) function was used
to export the triangulated structure to a file with the
standardized format. Both the smoothed model and
the one from the previous step are exported, so they
can be tested against each other, ensuring that this aes-
thetic alterations do not compromise the model. A
comparison of both can be found in figure 8.
2828
3030
Y AxisY Axis
3232
3434
X AxisX Axis
X AxisX Axis
28283030
Y AxisY Axis
32323434
11
2222
33
4444
Z AxisZ Axis
55
Z AxisZ Axis
66
66
77
88
88
99
1010
1010
2424
2424
3434 3232
Y AxisY Axis
3030
3030 3030
X AxisX Axis
3434 3232
Y AxisY Axis
X AxisX Axis
3030
11
22
22
33
44
44
55
Z AxisZ Axis
Z AxisZ Axis
66
66
77
88
88
99
1010
1010
Figure 8: Comparison between the original mesh on the left
and the smoothed on the right.
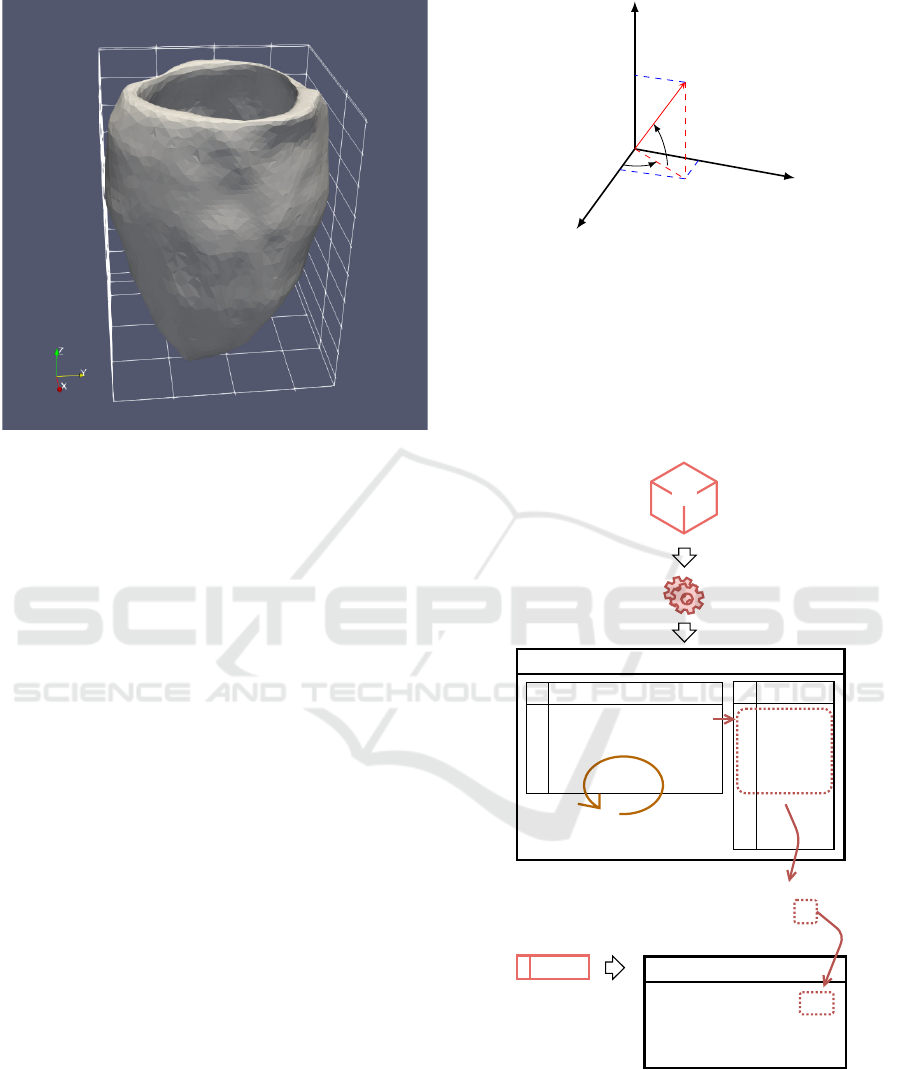
An example of a model build following the meth-
ods described, exported with the original resolution
and default alpha value, is presented in figure 9.
Left Ventricle Computational Model based on Patients Three-dimensional MRI
159
3434
3232
Y AxisY Axis
3030
3434
11
2626
3232
Y AxisY Axis
3030
22
11
33
X AxisX Axis
22
44
2828
33
55
Z AxisZ Axis
X AxisX Axis
44
66
3030
55
77
Z AxisZ Axis
66
88
3030
77
99
88
1010
99
1010
3232
Figure 9: Renderization of the resulting STL file.
2.4 Mesh Convertion
In order to use the model in the simulation software, it
is necessary to solve equations that describe the tissue
electrical properties for the whole mesh, therefore the
STL file must be converted to a tetrahedral mesh, to
define not only the surface but also the filling. This
was accomplished using the software TetGen. The
result are a set of files that list the elements .ele, the
respective nodes .node, facets .face and edges .edges.
The LV model is now made of pyramidal blocks.
2.5 Fibre Orientation
As stated previously there is no information available
about the fibres in the MRI dataset, nevertheless the
way fibres run in the heart muscle has been observed
in in vitro studies and also through Diffusion Tensor
Imaging, therefore is possible to estimate their posi-
tion in a given ventricle, analogously to the method
reported in (Bayer et al., 2012).
Our approach was to follow the geometry de-
scribed in (Khalique and Pennell, 2019), defining the
fibre orientation at each element(tetrahedron) using a
3D vector, that dictates the longitudinal direction the
fibres take from each element.
An example of a vector ~v with the origin at the
centroid of a given element is shown in figure 10. It is
known that the vector norm is one, therefore by defin-
ing θ and φ and applying trigonometry, it is possible
to determinate the values of v
x
, v
y
and v
z
that define
the vector.
At the time of the segmentation in MATLAB
x
y
z
~v
φ
θ
v
x
v
z
v
y
Figure 10: Vector ~v representing the fibre orientation at a
given point of the mesh.
when the myocardium is seen in the shape of an an-
nulus, the centre, minimum and maximum radius and
the slice(z value) are register for each slice in a text
file denominated ’Registration file’. The sketch of
figure 11, illustrates how the data necessary for the
following steps is obtained.
Mesh
Nodes
1
2
3
4
.
.
.
x
1
y
1
z
1
x
2
y
2
z
2
x
3
y
3
z
3
x
4
y
4
z
4
Repeat for
each element
Elements
1
.
.
.
node1 node2 node3 node4
TetGen
STL
Centroid
x
c
y
c
z
c
MatLab MatLab
Registration File
x
cm
y
cm
r
min
r
max
z
slice
.
.
.
Figure 11: Sketch of the steps required to get the spatial
information, for each element of the mesh.
The algorithm written in Python 2.6. starts by
reading the elements from the .ele file 11, and for each
tetrahedron gets the coordinates of its four nodes from
the .node file, and calculates the centroid, figure 12.
During the next steps the volume will have to be
BIOIMAGING 2020 - 7th International Conference on Bioimaging
160

a b
c
d
Centroid
Figure 12: Tetrahedron with nodes a,b,c,d and the calcu-
lated centroid.
Figure 13: The fibers closer to the outer border of the mus-
cle run.
imagined as a stack of slices as it was during the seg-
mentation. The closer 2D slice value from the regis-
tration file to the z coordinate of the centroid will be
attributed to this element. All the calculations will be
made considering the centroid a point in one of the
2D plans, placed at the annulus.
When the left ventricle contracts to pump the
blood it does it by torsion, due to the fact that the
muscle fibres run in opposite directions at the inner -
endocardium - and outer layers - epicardium - as ex-
emplified in figure 13.
In reality the fibres slope varies according to their
distance to the borders, the ones at the endocardium
do an angle with the horizontal plan that we will con-
sider approximately -60
o
and it increases along the
muscle till the last layer where we will consider the
angle to measure 60
o
.
Defined by an input variable, a number of circum-
ference radius between the minimum (r
min
) and max-
imum (r
max
) radius is saved in an array, imagining the
shape is layered as an onion, the scheme in figure 14
demonstrates a slice divided in six layers and a cen-
troid positioned at the outermost. At the bottom slices
of the ventricular apex, the minimum radius is consid-
ered zero, figure 15.
Knowing the angle the vector does with the xy
plan is a value in { -60 : 60 }, going from the out-
ermost circle to the inner, each radius is matched
with an angle θ, according to its position in the ar-
r
max
r
min
Figure 14: Left: Myocardium split in six layers defined by
five radius between r
min
and r
max
Right: tetrahedra with
centroid at the outermost layer.
Figure 15: At the apex r
min
is zero.
ray, which translates in its distance from the centre.
The distance between the centroid and the centre is
matched with the closer radius from the array, and
the θ angle from the orientation vector is therefore as-
signed. For an element which centroid is located at an
inner layer, the angle φthe vector does with the x axis
is the angle α between the line that connects the cen-
troid with the centre of mass and the vertical line that
crosses the centre of mass, plus 90
o
. As the scheme
of figure 16.
For the outer layers, that is when the respective
angle θ is positive, the logic is the same, changing
only the sum of 90
o
for a subtraction. Considering
the slice of the myocardium a perfect circle, the fibres
as in figure 13.
Now returning to the 3D system to calculate the
vector third coordinate we know that~v = 1 and both θ
and φ angles, applying the rules of trigonometry to the
scheme in figure 10 the three coordinates of the vector
can be calculated by the three equations bellow.
x = cos(θ) ∗ cos(φ) (1)
y = cos(θ) ∗ sin(φ) (2)
z = sin(θ) (3)
This logic is followed for every element of the
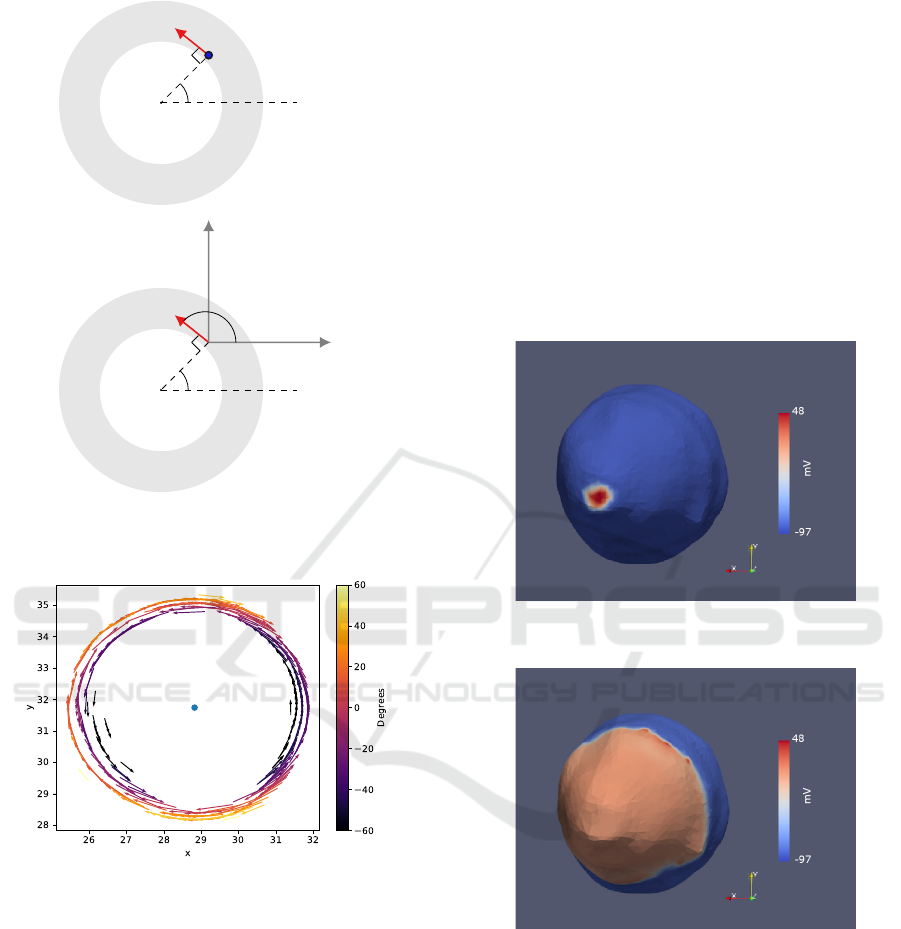
mesh, the plot of the the resulting orientation vectors
for one of the slices, considering only the 2D plan by
setting z = 0 is presented in figure 17.
Left Ventricle Computational Model based on Patients Three-dimensional MRI
161
element centroid
α
y = y
cm
z
α
y = y
cm
x
y
φ
Figure 16: Direction of the vector considering only the xy
plan for an element which centroid is located at an inner
layer.
Figure 17: 2D plot of the orientation vectors for one slice,
where is visible that that the vectors closer to the centre
(blue dot) run anticlockwise and the rest clockwise. From
the colormapping its understood that the inner vectors make
negative angles with the horizontal xy plan, in 3D, and that
this angle gets larger with the proximity to the outer border.
3 RESULTS
To test the model by running electrical simulations, it
was decided to work with the open source software
CHASTE, developed at Oxford University, as it in-
tegrates all the necessary libraries to solve electrical
activation problems on biological tissue, offers flex-
ibility and keeps being improved by the community.
The cardiac module is capable of recreating the exci-
tation signal propagation throughout adjacent neigh-
bouring cells, using a set of parameters that the user
can define (Mirams et al., 2013).
3.1 Simulation
A simulation ran in monodomain, using the Luo-
Rudy Backward Euler ionic model and intracellular
conductivities of be 3,75 mS/cm for the longitudi-
nal direction and 2.14 mS/cm for transversal and nor-
mal. Considering the tissue homogeneous, applying
a stimuli of -80.000uA at the apex, pictured in figure
18, after 80 ms the activation spread throughout the
ventricle, as shown in figure 19.
Figure 18: Spherical stimuli at the apex of the smoothed
model.
Figure 19: Extension of the activation wave after 90 ms.
4 CONCLUSION
Several research groups have been developing paral-
lel functional models of the heart, however most of
the solutions make use of high performance comput-
ers. The approach described in this work intends to
make use of tools available for free or with student
licences. We have proved that it is possible to manip-
ulate the medical images incorporating, in our code,
functions available in MATLAB, plus a couple shared
BIOIMAGING 2020 - 7th International Conference on Bioimaging
162

by the community, resulting in an accurate computa-
tional model of the LV. These are promising results
for the possibility of using non invasive imaging of
human cardiac electrical activity for risk assessments
in diagnostics.
The next logic step is to map different conduc-
tivities accordingly to the intensity levels visible on
contrast-enhanced MRI, allowing to perform virtual
electrophysiologic studies. In order to accomplish
this, the registration process during the segmentation
must be replaced by a more accurate method capa-
ble of mapping the velocities into a model that must
be already a filled volume, unlike the surface one de-
scribed here.
ACKNOWLEDGEMENTS
This work is part of a project co-funded by Fundac¸
˜
ao
para a Ci
ˆ
encia e Tecnologia and Compta SA Por-
tugal under the studentship PD/BDE/131399/2017
Ref.CRM:0008003
REFERENCES
Auricchio, Angelo, Singh, Jagmeet, Rademakers, F. E., ed-
itor (2012). Cardiac Imaging in Electrophysiology.
Bayer, J. D., Blake, R. C., Plank, G., and Trayanova,
N. A. (2012). A novel rule-based algorithm for as-
signing myocardial fiber orientation to computational
heart models. Annals of biomedical engineering,
40(10):2243–54.
De Maria, E., Aldrovandi, A., Borghi, A., Modonesi, L.,
and Cappelli, S. (2017). Cardiac magnetic resonance
imaging: Which information is useful for the arrhyth-
mologist? World journal of cardiology, 9(10):773–
786.
Di Yu, Dongping Du, Hui Yang, and Yicheng Tu (2014).
Parallel computing simulation of electrical excitation
and conduction in the 3D human heart. In 2014 36th
Annual International Conference of the IEEE Engi-
neering in Medicine and Biology Society, pages 4315–
4319. IEEE.
Holcombe, S. (2011). stlwrite - write ascii or binary stl files.
Retrieved May 4, 2018.
Khalique, Z. and Pennell, D. (2019). Diffusion tensor car-
diovascular magnetic resonance. Postgraduate medi-
cal journal, 95(1126):433–438.
Kroon, D.-J. (2010). stlwrite - write ascii or binary stl files.
Retrieved March 26, 2010.
Mirams, G. R., Arthurs, C. J., Bernabeu, M. O., Bordas,
R., Cooper, J., Corrias, A., Davit, Y., Dunn, S.-J.,
Fletcher, A. G., Harvey, D. G., Marsh, M. E., Osborne,
J. M., Pathmanathan, P., Pitt-Francis, J., Southern, J.,
Zemzemi, N., and Gavaghan, D. J. (2013). Chaste: An
Open Source C++ Library for Computational Phys-
iology and Biology. PLoS Computational Biology,
9(3):e1002970.
Tse, G. (2016). Mechanisms of cardiac arrhythmias. Jour-
nal of Arrhythmia, 32(2):75–81.
Left Ventricle Computational Model based on Patients Three-dimensional MRI
163
