
Properties of the Standard Genetic Code and Its Alternatives Measured
by Codon Usage from Corresponding Genomes
Małgorzata Wnetrzak, Paweł Bła
˙
zej and Paweł Mackiewicz
Department of Bioinformatics and Genomics, Faculty of Biotechnology, University of Wrocław,
Fryderyka Joliot-Curie 14a, 50-383 Wrocław, Poland
Keywords:
Alternative Genetic Code, Codon Usage, Error Minimization, Genetic Code, Mutation, Optimization.
Abstract:
The standard genetic code (SGC) and its modifications, i.e. alternative genetic codes (AGCs), are coding
systems responsible for decoding genetic information from DNA into proteins. The SGC is thought to be
universal for almost all organisms, whereas alternative genetic codes operate mainly in organelles and some
specific microorganisms containing usually reduced genomes. Previous analyzes showed that the AGCs mini-
mize the consequences of amino acid replacements due to point mutations better than the SGC. However, these
studies did not take into account the potential differences in codon usage between the genomes on which given
codes operate. The previous analyzes assumed a uniform distribution of codons, even though we can observe
significant codon bias in genomes. Therefore, we developed a new measure involving codon usage as an addi-
tional parameter, which allowed us to assess the quality of a given genetic code. We tested our approach on the
SGC and its 13 alternatives. For each AGC we applied an appropriate codon usage characteristic of a genome
on which this code operates. This approach is more reliable for testing the impact of codon reassignments
observed in the AGCs on their robustness to point mutations. The results indicate that the AGCs are generally
more robust to point mutation than the SGC, especially when we consider the codon usages characteristic
of their corresponding genomes. Moreover, we did not find a genetic code optimal for all considered codon
usages, which indicates that the alternative variants of the SGC evolved in specific conditions.
1 INTRODUCTION
There are many alternative genetic codes (AGCs)
which have emerged from the standard genetic
code (SGC). They are used mainly in mitochondrial
genomes (Abascal et al., 2012; Boore and Brown,
1994; Sengupta et al., 2007) but also in plastid (Cor-
tona et al., 2017; Janouskovec et al., 2013), some
nuclear genomes (Hoffman et al., 1995; Sanchez-
Silva et al., 2003; Santos et al., 1993) and bacterial
genomes (Bove, 1993; Campbell et al., 2013; Mc-
Cutcheon et al., 2009). Recent findings of AGCs in
various protists suggest that the number of these codes
can be strongly underestimated (Heaphy et al., 2016;
P
´
anek et al., 2017; Z
´
ahonov
´
a et al., 2016). Except for
the quite large genomes of ciliates, the AGCs operate
typically in small genomes encoding a limited number
of proteins. The small genome size facilitates the evo-
lution of genetic code variants because the potential
codon reassignments may not cause such a harmful
effect in synthesized proteins as in the case of large
nuclear genomes, in which even a single change in
the codon meaning may affect thousands of proteins
(Massey and Garey, 2007). Therefore, such reassign-
ments cannot be easily accepted.
The NCBI database includes currently 33 AGCs:
www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
The main differences between them and the SGC can
be classified into three categories: (i) reassignment of
a codon encoding one of the 20 typical amino acids
or stop translation signal, (ii) loss of codon meaning
induced by the disappearance of this codon from the
genome, and (iii) assignment of new amino acids
such as selenocysteine and pyrrolysine (Sengupta
et al., 2007). The most common are the reassign-
ments of stop codons to sense codons, e.g. codon
UGA to tryptophane (Bła
˙
zej et al., 2019). Changes
in sense codons occur less frequently and mainly in
mitochondrial genomes.
Furthermore, it was shown that the same reassign-
ments have often evolved independently in different
phylogenetic lineages (Sengupta et al., 2007). The
main mechanisms involved in the evolution of the
AGCs are: (i) deletion of tRNA genes, (ii) duplication
of tRNA genes and their mutations, (iii) editing and
post-transcriptional base modifications of tRNAs, (iv)
44
Wnetrzak, M., Bła
˙
zej, P. and Mackiewicz, P.
Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes.
DOI: 10.5220/0008981000440051
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 3: BIOINFORMATICS, pages 44-51
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

mutations in genes encoding translational release fac-
tors, or (v) loss of codons subjected to a strong muta-
tional pressure (Sengupta and Higgs, 2005; Sengupta
et al., 2007). Horizontal gene transfer could also have
played a certain role in the AGCs evolution (Devoto
et al., 2019).
It is commonly believed that the AGCs emerged
through neutral evolution in small populations sub-
jected to genetic drift and strong mutational pres-
sure leading to tiny AT-rich genomes (Freeland et al.,
2000; Sengupta et al., 2007; Swire et al., 2005). How-
ever, other hypotheses concerning the evolution of
the alternative genetic codes have also been proposed.
They assume that: (i) codon changes associated with
the deletion of tRNA genes are driven by selection
to minimize the genome size and the time of repli-
cation (Andersson and Kurland, 1995), (ii) the reas-
signment of codon AUA from isoleucine to methion-
ine results in accumulation of methionine at the inner
membrane of animal mitochondria, which plays anti-
oxidant and cytoprotective role (Bender et al., 2008),
(iii) codon ambiguity can facilitate phenotypic diver-
sity and adaptability, which may help, e.g. yeasts, to
cope with stressful environments (Santos et al., 1999;
Gomes et al., 2007), (iv) mitochondrial genetic codes
may have evolved to reduce protein synthesis costs by
reassigning amino acids that are less expensive in syn-
thesis (Swire et al., 2005), (v) some changes in the ge-
netic code were accepted because they minimized ef-
fects of point mutations at the amino acid level (Kur-
naz et al., 2010; Morgens and Cavalcanti, 2013). All
these hypotheses suggest that the changes in the SGC
leading to the AGCs evolved as an adaptive trait.
The latter explanation seems interesting because
the same hypothesis, postulating that the code evolved
to minimize the effects of amino acid replacements
and errors during translation, was put forward for the
SGC (Epstein, 1966; Haig and Hurst, 1991; Freeland
et al., 2003; Goodarzi et al., 2005). Thus, common
principles could have governed the evolution of ge-
netic codes in general.
To verify this hypothesis, the SGC was compared
with: (i) possible theoretical genetic codes differing
from the universal code in one, two, or three codon
assignments, as well as (ii) its alternatives, regarding
the minimization of the harmful effects of amino acid
replacements in synthesized proteins (Bła
˙
zej et al.,
2018; Bła
˙
zej et al., 2019). The results indicated that
the codon reassignments observed in the AGCs gen-
erally improve their robustness to amino acid replace-
ments in comparison with the SGC and such natural
reassignments are often almost as good as the best
theoretical ones.
However, the cost function used to assess the
properties of the genetic codes took into account only
the sum of changes in the polarity of encoded amino
acids induced by single-point mutations in all the
codons. This function did not include the effects of
any mutational pressure or codon usage but assumed
a simple model, in which each codon occurs with
the same frequency
1
64
and the probability of any nu-
cleotide mutation is
1
4
and does not depend on the
codon position. Such approach was useful only in
finding the general tendencies of genetic codes re-
garding the error minimization hypothesis.
Because the codon bias is an important factor
influencing the final mutational effect, it should be
taken into account. Therefore, in this work, we imple-
mented the codon frequencies observed in genomes
into the cost function formula. Then, we calculated
the cost values for the AGCs and for the SGC, in-
cluding the codon usage from the genomes of the or-
ganisms which use the alternative codes. The results
show that in almost all cases the AGCs tested on the
codon frequencies characteristic of the organisms that
use these codes outperform the SGC in terms of ro-
bustness to amino acid replacements.
2 METHODS
The examined alternative genetic codes were
downloaded from the NCBI taxonomy web page:
www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
From the whole set of described genetic codes we
chose 13 codes, which differ from the SGC by at
least one codon assignment and for which we were
able to easily obtain the codon frequencies of the
respective genomes that use these codes. The data
were extracted from the Codon Usage Database,
www.kazusa.or.jp/codon, (Nakamura et al., 2000)
and appropriate references (Perseke et al., 2011;
Swart et al., 2016) (Table 1). As we mentioned in
the Introduction section, we investigated the quality
of the SGC and its selected alternatives including the
respective codon usages in the applied cost function.
We tested all possible combinations of the SGC and
13 chosen genetic codes with 13 codons usages,
which gave us 14 × 13 = 182 cost values in total.
In order to test the properties of the genetic
codes, including specific codons usage observed in
the genomes on which a given code operate, we in-
troduced a new formula for the cost function F. This
parameter combines the differences between the prop-
erties of amino acids encoded by pairs of codons
< i, j > varying in one nucleotide substitution and
the probability of choosing codons < i, j > computed
from the given codon usage. According to these re-
Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes
45
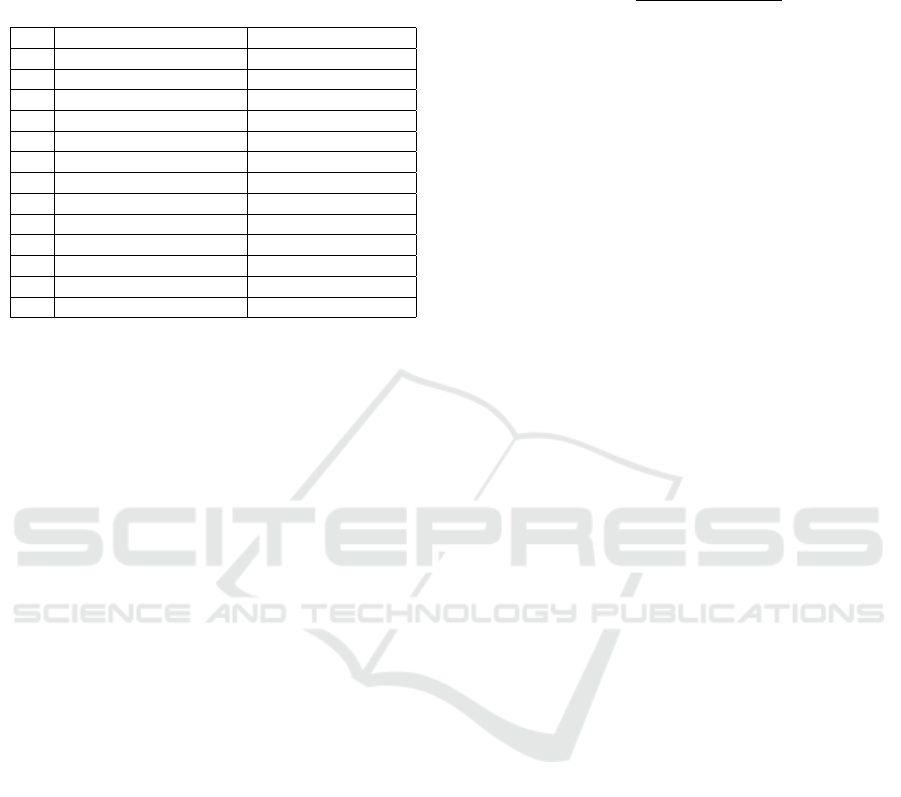
Table 1: The genetic code variants studied in this work and
the list of selected genomes from which we extracted re-
spective codon usages. All the codes are numbered (No.)
according to the notation in the NCBI taxonomy web page:
www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi.
No. Genetic code Genome
4 The Mold, Protozoan Mitoch. Aspergillus nidulans
5 Invertebrate Mitoch. Ascaris suum
6 Hexamita Nuclear Oxytricha falla
9 Flatworm Mitoch. Astropecten polyacanthus
10 Euploid Nuclear Euplotes octocarinatus
12 The Alternative Yeast Nuclear Candida albicans
13 Ascidian Mitoch. Halocynthia roretzi
16 Chlorophycean Mitoch. Spizellomyces punctatus
22 Scenedesmus Mitoch. Scenedesmus obliquus
23 Thraustochytrium Mitoch. Thraustochytrium aureum
24 Pterobranchia Mitoch. Rhabdopleura compacta
27 Karyorelict Nuclear Parduczia
28 Condylostoma Nuclear Condylostoma magnum
strictions, we defined F for a given genetic code and
codon usage, in the following way:
F =
∑
<i, j>∈D
P(< i, j >)[ f (i) − f ( j)]
2
, (1)
where D is the set of pairs of codons that differ in
one nucleotide substitution, whereas P(< i, j >) is the
probability of choosing the pair < i, j >, calculated
according to the total probability formula:
P(< i, j >) = P(i) · P( j|i) (2)
where P(i) is the probability of selecting the codon
i and P( j|i) is the conditional probability of choos-
ing the codon j, when we know that i has been se-
lected (the frequency of the codon j divided by the
sum of frequencies of all the codons differing from i
by one nucleotide substitution). Moreover, f (i) and
f ( j) are the polarity values of the amino acids, com-
monly used in the study of the genetic code optimal-
ity, (Woese, 1973) encoded by the codons i and j, re-
spectively. Therefore, F represents the total weighted
sum of the squared differences between physicochem-
ical properties of amino acids encoded by the codon
pairs differing in one nucleotide substitution. What
is more, all the single substitutions that lead to non-
sense mutations, i.e. replacements of any amino acid
by stop translation signal, are included in the calcula-
tion as the maximum of squared difference computed
for all possible changes between the chosen amino
acid property. Thus small F values indicate that the
given code shows a tendency to minimize the conse-
quences of amino acid replacements, whereas large
values mean that the code is poorly optimized in this
respect.
Similarly to (Bła
˙
zej et al., 2019), we calculated
the normalized percentage difference Pd between the
values of the function F for the SGC and the tested
code test for a fixed codon usage. This difference is
defined by
Pd(test,SGC) =
F(test) − F(SGC)
F(SGC)
· 100% . (3)
Clearly, Pd takes values in the range from −100 to
+∞. Particularly, negative values of Pd suggest that
the SGC is less robust to the consequences of point
mutations than the test code.
We would also like to point up the relationship
between F defined here and several quality measures
proposed by other authors (Di Giulio, 1989; Haig and
Hurst, 1991; Freeland and Hurst, 1998; Santos and
Monteagudo, 2010; Bła
˙
zej et al., 2016). The key dif-
ference lies in including the effect of different codon
frequencies on the potential costs of changes in amino
acid properties. In this case, for each codon i we as-
sume that this change is proportional to the probabil-
ity of choosing the second codon of the pair, j, from
the set of all codons differing from i in one nucleotide.
This requirement seems to be more realistic in com-
parison to the previous studies assuming that all pos-
sible pairs of codons D are equally probable.
Differences in the F values between the SGC and
its alternatives for various codon usages were as-
sessed statistically using the t-Student test because
the variables fulfilled the normal distribution require-
ment, as tested in the Shapiro-Wilk test. The re-
sulted p-values were corrected using the Benjamini-
Hochberg method to control the false discovery rate.
Differences between the compared groups were con-
sidered significant when the p-value was smaller than
0.05. The analyzes were performed in R package
3.5.1 (A language and environment for statistical
computing, R Core Team 2018, R Foundation for Sta-
tistical Computing, Vienna, Austria).
3 RESULTS
At the first stage of our study we calculated the val-
ues of the cost function F for the individual AGCs and
compared them with the result for the SGC. In this ap-
proach, we applied the codon usage corresponding to
the genome on which a selected alternative code op-
erates. In the Table 2, we presented the F function
values calculated for the chosen genetic codes and
the selected codon usages. It is clear that the qual-
ity of the SGC is strongly dependent on the assumed
codon usage because the F values change from 5.06
(codon usage 23) to 11.02 (codon usage 6). Because
the quality of the SGC depends on the codon usage,
we checked if the AGCs are better or worse than the
SGC regarding the codon usages gathered from their
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
46

corresponding genomes. Therefore, for every non-
standard genetic code, we calculated the values of the
cost function F and the measure Pd, which allowed us
to determine the difference in performance between
the SGC and the AGCs (Table 2). The calculated val-
ues of the Pd measure indicate that 11 out of 13 AGCs
perform from 11% to almost 39% better than the SGC
for the codon usages corresponding to the genomes on
which they operate. Only code 12 has its cost value
comparable with the SGC, and in just one case (code
23) the SGC outperforms the alternative genetic code
but only by less than 7%.
Table 2: The values of the F function calculated for the stan-
dard genetic code (F(SGC)) and its alternatives (F). For
each non-standard genetic code, we computed also the Pd
value as a measure of its distance from the SGC, which is
our reference point. The first column (Code) refers to the
record number of the considered alternative code according
to the annotation in the NCBI database. The second column
(Codon usage) includes the codon usages numbered accord-
ing to the numbers of alternative genetic codes operating on
genomes from which a given codon usage was extracted.
Code Codon usage F F(SGC) Pd
4 4 6.016 7.123 -15.54
5 5 5.086 5.733 -11.28
6 6 6.759 11.018 -38.65
9 9 4.136 6.255 -33.87
10 10 7.554 8.550 -11.64
12 12 6.580 6.558 0.334
13 13 4.30 6.027 -28.58
16 16 6.007 7.937 -24.31
22 22 4.895 7.784 -37.10
23 23 5.406 5.059 6.85
24 24 4.145 5.492 -24.51
27 27 6.621 9.823 -32.59
28 28 6.509 9.370 -30.52
Another interesting question which arose during
this investigation concerns the genetic code optimal-
ity in terms of minimizing the function F for ev-
ery considered codon usage. In order to answer this
question, we calculated the cost values of the cho-
sen genetic codes for every codon usage considered
in this study. Figure 1 presents the box-plots of these
values. Generally, 10 AGCs show smaller median
F values than the SGC (numbered as 1 in the fig-
ure). However, the difference is statistically signif-
icant only for the code 28, i.e. the Condylostoma
Nuclear Code (p=0.017). In turn, 3 alternative codes
have the median F value greater than the SGC but
the difference is statistically significant only for the
code 23, i.e. the Thraustochytrium Mitochondrial
Code (p=0.004), which substantially stands out from
the others and shows the greatest variation in terms
of F values. Nevertheless, 11 codes have the min-
imum value of the cost function smaller than that of
5
10
15
1 4 5 6 9 101213162223242728
Genetic code
F
Figure 1: The box-plots of the cost function F values cal-
culated for every considered codon usage and the given ge-
netic code numbered on the x-axis. The standard genetic
code is indicated as 1, whereas other numbers refer to its
alternative versions. The thick horizontal line indicates the
median, the box shows the range between the first and third
quartiles (IQR, the inter-quartile range) and the whiskers
determine the range without outliers for the assumption of
1.5 IQR.
the SGC. Only the codes 12 and 23 have the minimum
slightly greater than the SGC.
We also checked which codon usages generate the
smallest and the largest cost values for the SGC and
each of the AGCs. The results are presented in the
Table 3. It is interesting that only three out of 13
AGCs reached the minimum of the cost function F for
their respective codon usage. These genetic codes are:
Scenedesmus Mitochondrial Code (code 22), Thraus-
tochytrium Mitochondrial Code (code 23) and Pter-
obranchia Mitochondrial Code (code 24). What is
more, the codon usages characteristic of the organ-
isms using the codes 23 and 24 are also the best solu-
tions in terms of cost minimization for other genetic
codes, including the SGC. Generally, the minimal F
values are very similar to each other because they
range from 3.84 (Condylostoma Nuclear Code, code
28) to 5.41 (Thraustochytrium Mitochondrial Code,
code 23). The SGC does not seem to be well opti-
mized in terms of the minimal F value because there
are 11 AGCs that have this value smaller. In the
case of the AGCs, the minimal F values do not de-
viate substantially from the cost values calculated for
the codon usages typical of the corresponding codes.
The differences are from 0 to 3.4 (Euploid Nuclear
Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes
47

Code, code 10). These results agree with our observa-
tions presented in the Table 2, where we showed that
the AGCs are mostly better adapted than the SGC to
their respective codon usages. It suggests that an op-
timization process must have taken place between the
AGCs and the specific codon usages of the genomes
on which these codes operate.
Table 3: The codon usages for which the given genetic
codes reach the minimal and maximal cost values (F min
and F max). CU min - the codon usage for which F is
minimized; CU max - the codon usage for which F is max-
imized. The codon usages were numbered according to the
alternative genetic codes operating on the genomes from
which the given codon usage was extracted.
Code CU min F min CU max F max
1 23 5.05940 6 11.01860
4 24 4.15692 6 10.76410
5 24 4.05795 6 10.57890
6 23 4.84787 10 8.17872
9 24 4.06393 6 10.62270
10 24 4.14932 6 10.76050
12 23 5.08569 6 11.09380
13 24 4.06806 6 10.57490
16 22 4.55405 6 9.99753
22 22 4.89575 6 11.26960
23 23 5.40601 4 18.08380
24 24 4.14572 6 10.72980
27 23 4.84787 10 8.17872
28 24 3.84902 10 7.16863
In the case of maximization of F, there is no code
that is the least optimized for its specific codon us-
age (Table 3). The codon usage that produced the
maximal cost values for 12 out of 14 genetic codes
is typical of Oxytricha falla (codon usage 6). In this
case, the cost values varied from nearly 10 (code 16)
to 11.27 (code 22). For alternative genetic codes 6,
27 and 28, the worst codon usage proved to be that
of Euplotes octocarinatus (codon usage 10), with the
cost values ranging from 7.17 (code 28) to 8.18 (codes
6 and 27). The codon usage from Aspergillus nidu-
lans (codon usage 4) occurred the worst for Thraus-
tochytrium Mitochondrial Code (code 23). Its cost
function reached the largest cost value equal to 18.08.
Among the tested codes, the Condylostoma
Nuclear Code (code 28) minimized the F function for
almost all considered codon usages (Table 4). The
one exception was the Chlorophycean Mitochondrial
Code (code 16) that minimized the cost function
best for the codon usage typical of the mitochondrial
genome of Scenedesmus obliquus. The smallest of
these cost values is 3.85 and was reached for the code
28 and the codon usage 24 (corresponding to Rhabdo-
pleura compacta mitochondrial genome). The largest
value was 7.17, which was reached for the code 28
Table 4: The codon usages (Codon usage) and the genetic
codes (Code) which showed the minimum cost value (F) for
these usages. The codon usages were numbered according
to the alternative genetic codes operating on the genomes
from which the given codon usage was extracted.
Codon usage Code F
4 28 5.45865
5 28 4.71020
6 28 6.40637
9 28 3.85415
10 28 7.16863
12 28 6.32379
13 28 4.19715
16 28 5.59880
22 16 4.55405
23 28 4.82071
24 28 3.84802
27 28 6.48476
28 28 6.50975
and the codon usage 10 (corresponding to Eu-
plotes octocarinatus genome).
4 DISCUSSION
The main goal of this work was to test the quality
of the alternative genetic codes by using a more re-
alistic measure, which includes the codon usage and
the probabilities of codon substitution characteristic
of the genomes on which these coding systems oper-
ate. This approach can be justified because we found
out that minimizing the costs of amino acid replace-
ments by the genetic codes strongly depends on the
codon usage. The AGCs were generally more ef-
fective than the SGC in minimization of the conse-
quences of point mutations but the differences were
not statistically significant in most cases, when many
codon usages were tested. However, the AGCs mini-
mized the mutational costs substantially better (up to
39%) than the SGC for the codon usage correspond-
ing to the genomes on which these codes operate. The
cost function values calculated for the codon usages
specific for the AGCs were very similar or identical
with the minimal values found for any codon usage
and the given code. On the other hand, we could not
find any genetic code optimal for every considered
codon usage.
These results indicate that the tested AGCs are
generally optimized to the codon usages correspond-
ing to the genomes on which they function. These
codes evolved in specific conditions of the given
genomes, which could have promoted their optimiza-
tion. In contrast, the SGC evolved very early dur-
ing life evolution in primordial organisms that easily
transferred genetic information between themselves,
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
48

which could have made this code universal for many
organisms characterized by various codon biases.
Therefore, it could not have been fully optimized dur-
ing its evolution, as indicated also by simulation stud-
ies based on evolutionary algorithms (Bła
˙
zej et al.,
2018; Bła
˙
zej et al., 2016; Massey, 2008; Novozhilov
et al., 2007; Santos and Monteagudo, 2011; San-
tos and Monteagudo, 2017; Wnetrzak et al., 2018),
whereas the minimization of mutation errors could
have occurred by the direct optimization of the mu-
tational pressure around the established genetic code
(Dudkiewicz et al., 2005; Mackiewicz et al., 2008;
Bła
˙
zej et al., 2013; Bła
˙
zej et al., 2017; Bła
˙
zej et al.,
2015). It is also possible that the codon usage was
tuned in response to the changes in the genetic code.
The lack of the full SGC optimization could have also
resulted from its evolution driven by the expansion of
biosynthetic pathways of amino acids (Wong, 1975;
Di Giulio, 1999; Wong et al., 2016; Di Giulio, 2017).
The presented results may have an importance
for designing artificially modified organisms with al-
ternative codes, which produce peptides or proteins
with non-natural amino acids (Xie and Schultz, 2006;
Chin, 2014), by indicating that codon usage can sub-
stantially change the performance of the codes and
should be taken into account in the design. We are
planning to expand out model by including other
types of selection and codon measures to analyze the
optimality of the genetic codes.
ACKNOWLEDGEMENTS
We are very grateful to five anonymous reviewers for
their insightful comments and remarks, which signif-
icantly improved the manuscript. This work was sup-
ported by the National Science Centre, Poland (Nar-
odowe Centrum Nauki, Polska) under Grant number
2017/27/N/NZ2/00403.
REFERENCES
Abascal, F., Posada, D., and Zardoya, R. (2012). The evo-
lution of the mitochondrial genetic code in arthropods
revisited. Mitochondrial DNA, 23(2):84–91.
Andersson, S. and Kurland, C. (1995). Genomic evolution
drives the evolution of the translation system. Bio-
chemistry and Cell Biology, 73(11-12):775–787.
Bender, A., Hajieva, P., and Moosmann, B. (2008). Adap-
tive antioxidant methionine accumulation in respira-
tory chain complexes explains the use of a deviant ge-
netic code in mitochondria. Proc Natl Acad Sci U S A,
105(43):16496–16501.
Bła
˙
zej, P., Mackiewicz, D., Grabinska, M., Wnetrzak, M.,
and Mackiewicz, P. (2017). Optimization of amino
acid replacement costs by mutational pressure in bac-
terial genomes. Scientific Reports, 7:1061.
Bła
˙
zej, P., Mackiewicz, P., Cebrat, S., and Wanczyk, M.
(2013). Using evolutionary algorithms in finding of
optimized nucleotide substitution matrices. In Genetic
and Evolutionary Computation Conference, GECCO
’13, Amsterdam, The Netherlands, July 6-10, 2013,
Companion Material Proceedings, pages 41–42.
Bła
˙
zej, P., Miasojedow, B., Grabinska, M., and Mack-
iewicz, P. (2015). Optimization of mutation pressure
in relation to properties of protein-coding sequences
in bacterial genomes. PLoS One, 10:e0130411.
Bła
˙
zej, P., Wnetrzak, M., Mackiewicz, D., Gagat, P., and
Mackiewicz, P. (2019). Many alternative and theoret-
ical genetic codes are more robust to amino acid re-
placements than the standard genetic code. Journal of
Theoretical Biology, 464:21–32.
Bła
˙
zej, P., Wnetrzak, M., and Mackiewicz, P. (2016). The
role of crossover operator in evolutionary-based ap-
proach to the problem of genetic code optimization.
Biosystems, 150:61–72.
Bła
˙
zej, P., Wnetrzak, M., and Mackiewicz, P. (2018). The
importance of changes observed in the alternative ge-
netic codes. Proceedings of the 11th International
Joint Conference on Biomedical Engineering Systems
and Technologies - Volume 4: BIOINFORMATICS,
pages 154–159.
Boore, J. L. and Brown, W. M. (1994). Complete DNA
sequence of the mitochondrial genome of the black
chiton, Katharina tunicata. Genetics, 138(2):423–43.
Bove, J. M. (1993). Molecular features of mollicutes. Clin
Infect Dis, 17 Suppl 1:S10–S31.
Campbell, J. H., O’Donoghue, P., Campbell, A. G.,
Schwientek, P., Sczyrba, A., Woyke, T., S
¨
oll, D., and
Podar, M. (2013). UGA is an additional glycine codon
in uncultured SR1 bacteria from the human micro-
biota. Proc Natl Acad Sci U S A, 110:5540–5545.
Chin, J. W. (2014). Expanding and reprogramming the ge-
netic code of cells and animals. Annu Rev Biochem,
83:379–408.
Cortona, A. D., Leliaert, F., Bogaert, K. A., Turmel, M.,
Boedeker, C., Janouskovec, J., Lopez-Bautista, J. M.,
Verbruggen, H., Vandepoele, K., and Clerck, O. D.
(2017). The plastid genome in cladophorales green
algae is encoded by hairpin chromosomes. Current
Biology, 27(24):3771–3782.
Devoto, A. E., Santini, J. M., Olm, M. R., Anantharaman,
K., Munk, P., Tung, J., Archie, E. A., Turnbaugh, P. J.,
Seed, K. D., Blekhman, R., Aarestrup, F. M., Thomas,
B. C., and Banfield, J. F. (2019). Megaphages infect
Prevotella and variants are widespread in gut micro-
biomes. Nature Microbiology, 4:693–700.
Di Giulio, M. (1989). The extension reached by the mini-
mization of the polarity distances during the evolution
of the genetic code. J Mol Evol, 29(4):288–93.
Di Giulio, M. (1999). The coevolution theory of the origin
of the genetic code. J Mol Evol, 48(3):253–5.
Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes
49

Di Giulio, M. (2017). Some pungent arguments against
the physico-chemical theories of the origin of the ge-
netic code and corroborating the coevolution theory. J
Theor Biol, 414:1–4.
Dudkiewicz, A., Mackiewicz, P., Nowicka, A., Kowalezuk,
M., Mackiewicz, D., Polak, N., Smolarczyk, K., Ba-
naszak, J., Dudek, M. R., and Cebrat, S. (2005).
Correspondence between mutation and selection pres-
sure and the genetic code degeneracy in the gene
evolution. Future Generation Computer Systems,
21(7):1033–1039.
Epstein, C. J. (1966). Role of the amino-acid ”code” and of
selection for conformation in the evolution of proteins.
Nature, 210(5031):25–8.
Freeland, S. J. and Hurst, L. D. (1998). The genetic code is
one in a million. J Mol Evol, 47(3):238–248.
Freeland, S. J., Knight, R. D., Landweber, L. F., and Hurst,
L. D. (2000). Early fixation of an optimal genetic
code. Mol Biol Evol, 17(4):511–8.
Freeland, S. J., Wu, T., and Keulmann, N. (2003). The
case for an error minimizing standard genetic code.
Origins of Life and Evolution of the Biosphere, 33(4-
5):457–477.
Gomes, A. C., Miranda, I., Silva, R. M., Moura, G. R.,
Thomas, B., Akoulitchev, A., and Santos, M. A.
(2007). A genetic code alteration generates a pro-
teome of high diversity in the human pathogen Can-
dida albicans. Genome Biol, 8(10):R206.
Goodarzi, H., Najafabadi, H. S., Hassani, K., Nejad, H. A.,
and Torabi, N. (2005). On the optimality of the ge-
netic code, with the consideration of coevolution the-
ory by comparison of prominent cost measure matri-
ces. J Theor Biol, 235(3):318–25.
Haig, D. and Hurst, L. D. (1991). A quantitative measure
of error minimization in the genetic-code. J Mol Evol,
33(5):412–417.
Heaphy, S. M., Mariotti, M., Gladyshev, V. N., Atkins, J. F.,
and Baranov, P. V. (2016). Novel ciliate genetic code
variants including the reassignment of all three stop
codons to sense codons in Condylostoma magnum.
Mol Biol Evol, 33:2885–2889.
Hoffman, D. C., Anderson, R. C., DuBois, M. L.,
and Prescott, D. M. (1995). Macronuclear gene-
sized molecules of hypotrichs. Nucleic Acids Res,
23:1279–1283.
Janouskovec, J., Sobotka, R., Lai, D.-H., Flegontov, P.,
Kon
´
ık, P., Komenda, J., Ali, S., Pr
´
a
ˇ
sil, O., Pain, A.,
Oborn
´
ık, M., Luke
ˇ
s, J., and Keeling, P. J. (2013). Split
photosystem protein, linear-mapping topology, and
growth of structural complexity in the plastid genome
of Chromera velia. Molecular Biology and Evolution,
30(11):2447–2462.
Kurnaz, M. L., Bilgin, T., and Kurnaz, I. A. (2010). Certain
non-standard coding tables appear to be more robust
to error than the standard genetic code. J Mol Evol,
70(1):13–28.
Mackiewicz, P., Biecek, P., Mackiewicz, D., Kiraga, J.,
Baczkowski, K., Sobczynski, M., and Cebrat, S.
(2008). Optimisation of asymmetric mutational pres-
sure and selection pressure around the universal ge-
netic code. Computational Science - Iccs 2008, Pt 3,
5103:100–109.
Massey, S. E. (2008). A neutral origin for error minimiza-
tion in the genetic code. J Mol Evol, 67(5):510–516.
Massey, S. E. and Garey, J. R. (2007). A comparative ge-
nomics analysis of codon reassignments reveals a link
with mitochondrial proteome size and a mechanism
of genetic code change via suppressor tRNAs. J Mol
Evol, 64(4):399–410.
McCutcheon, J. P., McDonald, B. R., and Moran, N. A.
(2009). Origin of an alternative genetic code in the
extremely small and GC-rich genome of a bacterial
symbiont. PLoS Genetics, 5(7):e1000565.
Morgens, D. W. and Cavalcanti, A. R. (2013). An alterna-
tive look at code evolution: using non-canonical codes
to evaluate adaptive and historic models for the origin
of the genetic code. J Mol Evol, 76(1-2):71–80.
Nakamura, Y., Gojobori, T., and Ikemura, T. (2000). Codon
usage tabulated from the international DNA sequence
databases: status for the year 2000. Nucleic Acids Res,
28:292.
Novozhilov, A. S., Wolf, Y. I., and Koonin, E. V. (2007).
Evolution of the genetic code: partial optimization of
a random code for robustness to translation error in a
rugged fitness landscape. Biol Direct, 2:24.
Perseke, M., Hetmank, J., Bernt, M., Stadler, P. F., Schlegel,
M., and Bernhard, D. (2011). he enigmatic mito-
chondrial genome of Rhabdopleura compacta (Pter-
obranchia) reveals insights into selection of an effi-
cient tRNA system and supports monophyly of Am-
bulacraria. BMC Evolutionary Biology, 11:134.
P
´
anek, T.,
ˇ
Zihala, D., Sokol, M., Derelle, R., Klime
ˇ
s, V.,
Hradilov
´
a, M., Zadrob
´
ılkov
´
a, E., Susko, E., Roger,
A. J.,
ˇ
Cepi
ˇ
cka, I., and Eli
´
a
ˇ
s, M. (2017). Nuclear ge-
netic codes with a different meaning of the UAG and
the UAA codon. BMC Biology, 15:8.
Sanchez-Silva, R., Villalobo, E., Morin, L., and Torres, A.
(2003). A new noncanonical nuclear genetic code:
Translation of UAA into glutamate. Current Biology,
13(5):442–447.
Santos, J. and Monteagudo, A. (2010). Study of the genetic
code adaptability by means of a genetic algorithm. J
Theor Biol, 264(3):854–865.
Santos, J. and Monteagudo, A. (2011). Simulated evolution
applied to study the genetic code optimality using a
model of codon reassignments. BMC Bioinformatics,
12.
Santos, J. and Monteagudo,
´
A. (2017). Inclusion of the fit-
ness sharing technique in an evolutionary algorithm
to analyze the fitness landscape of the genetic code
adaptability. BMC Bioinformatics, 18(1):195.
Santos, M. A., Cheesman, C., Costa, V., Moradas-Ferreira,
P., and Tuite, M. F. (1999). Selective advantages cre-
ated by codon ambiguity allowed for the evolution of
an alternative genetic code in Candida spp. Molecular
Microbiology, 31:937–947.
Santos, M. A., Keith, G., and Tuite, M. F. (1993). Non-
standard translational events in Candida albicans me-
diated by an unusual seryl-tRNA with a 5’-CAG-3’
(leucine) anticodon. The EMBO Journal, 12:607–616.
BIOINFORMATICS 2020 - 11th International Conference on Bioinformatics Models, Methods and Algorithms
50

Sengupta, S. and Higgs, P. G. (2005). A unified model of
codon reassignment in alternative genetic codes. Ge-
netics, 170(2):831–40.
Sengupta, S., Yang, X., and Higgs, P. G. (2007). The mech-
anisms of codon reassignments in mitochondrial ge-
netic codes. J Mol Evol, 64(6):662–88.
Swart, E. C., Serra, V., Petroni, G., and Nowacki, M.
(2016). Genetic codes with no dedicated stop codon:
Context-dependent translation termination. Cell,
166:691–702.
Swire, J., Judson, O. P., and Burt, A. (2005). Mitochon-
drial genetic codes evolve to match amino acid re-
quirements of proteins. J Mol Evol, 60(1):128–39.
Wnetrzak, M., Bła
˙
zej, P., Mackiewicz, D., and Mackiewicz,
P. (2018). The optimality of the standard genetic code
assessed by an eight-objective evolutionary algorithm.
BMC Evolutionary Biology, 18:192.
Woese, C. R. (1973). Evolution of the genetic code. Natur-
wissenschaften, 60(10):447–59.
Wong, J. T. (1975). A co-evolution theory of the genetic
code. Proc Natl Acad Sci U S A, 72(5):1909–12.
Wong, J. T., Ng, S. K., Mat, W. K., Hu, T., and Xue, H.
(2016). Coevolution theory of the genetic code at age
forty: Pathway to translation and synthetic life. Life
(Basel), 6(1):E12.
Xie, J. M. and Schultz, P. G. (2006). Innovation: A chemical
toolkit for proteins - an expanded genetic code. Nat
Rev Mol Cell Biol, 7(10):775–782.
Z
´
ahonov
´
a, K., Kostygov, A. Y.,
ˇ
Sev
ˇ
c
´
ıkov
´
a, T., Yurchenko,
V., and Eli
´
a
ˇ
s, M. (2016). An unprecedented non-
canonical nuclear genetic code with all three termi-
nation codons reassigned as sense codons. Curr Biol,
26:2364–2369.
Properties of the Standard Genetic Code and Its Alternatives Measured by Codon Usage from Corresponding Genomes
51
