
A Computational Platform for Heart Failure Cases Research
Jo
˜
ao Rafael Almeida
1,2 a
, Pedro Freire
1 b
, Olga Fajarda
1 c
and Jos
´
e Lu
´
ıs Oliveira
1 d
1
DETI/IEETA, University of Aveiro, Aveiro, Portugal
2
Department of Information and Communications Technologies, University of A Coru
˜
na, A Coru
˜
na, Spain
Keywords:
Health Data, Clinical Studies, Cohorts, Heart Failure.
Abstract:
Heart failure is a global health issue that affects millions of people worldwide, and is the main cause of disabil-
ity and hospitalisation of elderly people. Approximately half of these have heart failure with preserved ejection
fraction (HFpEF) and this proportion is increasing as the population ages. There is still no efficient treatment
for HFpEF and today’s existing therapies only aim at relieving symptoms. With the aim to unravelling the
pathophysiology of HFpEF and identify new therapeutic targets, ongoing long-time studies are collecting pa-
tient’s data, including the genomic information. This procedure is complex and requires electronically-stored
health information to keep the patient’s information centralised to simplify the following up. In this paper,
we present an computational system to support researchers in the different stages of a clinical study, and we
describe its use in the management and analyse of HFpEF cohorts.
1 INTRODUCTION
Heart failure (HF) is a global scourge that affects over
26 million people worldwide (Savarese and Lund,
2017). This condition is associated with a high risk
of morbidity and mortality, as well as a large health
care resource consumption (Bui et al., 2011). HF is,
also, the main cause of disability and hospitalisation
of elderly people (Farmakis et al., 2015) and is ex-
pected to aggravate as the population ages (Gaggin
and Januzzi Jr, 2013).
Approximately 50% of all the cases are related
to heart failure with preserved ejection fraction (HF-
pEF) (Dunlay et al., 2017), and these patients have
typically higher rates of hospitalisations then the ones
suffering from heart failure with reduced ejection
fraction (HFrEF) (Steinberg et al., 2012).
Unlike HFrEF, whose prognosis has been improv-
ing, the prognosis of HFpEF has remained mostly un-
changed over the last decades (Bonsu et al., 2018).
No therapy for HFpEF has demonstrated improve-
ment in prognosis and several pharmacological agents
applied in clinical trials have shown no benefit (Fer-
rari et al., 2015). The existing therapies only aim
a
https://orcid.org/0000-0003-0729-2264
b
https://orcid.org/0000-0001-5663-3403
c
https://orcid.org/0000-0003-1957-4947
d
https://orcid.org/0000-0002-6672-6176
at relieving symptoms. Even the diagnosis of HF-
pEF is not consensual. The American College
of Cardiology Foundation/American Heart Associa-
tion (ACCF/AHA) and the European Society of Car-
diology (ESC) use different criteria to diagnose HF-
pEF. ACCF/AHA uses a diagnosis of exclusion in
compliance with the Framingham HF criteria, while
ESC considers that impairment of diastolic function is
the key diagnosis criteria (Paulus et al., 2007; Yancy
et al., 2013). The ACCF/AHA diagnosis criteria
recognise close to 30% more patients consider to have
HFpEF than the ESC criteria and so the patients en-
rolled in clinical trials are different depending on the
criteria used (Persson et al., 2007).
HFpEF patient population is mostly elderly, het-
erogeneous and with numerous comorbidities includ-
ing systemic arterial hypertension (SAH), obesity,
and diabetes mellitus (DM) (Shah, 2017). The vari-
ations of comorbidities have a significant impact on
the symptoms and the therapy responses, as well as
the mortality. The underlying pathophysiologies are,
therefore, variable and remain unclear (Sharma and
Kass, 2014).
During the last decade, several projects emerged
to study several aspects of HF, some of which targeted
specifically HFpEF.
Heart OMics in AGEing (HOMAGE) (Jacobs
et al., 2014) was a project launched with the pur-
pose to identify and validate predictive ‘omics’-based
Almeida, J., Freire, P., Fajarda, O. and Oliveira, J.
A Computational Platform for Heart Failure Cases Research.
DOI: 10.5220/0009104706010608
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 5: HEALTHINF, pages 601-608
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
601

biomarker for HF. These biomarkers should help to
identify patients at risk to develop HF in order to pre-
vent the development of this disease which affects
mostly the elderly population. During the project,
a centralised database of existing cohorts, which in-
clude patients with heart failure, patients at risk to de-
velop heart failure and healthy individuals, was cre-
ated.
The INTERnational Congestive Heart Failure
Study (INTER-CHF) (Dokainish et al., 2015), was a
prospective study conducted from 2012 to 2014. The
purpose of this project was to document sociodemo-
graphics, HF etiologies, treatment and mortality of
patients, from low and middle-income countries in
Africa, Asia, the Middle East, and South America,
with HP.
The OPTIMEX project studied the impact of ex-
ercise training as primary and secondary prevention
of HFpEF (Suchy et al., 2014). The objective of this
project was to define the optimal dose of exercise
training in patients with HFpEF in order to prevent
the disease development and improve the pathophysi-
ology.
NETDIAMOND is another project that joins a
network of referenced centres with complementary
expertise to unravel the pathophysiology of HFpEF
and develop evidence-based therapeutics strategies
directed at HFpEF (Lourenco et al., 2018). By cor-
relating and integrating transcriptomics, proteomics
and lipidomics studies with clinical data, this project
aims to achieve a holistic view of HFpEF and the role
of comorbidities. Furthermore, this project intends to
determine gene variants that may predispose to HF-
pEF.
Due to the complexity of HFpEF and the multifar-
ious pathophysiology, this disease is highly challeng-
ing and only a deep omics analysis integrated with
clinical data and mixed with data mining will pro-
vide precise patient-directed treatments. To address
this issues, we present a platform composed of several
tools to support the research in HFpEF. This ecosys-
tem provides the following features:
• A centralised system following standard practices
of clinical, functional and follow-up procedures
to gather clinical and omics data from HFpEF pa-
tients;
• A secure central large data repository for all pa-
tients and animal model data;
• Analytic features to design and explore cohorts
over the collected data;
• And algorithms to automatically analyse the pa-
tients’ data, including the omics records.
2 GATHERING DATA IN HFpEF
STUDIES
Clinical studies are typically performed using a com-
putational system to support the research. Frequently
the institutional Electronic Health Record (EHR) sys-
tems are used to gather the patient clinical data. How-
ever, there are studies with a very focused purpose
in which the EHR is too generic for storing all the
patient’s information. In these scenarios, the use of
spreadsheets has been the most common solution, but
this is not the best procedure for several reasons. This
approach fails by not following a standard structure,
leading to issues in future reuse of the collected data.
Moreover, it also does not have any control over who
can edit or see the data, complicating the study man-
agement.
Regarding these issues, we propose the NETDIA-
MOND Platform
1
, which is a web-based system to
record patient information in clinical studies focused
on HFpEF patients. The system was designed on top
of the MONTRA framework, which relies on an easy-
to-use tabular skeleton, intended for data integration,
with emphasis on biomedical data (Silva et al., 2018).
The developed platform was targeted to groups of
researchers, mainly because the studies are conducted
in different organisations, by distinct researchers, and
with different roles. Therefore, for the same patient,
various entities are involved in the data collection, for
instance, one group could be responsible for inquir-
ing the patient’s habits and another for recording the
omics information extracted from samples of the heart
tissue. This led to the creation of Role-Based Access
Control (RBAC) policies at the user and group levels.
The patient’s clinical data is stored in a data struc-
ture created by consensus among all the entities in-
volved in the study. This data structure keeps all the
information centralised and in accordance with all of
the study’s needs. This structure consists of differ-
ent input components, such as numeric type, free-text,
multi-choice, and others. This reduces insertion er-
rors by reducing the flexibility in the inputs available
for the specified data type. The data schema is also
composed of rules, i. e., some fields are mandatory
and others are only mandatory depending on the out-
come of other variables. The system’s data structure
has also flexible management features, so that it can
be adapted to the future data collection needs of each
study.
This platform was designed to gather phenotype-
genotype data in HFpEF studies, but the omics data
are stored in a different system. However, there is a
1
https://bioinformatics.ua.pt/netdiamond
HEALTHINF 2020 - 13th International Conference on Health Informatics
602

direct connection between the two systems, so as not
to lose context.
3 DATA REPOSITORY
This project sets forth to address the HFpEF issue
through comprehensive multi-omics studies in plasma
and tissues from HFpEF patients and animal mod-
els, which generates large and dissimilar volumes of
data. The collected omics data are widely spread in a
vast variety of unstructured formats, making it harder
for researchers to manage and create cohorts. They
hold valuable information on both metadata and con-
tent planes that could be used to enhance data discov-
ery. Thus, a data repository with enhanced search fea-
tures, able to execute full-text searches through meta-
data and raw data are desirable, in such a way that
allows researchers to manage and create cohorts more
effectively.
Several open-source projects for data management
were considered for analysis: Girder
2
, CKAN
3
,
DSpace
4
, Islandora
5
and Alfresco
6
. After analysing
each one, it was possible to conclude that Alfresco
was far better than its competitors, due to its ability
to extract and index metadata and to its flexibility to
add custom support to other files types. Alfresco is an
open-source Enterprise Content Management (ECM)
system that provides several services and controls for
data management, offering tools to index metadata
content and execute full-text searches through files’
content (Sladi
´
c et al., 2011).
Alfresco does not support genomic files by de-
fault, which led to the adaptation of the software to
the biomedical research scenario, by adding support
for FASTA and Genbank files. The adaptation was
achieved by developing a metadata extractor featured
by BioJava library (Lafita et al., 2019) and a custom
model to store the extracted data.
The custom model was designed taking in mind
the properties that could be extracted from FASTA
and Genbank files. After analysing both file formats,
it was possible to conclude that the following prop-
erties should be considered: accession, description,
keywords, gene, organisms, number of sequences and
sequences. It’s important to notice that, in our so-
lution, not every sequence is indexed. Indexing an
entire genome is not reliable due to the huge size of
2
https://girder.readthedocs.io/en/stable/
3
https://ckan.org/
4
https://duraspace.org/dspace/
5
https://islandora.ca/
6
https://www.alfresco.com
its sequences. As a workaround to this problem, only
sequences listed on a configuration file are indexed.
Furthermore, the metadata extractor extracts and
saves target properties into the created custom model.
In Addiction, BioJava library is used on the extractor
to parse genomic files and extract target properties.
The extraction is triggered right after a successful up-
load.
Researchers, by using Alfresco with the extension
presented here, can store, manage and share genomic
files in a more efficient way, having the possibility to
search for certain genes or even sequences.
4 PATIENT COHORT
DEFINITION
The previous two sections described the two pieces
of the ecosystem responsible for gathering patients’
data in HFpEF studies. The next stage in the proposed
ecosystem is patient cohort selection. This step is es-
sential to filter patients of interest in specific scenar-
ios. Therefore, aiming the cohort definition and data
filtering, we used TranSMART, which is a platform
with analytical web applications to aggregate differ-
ent data sets. In this platform, the researchers can se-
lect the variables of interest through a friendly user
interface, without interacting directly with the data.
This tool enables patient and variable selection fea-
tures over the data set, as well as the aggregation of
several data sets, if necessary.
However, to employ this tool in the ecosystem,
we needed to map the recorded data into the tool’s
database. Therefore, we defined a semantic ontology
and used a migration pipeline to extract, transform
and load (ETL) the data from the NETDIAMOND
platform to the TranSMART tool. This ontology was
built on top of the initial data structure detailing the
recorded concepts. This new extra information allows
us to validate clinical data with ranges of values, vari-
able types and options for questions. For instance,
in this migration, all the data is processed, and when
a variable is out-of-range, we will be notified and re-
quest for rectification in the record, thereby increasing
the quality of the data.
Figure 1 shows the detailed migration workflow
divided into the three ETL stages: Extracting, Trans-
formation and Loading. The workflow starts by ex-
porting the collected data from the NETDIAMOND
Platform to CSV files, which are then read (Extract-
ing stage). Afterwards, the collected data is trans-
formed into a new structure, that simplifies the har-
monisation procedure. The Cohort harmoniser builds
a new cohort using that structure and validates the
A Computational Platform for Heart Failure Cases Research
603

Extracting Transformation Loading
Cohort in
raw data
TranSMART
Loader
WebProtégé Output:
Ontology rules
Cohort
Reader
Structure
Transformer
Cohort
Harmoniser
Migration Report
Figure 1: The migration workflow from NETDIAMOND Platform to TranSMART divided into the three stages.
collected data applying the ontology rules. Addition-
ally, those rules are also used to calculate new patient
information (Transformation stage). Finally, the co-
hort is loaded into TranSMART, and the migration re-
port is generated (Loading stage). This report has all
the warning and errors produced during the migration,
which helps to validate the data in both platforms.
Another positive aspect of performing this transi-
tion is the possibility to insert new knowledge about
patients that are already present in the data but in a
roundabout way. Rules and conditions, that are not
typically recorded during the study, can be defined in
the ontology. This occurs mainly because those data
can be calculated during the analysis. Moreover, this
kind of information could help to define a more fo-
cused cohort. For instance, since obese patients are
considered patients with a cardiovascular risk factor,
researchers may want to build a cohort composed of
patients who are obese and if the concept obese was
not recorded during the clinical visits, they need to
analyse the raw data to understand which patients be-
long to this group. To know if the patient is obese,
researchers only need the patients’ height and weight,
and then during the migration, the body mass index
can be calculated. If this value is higher than 30, then
the patient is considered obese (James, 2004).
Although the TranSMART was designed to per-
form more complex tasks such as data aggregation of
different data sources, we decided to use it only for
cohort definition. Using TranSMART, the researchers
can select the desired data sets to follow or process,
by defining a set of conditions. These conditions
are mainly inclusion and exclusion criteria, which are
then applied to the study data set. Moreover, these
criteria can also be applied to two subsets aiming the
comparison between them.
5 DATA ANALYSIS
The analysis of the genomic data collected, by com-
paring the measurements in different conditions, is
useful for diagnostic, prognostic, subdivision of pa-
tients in clinical studies and prediction of therapeu-
tic response (Haury et al., 2011). Statistical methods
are commonly used to do the analysis, however, these
rely on the definition of an arbitrary threshold, whose
value to use is not consensual (Cui and Churchill,
2003). The analysis is usually done by comparing the
expressions of genes in order to identify differentially
expressed genes (DEGs) in different conditions.
The main technologies used to measure gene ex-
pressions are microarray and the more recently de-
veloped RNA-Sequencing (RNA-seq) (Wang et al.,
2009). RNA-seq is technologically more advanced,
however, microarray technology continues to be
widely used because it is cheaper and there are, freely
available, robust, mature and reliable tools to pro-
cess and analyse the data obtained (Thompson et al.,
2016). Different platforms are used by researchers
to measure the genes’ expression. These platforms
are composed of diverse sets of genes and merging
data sets from different platforms is challenging (Ku-
mar Sarmah and Samarasinghe, 2010).
We developed a pipeline to analyse genomic data,
namely gene expressions obtained using microarray
technology. Figure 2 presents the pipeline.
The first step is to pre-process the data, which con-
sists of performing background correction, normali-
sation and probe summarisation of the raw data. The
raw data originated using the same platform are joint
before the pre-processing.
Before merging the several data sets the common
genes across the different platforms must be identi-
fied. To not reduce significantly the number of genes,
instead of the gene identifier, the GenBank sequence
HEALTHINF 2020 - 13th International Conference on Health Informatics
604

Pre-process the data
↓
Merge the data
↓
Determine the adjusted p-value and fold change
↓
Obtain different sets of features by using different
thresholds
↓
Batch-adjust the data
↓
Use a supervised learning algorithm to select the set
of features with the highest accuracy
Figure 2: Pipeline to analyse genomic data.
accession identifier, which uniquely identifies a bio-
logical sequence, should be used. After merging the
several data sets using the common GenBank identi-
fiers, a gene expressions data set is obtained.
Thereafter a statistical method is used to deter-
mine the adjusted p-value and fold change for the
different features (the GenBank identifiers) by us-
ing the expressions in one condition and the expres-
sions in another condition, e.g. diseased vs. con-
trol. By choosing different thresholds for the ad-
justed p-value and fold change, several sets of features
are obtained. The next step is to select the one set
with the best predictive accuracy and this can be done
by using supervised learning algorithms, e.g. Ran-
dom Forest (Breiman, 2001) or Support Vector Ma-
chines (Cortes and Vapnik, 1995). However, before
using a supervised learning algorithm, the data must
be batch-adjusted.
The batch effect is the non-biological variation
that affects the gene expressions (Leek et al., 2010).
To account for the batch effect, two approaches can be
used: include the batch variable in the statistical anal-
ysis or adjust the data for batch effects before using
it (Nygaard et al., 2016). In this pipeline, these two
approaches are used. Thus, the batch variables are
included in the statistical method used to determine
the adjusted p-value and fold change and the gene ex-
pression data are batch-adjusted before using the su-
pervised learning algorithm. Several methods exist to
batch-adjust the data and a survey of these methods
can be found in (Lazar et al., 2012).
The evaluation of the pipeline was done using nine
publicly available microarray data sets from studies
about heart diseases. The pre-processement of the raw
data was done using the oligo package (Carvalho
and Irizarry, 2010) of the R/Bioconductor software
packager (Gentleman et al., 2004), which implements
the robust multichip average (RMA) pre-processing
method (Irizarry et al., 2003). The microarray data
sets were generated using four different platforms that
have 8354 GenBank sequence accession identifier in
common and so after merging the nine data set, a data
set with 689 samples and 8354 features was obtained.
To determine the adjusted p-value and fold change
the R/Bioconductor software package limma (Ritchie
et al., 2015) was used. The thresholds used for the
adjusted p-value are 0.01 and 0.05 and for the fold
change the values from 1.5 to 3. Using the threshold
0.01 or 0.05 for the p-value in conjunction with the
thresholds of the fold change resulted in the same sets
of features. The fold change thresholds of 2.7 and 2.8
resulted in the same set of features and the same hap-
pened for the fold change thresholds 2.9 and 3. So
fourteen different set of features were obtained. To
select the set of features with the best predictive accu-
racy we used the R package caret (Kuhn et al., 2008)
implementation of the Random Forest algorithm. The
evaluation of each model was done using repeated 10-
fold cross-validation with 3 repeats. We identified a
set of 86 differentially expressed genes that correctly
classifies samples with a heart disease and samples
with no heart condition with an accuracy of approxi-
mately 96%.
6 RESULTS AND DISCUSSION
The proposed ecosystem provides support for all the
main stages in HFpEF clinical studies. Figure 3
presents an overview of the workflow from data col-
lection to the discovery of new biomarkers. This
workflow is divided into three important stages, usu-
ally performed by different entities. Patients’ data
collection is done at health care facilities by physi-
cians during patients’ visits. In the NETDIAMOND
project, different groups of physicians from several
Portuguese health institutions are collecting those
data.
Both tools used in the data collection where used
and validated in different contexts. The MONTRA
framework, which is the most important piece of the
NETDIAMOND Platform, was already used to sup-
port the creation of catalogues of distributed health
databases. The base features were the same but us-
ing a different entity to categorise, while in one it is
the patients, in the other the data owners recorded the
metadata from their databases. The Alfresco was used
in countless projects for different purposes. However,
for our scenario, we needed to add some features to
increase the usability in the proposed context.
The second stage of this workflow can be executed
almost in parallel with the data collection as long as
A Computational Platform for Heart Failure Cases Research
605
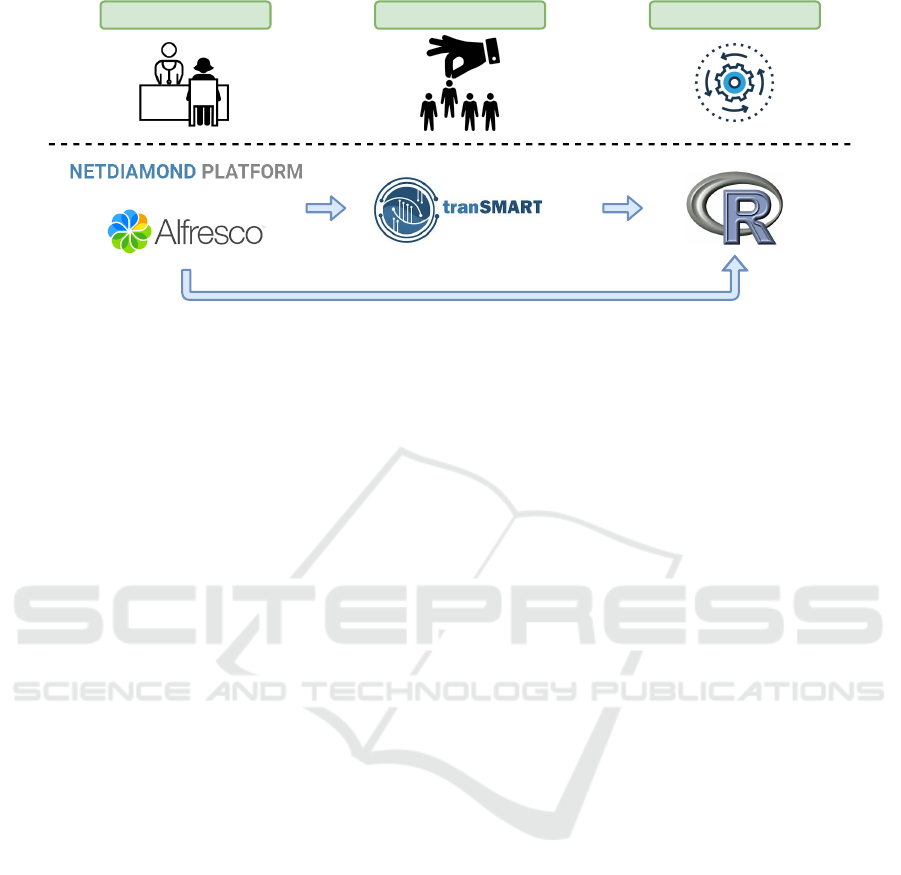
Patient data collect Cohort Selection Data processing
Figure 3: Workflow overview with all the steps and all the components’ interconnection.
data is migrated in the TranSMART. In this stage,
the medical research teams can define their cohorts
by selecting variables of interest to explore specific
biomarkers. These selections are essential to filter the
data on the omics repository.
The TranSMART is a tool already validated in the
clinical context. Our main contribution to this part
of the workflow was primarily the data migration and
aggregation in a centralised data warehouse. This mi-
gration required the creation of an ontology for HF-
pEF and harmonisation of concepts. At the end of
this stage, we validated the data using the ontology
rules introduced during the migration process.
In the final stage, the data can be analysed using
the methodology presented in Section 5, or using oth-
ers. At this stage, researchers can define cohorts by
selecting the right inclusion and exclusion criteria in
order to discover new biomarkers or detect the effects
of therapies applied to patients.
This workflow allows the allocation of roles over
different institutions and improves their cooperation.
The ecosystem was designed to fulfil the technical
needs of the NETDIAMOND project by supporting
all the study pipeline, and to help unravelling patho-
physiology and identifying new therapeutic targets in
HFpEF. With this ecosystem and its tools we expect
that the medical community will be able to:
• Implement a standard clinical practice distributed
at a national level. This contemplates the collec-
tion of myocardial samples from HFpEF patients
and an organised registry for this disease.
• Examine cohorts of HFpEF patients composed of
their blood samples paired with samples of the
myocardium and adipose tissues.
• Use a centralised data repository for all the pa-
tients’ clinical data related to this disease.
• Evaluate the genetic variants by developing cell
and mouse models of this disease that rehash the
human gene variants amenable to high throughput
pharmacological screening.
• Increase public awareness of the HFpEF disease
and develop preventive design strategies to reduce
its impact on the elderly population.
7 CONCLUSION
HFpEF is becoming the predominant form of HF
which leads to new research initiatives aiming to dis-
cover new treatments and therapies. Regarding this
problem, we developed a computational ecosystem to
help conducting and supporting clinical studies. The
proposed solution was applied in the NETDIAMOND
project and can be reused in other projects that study
a disease-specific group. As a result, this multidisci-
plinary approach ensures the cross-checking of omics
data with clinical information, which promotes the
prototyping and application of new therapies with rel-
evance in the clinical practices. Moreover, it may help
gathering new knowledge about the genomic variants
that may predispose to HFpEF patients, leading to the
development of improved therapies.
ACKNOWLEDGEMENTS
This work was funded by the NETDIAMOND
project, grant number POCI-01-0145-FEDER-
016385. Jo
˜
ao Rafael Almeida is supported by FCT
- Foundation for Science and Technology (national
funds), grant SFRH/BD/147837/2019.
HEALTHINF 2020 - 13th International Conference on Health Informatics
606

REFERENCES
Bonsu, K. O., Arunmanakul, P., and Chaiyakunapruk, N.
(2018). Pharmacological treatments for heart fail-
ure with preserved ejection fraction—a systematic re-
view and indirect comparison. Heart failure reviews,
23(2):147–156.
Breiman, L. (2001). Random forests. Machine learning,
45(1):5–32.
Bui, A. L., Horwich, T. B., and Fonarow, G. C. (2011). Epi-
demiology and risk profile of heart failure. Nature
Reviews Cardiology, 8(1):30.
Carvalho, B. S. and Irizarry, R. A. (2010). A framework
for oligonucleotide microarray preprocessing. Bioin-
formatics, 26(19):2363–2367.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine learning, 20(3):273–297.
Cui, X. and Churchill, G. A. (2003). Statistical tests for dif-
ferential expression in cdna microarray experiments.
Genome biology, 4(4):210.
Dokainish, H., Teo, K., Zhu, J., Roy, A., Al-Habib, K., El-
Sayed, A., Palileo, L., Jaramillo, P. L., Karaye, K., Yu-
soff, K., et al. (2015). Heart failure in low-and middle-
income countries: background, rationale, and de-
sign of the international congestive heart failure study
(inter-chf). American heart journal, 170(4):627–634.
Dunlay, S. M., Roger, V. L., and Redfield, M. M. (2017).
Epidemiology of heart failure with preserved ejection
fraction. Nature Reviews Cardiology, 14(10):591.
Farmakis, D., Parissis, J., Lekakis, J., and Filippatos, G.
(2015). Acute heart failure: epidemiology, risk fac-
tors, and prevention. Revista Espa
˜
nola de Cardiolog
´
ıa
(English Edition), 68(3):245–248.
Ferrari, R., B
¨
ohm, M., Cleland, J. G., Paulus, W. J., Pieske,
B., Rapezzi, C., and Tavazzi, L. (2015). Heart fail-
ure with preserved ejection fraction: uncertainties
and dilemmas. European journal of heart failure,
17(7):665–671.
Gaggin, H. K. and Januzzi Jr, J. L. (2013). Biomark-
ers and diagnostics in heart failure. Biochimica et
Biophysica Acta (BBA)-Molecular Basis of Disease,
1832(12):2442–2450.
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B.,
Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y.,
Gentry, J., et al. (2004). Bioconductor: open software
development for computational biology and bioinfor-
matics. Genome biology, 5(10):R80.
Haury, A.-C., Gestraud, P., and Vert, J.-P. (2011). The influ-
ence of feature selection methods on accuracy, stabil-
ity and interpretability of molecular signatures. PloS
one, 6(12):e28210.
Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D.,
Antonellis, K. J., Scherf, U., and Speed, T. P. (2003).
Exploration, normalization, and summaries of high
density oligonucleotide array probe level data. Bio-
statistics, 4(2):249–264.
Jacobs, L., Thijs, L., Jin, Y., Zannad, F., Mebazaa, A.,
Rouet, P., Pinet, F., Bauters, C., Pieske, B., Tomas-
chitz, A., et al. (2014). Heart ‘omics’ in ageing
(homage): design, research objectives and character-
istics of the common database. Journal of biomedical
research, 28(5):349.
James, P. T. (2004). Obesity: the worldwide epidemic. Clin-
ics in dermatology, 22(4):276–280.
Kuhn, M. et al. (2008). Building predictive models in r us-
ing the caret package. Journal of statistical software,
28(5):1–26.
Kumar Sarmah, C. and Samarasinghe, S. (2010). Microar-
ray data integration: frameworks and a list of underly-
ing issues. Current Bioinformatics, 5(4):280–289.
Lafita, A., Bliven, S., Prli
´
c, A., Guzenko, D., Rose, P. W.,
Bradley, A., Pavan, P., Myers-Turnbull, D., Valasa-
tava, Y., Heuer, M., et al. (2019). Biojava 5: A
community driven open-source bioinformatics library.
PLoS computational biology, 15(2):e1006791.
Lazar, C., Meganck, S., Taminau, J., Steenhoff, D., Coletta,
A., Molter, C., Weiss-Sol
´
ıs, D. Y., Duque, R., Bersini,
H., and Now
´
e, A. (2012). Batch effect removal meth-
ods for microarray gene expression data integration: a
survey. Briefings in bioinformatics, 14(4):469–490.
Leek, J. T., Scharpf, R. B., Bravo, H. C., Simcha, D., Lang-
mead, B., Johnson, W. E., Geman, D., Baggerly, K.,
and Irizarry, R. A. (2010). Tackling the widespread
and critical impact of batch effects in high-throughput
data. Nature Reviews Genetics, 11(10):733.
Lourenco, A. P., Leite-Moreira, A. F., Balligand, J.-L.,
Bauersachs, J., Dawson, D., de Boer, R. A., de Windt,
L. J., Falc
˜
ao-Pires, I., Fontes-Carvalho, R., Franz, S.,
et al. (2018). An integrative translational approach to
study heart failure with preserved ejection fraction: a
position paper from the working group on myocardial
function of the european society of cardiology. Euro-
pean journal of heart failure, 20(2):216–227.
Nygaard, V., Rødland, E. A., and Hovig, E. (2016). Meth-
ods that remove batch effects while retaining group
differences may lead to exaggerated confidence in
downstream analyses. Biostatistics, 17(1):29–39.
Paulus, W. J., Tsch
¨
ope, C., Sanderson, J. E., Rusconi,
C., Flachskampf, F. A., Rademakers, F. E., Marino,
P., Smiseth, O. A., De Keulenaer, G., Leite-Moreira,
A. F., et al. (2007). How to diagnose diastolic heart
failure: a consensus statement on the diagnosis of
heart failure with normal left ventricular ejection frac-
tion by the heart failure and echocardiography asso-
ciations of the european society of cardiology. Euro-
pean heart journal, 28(20):2539–2550.
Persson, H., Lonn, E., Edner, M., Baruch, L., Lang, C. C.,
Morton, J. J.,
¨
Ostergren, J., McKelvie, R. S., et al.
(2007). Diastolic dysfunction in heart failure with pre-
served systolic function: need for objective evidence:
results from the charm echocardiographic substudy–
charmes. Journal of the American College of Cardiol-
ogy, 49(6):687–694.
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi,
W., and Smyth, G. K. (2015). limma powers differ-
ential expression analyses for rna-sequencing and mi-
croarray studies. Nucleic acids research, 43(7):e47–
e47.
Savarese, G. and Lund, L. H. (2017). Global public health
burden of heart failure. Cardiac failure review, 3(1):7.
A Computational Platform for Heart Failure Cases Research
607

Shah, S. J. (2017). Precision medicine for heart failure
with preserved ejection fraction: an overview. Journal
of cardiovascular translational research, 10(3):233–
244.
Sharma, K. and Kass, D. A. (2014). Heart failure with
preserved ejection fraction: mechanisms, clinical fea-
tures, and therapies. Circulation research, 115(1):79–
96.
Silva, L. B., Trifan, A., and Oliveira, J. L. (2018). Montra:
An agile architecture for data publishing and discov-
ery. Computer methods and programs in biomedicine,
160:33–42.
Sladi
´
c, G., Gostoji
´
c, S., Milosavljevi
´
c, B., and Konjovi
´
c, Z.
(2011). Handling structured data in the alfresco sys-
tem. In Proceedings of the International Conference
on Information Society Technology and Management
(ICIST), pages 78–82.
Steinberg, B. A., Zhao, X., Heidenreich, P. A., Peterson,
E. D., Bhatt, D. L., Cannon, C. P., Hernandez, A. F.,
and Fonarow, G. C. (2012). Trends in patients hos-
pitalized with heart failure and preserved left ventric-
ular ejection fraction: prevalence, therapies, and out-
comes. Circulation, 126(1):65–75.
Suchy, C., Massen, L., Rognmo, Ø., Van Craenenbroeck,
E. M., Beckers, P., Kraigher-Krainer, E., Linke, A.,
Adams, V., Wisløff, U., Pieske, B., et al. (2014).
Optimising exercise training in prevention and treat-
ment of diastolic heart failure (optimex-clin): ratio-
nale and design of a prospective, randomised, con-
trolled trial. European journal of preventive cardiol-
ogy, 21(2 suppl):18–25.
Thompson, J. A., Tan, J., and Greene, C. S. (2016). Cross-
platform normalization of microarray and rna-seq data
for machine learning applications. PeerJ, 4:e1621.
Wang, Z., Gerstein, M., and Snyder, M. (2009). Rna-seq: a
revolutionary tool for transcriptomics. Nature reviews
genetics, 10(1):57.
Yancy, C. W., Jessup, M., Bozkurt, B., Butler, J., Casey,
D. E., Drazner, M. H., Fonarow, G. C., Geraci, S. A.,
Horwich, T., Januzzi, J. L., et al. (2013). 2013
accf/aha guideline for the management of heart fail-
ure: a report of the american college of cardiology
foundation/american heart association task force on
practice guidelines. Journal of the American College
of Cardiology, 62(16):e147–e239.
HEALTHINF 2020 - 13th International Conference on Health Informatics
608
