
Detection and Categorisation of Multilevel High-sensitivity
Cardiovascular Biomarkers from Lateral Flow Immunoassay Images via
Recurrent Neural Networks
Min Jing
1
, Donal McLaughlin
2
, David Steele
3
, Sara McNamee
1
, Brian MacNamee
4
, Patrick Cullen
1
,
Dewar Finlay
1
and James McLaughlin
1
1
Nanotechnology and Integrated BioEngineering Centre (NIBEC), Ulster University, U.K.
2
Department of Physics, University College London, U.K.
3
Biocolor Ltd, U.K.
4
School of Computer Science, University College Dublin, Republic of Ireland
Keywords:
Lateral Flow Immunoassays (LFA) Image, High-sensitivity Cardiovascular Biomarkers, Classification, Long
Short-Term Memory (LSTM), Point-of-Care (PoC).
Abstract:
Lateral Flow Immunoassays (LFA) have the potential to provide low cost, rapid and highly efficacious Point-
of-Care (PoC) diagnostic testing in resource limited settings. Traditional LFA testing is semi-quantitative
based on the calibration curve, which faces challenges in the detection of multilevel high-sensitivity biomark-
ers due its low sensitivity. This paper proposes a novel framework in which the LFA images are acquired
from a designed CMOS reader system under controlled lighting. Unlike most existing approaches based on
image intensity, the proposed system does not require detection of region of interest (ROI), instead each row
of the LFA image was considered as time series signals. The Long Short-Term Memory (LSTM) network was
deployed to classify the LFA data obtained from cardiovascular biomarker, C-Reactive Protein (CRP), at eight
concentration levels (within the range 0-5mg/L) that are aligned with clinically actionable categories. The per-
formance under different arrangements for input dimension and parameters were evaluated. The preliminary
results show that the proposed LSTM outperforms other popular classification methods, which demonstrate
the capability of the proposed system to detect high-sensitivity CRP and suggests the potential of applications
for early risk assessment of cardiovascular diseases (CVD).
1 INTRODUCTION
Cardiovascular disease (CVD) is considered as a ma-
jor threat to global health. There is a growing demand
for a range of portable, rapid and low cost biosens-
ing devices for the early detection of CVD. Lateral
Flow Immunoassays (LFA) are effective Point-of-
Care (PoC) devices that have attracted increased at-
tention recently because they can provide low cost,
rapid and highly efficacious PoC diagnostic testing
in resource limited settings. Although LFA have
found widespread applications in POC diagnostics,
the low sensitivity of LFA limits their ability to de-
tect biomarkers, such as C-Reactive Protein (CRP),
which are normally present in low concentration in
blood (Ridker, 2003). CRP is a protein that increases
in the blood with inflammation or infection as well as
following a heart attack, surgery, or trauma. Accord-
ing to the National Institute of Health and Care Ex-
cellence’s (NICE) guidelines measuring CRP quanti-
tatively over concentration levels between 10mg/L to
100 mg/L can assess the severity of bacterial infec-
tion. High-sensitivity CRP (hs-CRP) tests performed
over a lower range (from 0.5mg/L to 10 mg/L) can be
used for early risk assessment of cardiovascular dis-
eases (CVD) (Ridker et al., 2000).
Detection of multilevel high sensitivity biomark-
ers via LFA testing is a challenging task. Studies have
been carried out to improve the detection sensitivity to
allow the development of high sensitivity assays (Tor-
res and Ridker, 2003), improve the labeling strate-
gies, enhance the optical and electrochemical trans-
ducers and explore the evolution of recognition (Mak
et al., 2016). Several methods have been developed to
enhance the sensitivity of LFA, such as sample con-
centration (Moghadam et al., 2015), fluidic control
(Rivas et al., 2014), temperature–humidity technique
(Choi et al., 2016), probe-based signal enhancement
(Hu et al., 2013), enzyme-based signal amplification
(Hu et al., 2014) and electrochemical devices-based
enhancement (Cheung et al., 2015). However, these
approaches require either external equipment, high-
Jing, M., McLaughlin, D., Steele, D., McNamee, S., MacNamee, B., Cullen, P., Finlay, D. and McLaughlin, J.
Detection and Categorisation of Multilevel High-sensitivity Cardiovascular Biomarkers from Lateral Flow Immunoassay Images via Recurrent Neural Networks.
DOI: 10.5220/0009117901770183
In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2020) - Volume 2: BIOIMAGING, pages 177-183
ISBN: 978-989-758-398-8; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
177

cost reagents, or complicated fabrication with multi-
step procedure. To date, a low-cost, convenient and
equipment-free sensitivity enhancement method has
not been fully explored.
The use of smartphones has been reported for LFA
tests recently (Eltzov et al., 2015) (Quesada-Gonz
´
alez
and Merkoc¸i, 2017). However most of these applica-
tions are focused on simple binary classification based
on image intensity. In most cases, the LFA image
data are obtained from scanners or smartphone cam-
eras, in which the performance can be affected by im-
age quality due to data being acquired under ambi-
ent lights. Most smartphone-based LFA testing are
based on popular machine learning approaches such
as Support Vector Machine (SVM). Very limited stud-
ies have attempted to apply neural networks to LFA
scenarios with a different focus to this study. For ex-
ample, Deep Belief Networks (DBN) were applied in
(Zeng et al., 2016) to improve the efficiency of ROI
detection rather than classification. Multi-Layer Per-
ceptron (MLP) neural network was used in (Carrio
et al., 2015) for drugs-of-abuse detection based on av-
erage image intensity from the ROI, which assessed
saliva content but not blood based biomarkers.
Recurrent Neural Networks (RNN) are a class
of artificial neural networks that are capable of ex-
hibiting dynamic behaviour along a temporal se-
quence. Long Short-Term Memory (LSTM) networks
(Hochreiter and Schmidhuber, 1997) are a special
kind of RNN that are able to learn long-term depen-
dencies in time series data that have been success-
fully applied to speech recognition (Fern
´
andez et al.,
2007), language modelling (Jozefowicz et al., 2016)
and ECG arrhythmia detection (Picon et al., 2019)
(Xiong et al., 2018). In this study, we explore the po-
tential of applying LSTM to detect multilevel hs-CRP
by considering the LFA image data as time series sig-
nals.
Most existing approaches are based on the average
of image intensity from the LFA test line area, there-
fore the performance can be affected by the detection
of ROI. This study treats the LFA data along the sam-
ple flow direction as time series signals therefore no
ROI detection is needed. Note that the purpose of this
study is not to analyse the LFA strip at discrete or
continuous time points as the assay proceeds. Instead
the LFA images were captured at a fixed time point
(also known as an endpoint assay), following ‘com-
pletion’ of the lateral flow assay. The LFA image is a
final snapshot of the assay which contains a particular
spatial phenomenon, i.e., the leading edge is stronger
than the trailing edge, which also contains valuable
time-dependent information. Considering the LFA
as time series signals provides a novel perspective to
analyse the LFA data and helps to explore richer infor-
mation than image intensity. This study investigated
the dependence of data length, input dimension and
hidden layers in LSTM and the results demonstrate
the advantages and potential of the proposed frame-
work.
The rest of paper is organised as follows. In Sec-
tion 2 the structure of LFA is explained and an ex-
ample of LFA data is presented. In Section 3, the
proposed framework is described before the LSTM
network is explained, followed by the arrangement of
LFA data for the input sequence. The experimental
results are given in Section 4 with the conclusion and
future work presented subsequently.
2 LFA & IMAGE DATA
A schematic illustration of LFA is given in Figure
1, which shows the sample pad, sample flow direc-
tion, conjugate pad, test line (T-line), control line (C-
line) and absorbent pad. The conjugate pad, usually
colloidal gold, is labelled with antibodies specific to
the target analyte. When the sample is placed on the
sample pad, it flows by capillary action to the conju-
gate pad where the target analyte can interact (bind)
with these labelled conjugate antibodies. Conjugate
and any conjugate-sample complexes then travel lat-
erally along the strip. Upon reaching the T-line, any
formed conjugate-sample complexes are captured and
may begin to accumulate over the remainder of the
assay time. Once these reach sufficient density a vi-
sual change occurs on the T-line. The C-line captures
any particle so therefore always appears regardless of
presence of the target analyte.
Figure 1: The schematic illustration of the LFA structure.
For traditional devices like an image scanner and
smartphone cameras, the image quality can be af-
fected by lighting conditions. In this study a CMOS
reader system has been designed in which the LFA
image data were acquired from an opaque box under
controlled lighting condition. Figure 2 gives exam-
ples of LFA strip images obtained at eight hs-CRP
concentration levels using the designed CMOS reader
system. The eight levels (in mg/L) are: 0, 0.05, 0.1,
0.2, 0.5, 1, 2.5 and 5. It can be seen that each strip
contains the C-line and T-line, in which the intensity
of the T-line changes according to the concentration
BIOIMAGING 2020 - 7th International Conference on Bioimaging
178
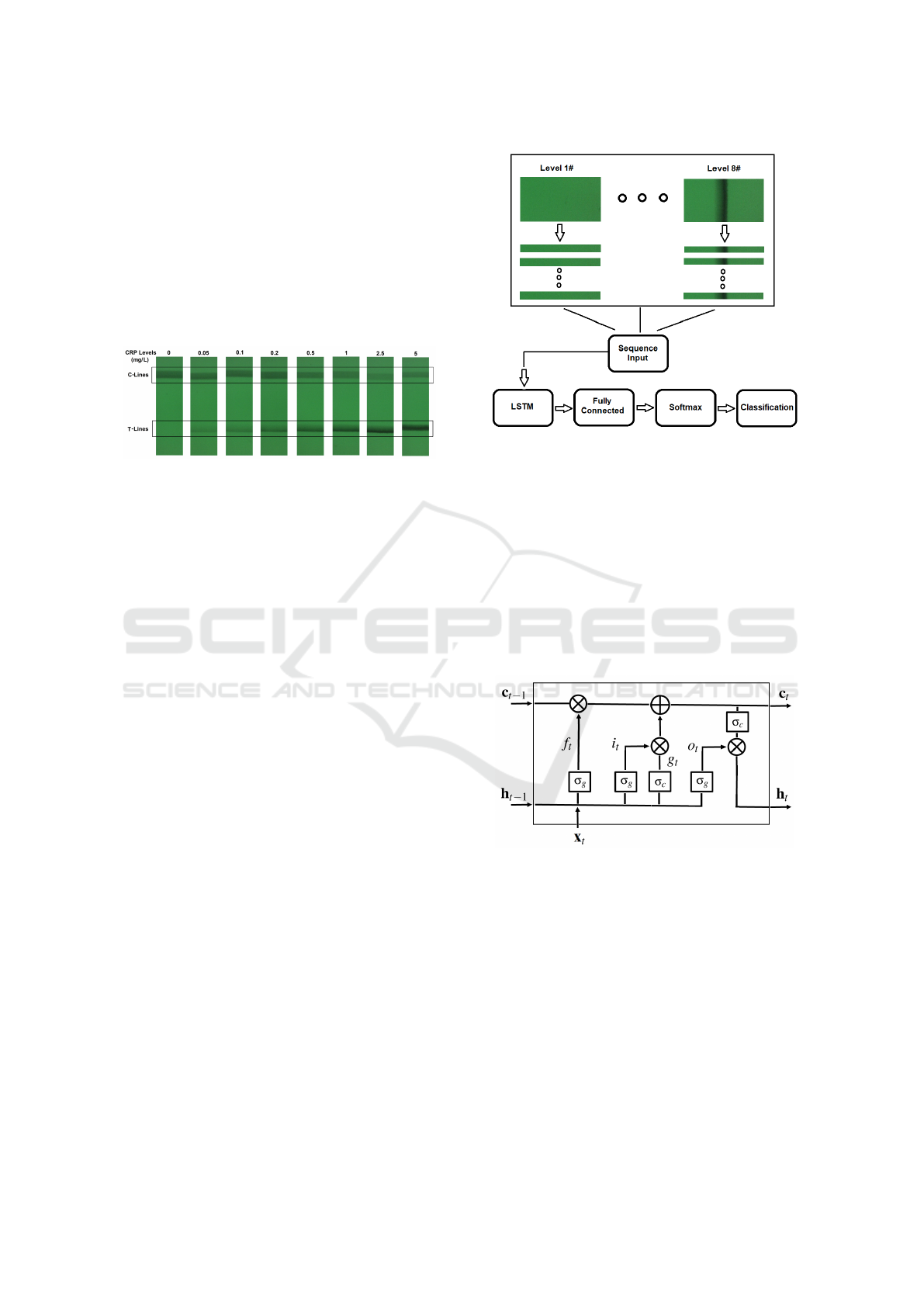
levels. It is also noticed that the position of the T-
line varies in each image. For those approaches based
on image intensity from the T-line area, the perfor-
mance is highly dependent on the accurate detection
of the T-line area. Features based on image intensity
works for simple binary classification but a more so-
phisticated approach is needed for detection of high
sensitivity biomarkers, such as hs-CRP with a con-
centration level range lower than 5mg/L as outlined
in this study.
Figure 2: Examples of LFA strip images obtained from
eight hs-CRP concentration levels.
3 PROPOSED METHODS
3.1 System Overview
The proposed framework is shown in a block diagram
in Figure 3. For the purposes of this study, only the
area containing the T-line is needed for testing be-
cause the C-line does not reflect the change of con-
centration level in the sample. In a fully integrated
analysis system the formation of the C-line would be
used as a quality control check to ensure the assay
has performed correctly. Therefore, we directly select
the half LFA image containing the T-line without fur-
ther detection of T-line area. The half LFA image is
then divided into a number of mini LFA strips, which
are then used as the input sequences for the LSTM
network. The total number of input sequences is de-
pendent on the dimension of the mini strips. Once the
data has been arranged, they are fed into an LSTM
layer followed by a fully connected layer, a softmax
layer and an output layer for sequence classification.
3.2 Long Short-Term Memory Network
LSTM is one type of RNN that can learn and remem-
ber over long sequences of input data, such as data
up to 200 to 400 time steps. Like most neural net-
works, one benefit of LSTM is that it can learn from
the raw time series data directly and therefore does
not require feature extraction. In the LSTM, the sys-
tem updates the information for current state based on
the previous state via different gates. A block diagram
Figure 3: The block diagram for the proposed framework
including arrangement of LFA data for LSTM.
of the LSTM is given in Figure 4, which includes the
cell state c
t
and the hidden state (output state) h
t
at
time t. At each time step t, the LSTM updates the
output and cell state by considering the cell state and
output at previous time step (c
t−1
, h
t−1
). Also seen
from Figure 4, there are four components to control
the system, i
t
, f
t
, g
t
and o
t
, which denote the input
gate, forget gate, cell candidate and output gate, re-
spectively.
Figure 4: Illustration of LSTM model.
Given a time series sequence X with k features
(channels) of length N, the input sequence for LSTM
at the current state t can be presented as a vector
x(t) = [x
1
(t), x
2
(t), ...,x
k
(t)]
T
where T denotes the
transpose operation. Mathematically, the formulas for
the network in forward direction can be presented as
(Hochreiter and Schmidhuber, 1997):
c
t
= f
t
K
c
t−1
+ i
t
K
g
t
(1)
where
J
denotes the Hadamard product (element-
wise multiplication of vectors).
h
t
= o
t
K
σ
c
(c
t
) (2)
Detection and Categorisation of Multilevel High-sensitivity Cardiovascular Biomarkers from Lateral Flow Immunoassay Images via
Recurrent Neural Networks
179

where σ
c
is the state activation function, which the
hyperbolic tangent function (tanh) is used. For each
gate, the network is updated by:
i
t
= σ
g
(W
i
x
t
+ R
i
h
t−1
+ b
i
) (3)
f
t
= σ
g
(W
f
x
t
+ R
f
h
t−1
+ b
f
) (4)
g
t
= σ
c
(W
g
x
t
+ R
g
h
t−1
+ b
g
) (5)
o
t
= σ
g
(W
o
x
t
+ R
o
h
t−1
+ b
o
) (6)
where the parameters W = [W
i
,W
f
,W
g
,W
o
]
T
, R =
[R
i
,R
f
,R
g
,R
o
]
T
and b = [b
i
,b
f
,b
g
,b
o
]
T
are the input
weights, recurrent weights and bias, respectively.
3.3 Data Preparation for LSTM
As mentioned earlier, only half of the LFA image con-
taining the T-line was used. The image dimension is
450 pixels × 800 pixels, in which 450 is the width
of LFA strip and 800 is the length from left to right
across the strip (along the flow direction). We con-
sider each row of images as a time series (with 800
time-steps) as they contain the information that arises
as a result of temporo-spatial interactions throughout
the assay time (via the gradual accumulation of label
conjugate particles).
The sequence input for the LSTM is made by a
number of mini LFA strip images, and each sequence
can be presented as:
X =
x
11
x
12
... x
1N
x
21
x
22
... x
2N
.
.
.
.
.
.
.
.
.
.
.
.
x
k1
x
k2
... x
kN
(7)
Here the length of time steps N is defined by the
length of the LFA strip which is 800. The dimension
k is determined by the width of the mini strip, which
may vary depending how the experiment is designed.
Therefore, the total number of input sequences is de-
pendent on the dimension (width) of the mini strips,
which is 450/k. The dependence between the dimen-
sion of mini strips and the number of sequences was
investigated by changing the width from 10 pixels to
90 pixels. The performances were evaluated and the
results are presented in Section 4.4.
4 EXPERIMENTS & RESULTS
4.1 Setup
This study contains the LFA data obtained based on
eight hs-CRP concentration levels. For each level
(class), we have 30 LFA images in total. Note the
number of images is not the number of data samples
used for the network training and testing because we
treat each row of the LFA image as one time series
signal. As described in the method section, one LFA
image contains 450 time series, so the total number
of time series for each class is 450 × 30 = 13500 and
total number of time series signals available for eight
classes is 108,000. The number of input sequences is
dependent on the dimension of the mini strips.
For all experiments, a holdout data partition was
used, in which 90% (27 images) were randomly se-
lected for training and the remaining 10% (3 images)
for testing. The accuracy was defined as: sum (Pre-
dict = Test)/(number of Test). The number of epochs,
batch size and iteration rate was empirically set to
30, 32 and 0.001 respectively. All numerical as-
pects of experimentation were conducted using MAT-
LAB2019a.
4.2 Preprocessing
To reduce the high-frequency noise, the LFA time se-
ries were first smoothed by moving average via slid-
ing window method. Different window lengths, 2,
5 and 10 were tested in the experiment described in
Section 4.3. All data were normalised by z-score to
remove the mean value and set variance to unit. An
example of normalised LFA time series from eight hs-
CRP levels is given in Figure 5, in which the moving
window length is 5 and the time steps were down sam-
pled from 800 to 200. The y-axis gives the normalised
intensity and the x-axis is the time steps. It can be ob-
served that the signal contains a ‘time element’ that
arises from the interplay between biomarker and la-
bel as it develops over time, which provides valu-
able time-dependent information. Such information
is usually ignored in the approaches based on averag-
ing the image intensity within T-line area.
4.3 Dependence with Data Length
To examine the impact of data length on the perfor-
mance, different data lengths were tested by down
sampling the original data (length of 800) to 400 and
200. The input dimension was 45 and the hidden layer
for the LSTM was 150 (based on results from Section
4.4). The performances based on different data length
and filter length were evaluated and the results are
shown in Figure 6. It appears that for filter window
length 2, time steps of 400 works better than others.
For other cases, a data length of 800 performs better
than the other two, however, it takes a much longer
time for training compared to the shorter data length.
BIOIMAGING 2020 - 7th International Conference on Bioimaging
180
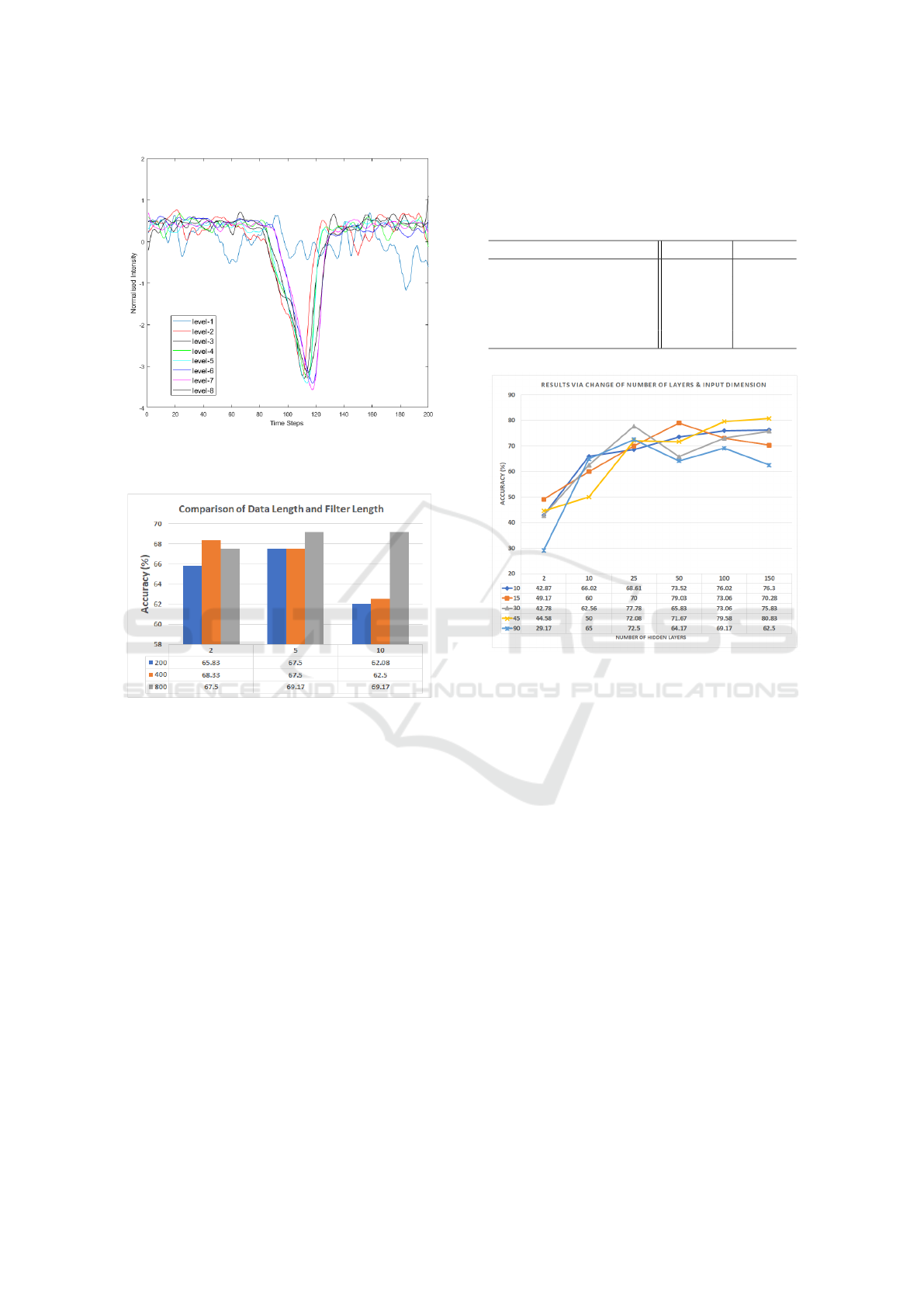
Figure 5: Examples of the normalised LFA time series data
obtained at eight hs-CRP concentration levels.
Figure 6: Comparison of results based on different data
length and filter window length.
4.4 Dependence with Input Dimension
It was found from the initial testing that the perfor-
mance can be affected by different settings for the
dimension of the mini strips and the number of hid-
den layers. Therefore, we evaluated the performance
by changing these parameters. The dimension of the
mini strip were set as 10, 15, 30, 45 and 90. The
number of input sequences for the LSTM network is
dependent on the dimension of the strip. For exam-
ple, when we treat the mini strip with a dimension 45
as one sequence, then each LFA strip image can be
considered to have 10 sequences. Therefore the total
data for training set is 10 (sequences) × 8 (classes) ×
27 (images) = 2160 and the data size for testing is 10
× 8 × 3 = 240. The number of data samples used for
training and testing under different input arrangement
are given in Table 1. To reduce training time, a data
length of 200 and filter length of 5 were used. The
performances based on alternative settings are shown
in Figure 7, from which it can be seen that the best
performance is achieved when the input dimension is
45 and number of hidden layers is 150.
Table 1: Data size based on different input dimension.
(Dimension, Sequences) Training Testing
(10, 45) 9720 1080
(15, 30) 6480 720
(30, 15) 3240 360
(45, 10) 2160 240
(90, 5) 1080 120
Figure 7: Comparison of results based on different input
dimension and number of hidden layers.
4.5 Results for Classification
The performance of the LSTM for classification was
compared to six classifiers including SVM, K Near-
est Neighbours (KNN), Linear Discriminant Analysis
(LDA), Decision Tree (DT), Naive Bayes (NB) and
Ensemble. The evaluation was based on different ar-
rangements for input dimensions as shown in Table
1. For fair comparison the same data (partition) was
used to test all algorithms. For LSTM, the data length
was 200 and filter length was 5. The number of hid-
den layers was selected based on the best performance
from the results in Section 4.4. Since the focus of this
study was to investigate the proposed method based
on LSTM, the default settings in MATLAB were used
for other classifiers, such as for Ensemble, the adap-
tive boosting was used for multiclass classification.
For KNN the number of neighbours was determined
based on testing the number of neighbours in a range
of 2-8 and the one that gave the best performance was
selected.
The comparison of performance for classification
based on different input dimension and number of
Detection and Categorisation of Multilevel High-sensitivity Cardiovascular Biomarkers from Lateral Flow Immunoassay Images via
Recurrent Neural Networks
181

sequences are promising as seen from the Table 2,
which shows that the LSTM outperforms the other
methods in all cases. An example confusion matrix
based on the LSTM (dimension 45, sequence 10) is
given in Figure 8. An example of the Receiver Oper-
ating Characteristic (ROC) curve based on the perfor-
mance from all algorithms is given in Figure 9, which
shows that the LSTM performs better than other clas-
sification algorithms. The performances for all meth-
ods can be improved, such as by fine tuning parame-
ters via cross validation, or including a third indepen-
dent dataset to chose the optimal number of neigh-
bours in KNN, which will be considered in the next
stage of this work.
Table 2: Comparison of Performance for Classification.
Accuracy (%)
Classifiers (10,45) (15,30) (30,15) (45,10) (90,5)
SVM 49.35 38.61 58.61 43.33 44.17
KNN 53.43 42.36 49.17 37.92 39.17
LDA 52.50 39.31 53.33 39.58 49.17
DT 54.07 47.92 50.28 45.00 43.33
NB 66.20 58.33 62.60 60.42 57.50
Ensemble 56.30 55.83 65.83 55.42 54.17
LSTM 76.30 73.30 73.33 78.75 76.67
5 DISCUSSION
Over the last decade, many biosensors have been de-
veloped to detect and quantify cardiac biomarkers
in medical diagnostics, in which the signals can be
electrochemical, optical, mass change (piezoelectric /
acoustic wave) or magnetic in nature (Qureshi et al.,
2012). Some of there techniques are relatively de-
manding in terms of sample preparation, costs, analy-
sis times, and skill levels. In contrast, immunoassays
are effective alternatives for rapid screening of sam-
ples, such as the enzyme-linked immunosorbent as-
say (ELISA) and LFA. Main advantages of ELISA are
the possibility to analyse multiple samples simultane-
ously, sensitivity and the relative simplicity. However,
ELISA testing is more time consuming than LFA due
to the operations like repeated incubation, washing
steps and enzyme reaction for final signal generation
(Fojt
´
ıkov
´
a et al., 2018). The outcomes from this study
are promising which show the improvement of detec-
tion of low concentration biomarkers by LFA using
proposed LSTM networks. For the future work, a
cross-validation can be considered in training to find
the optimum parameters for the networks. Techniques
based on the time series analysis or image processing
can be explored for feature extraction before apply-
ing the network, which will potentially improve the
performance.
Figure 8: Confusion matrix for classification of eight hs-
CRP concentration levels using LSTM.
Figure 9: Comparison of ROC curve for classification.
6 CONCLUSIONS
In this study, a novel method for detection of multi-
level high-sensitivity CRP biomarkers via LFA testing
using LSTM recurrent neural networks is presented.
The proposed methods were evaluated using eight hs-
CRP levels below 5mg/L which is the CRP range for
early risk assessment of CVD. The LFA strip images
were collected from a designed CMOS reader system
under controlled lighting. Each row of an LFA image
is considered as a time series which can be fed to an
LSTM model for classification. The dependence be-
tween data length, filter window length, input dimen-
sion and hidden layers were investigated. The results
show that the proposed LSTM approach achieves bet-
ter performance than other popular machine learning
algorithms although the performance can be further
improved in future work. The preliminary outcomes
BIOIMAGING 2020 - 7th International Conference on Bioimaging
182

are encouraging and suggest the potential of apply-
ing the proposed method for early risk assessment for
CVD.
ACKNOWLEDGEMENTS
This research is carried out under the project of East-
ern Corridor Medical Engineering Centre (ECME)
and funded by the European Unions INTERREG VA
Programme, managed by the Special EU Programmes
Body (SEUPB).
REFERENCES
Carrio, A., Sampedro, C., Sanchez-Lopez, J., Pimienta,
M., and Campoy, P. (2015). Automated low-cost
smartphone-based lateral flow saliva test reader for
drugs-of-abuse detection. Sensors, 15(11):29569–
29593.
Cheung, S. F., Cheng, S. K., and Kamei, D. T. (2015). based
systems for point-of-care biosensing. Journal of lab-
oratory automation, 20(4):316–333.
Choi, J. R., Hu, J., Feng, S., Abas, W. A. B. W.,
Pingguan-Murphy, B., and Xu, F. (2016). Sen-
sitive biomolecule detection in lateral flow assay
with a portable temperature–humidity control device.
Biosensors and Bioelectronics, 79:98–107.
Eltzov, E., Guttel, S., Low Yuen Kei, A., Sinawang, P. D.,
Ionescu, R. E., and Marks, R. S. (2015). Lateral flow
immunoassays–from paper strip to smartphone tech-
nology. Electroanalysis, 27(9):2116–2130.
Fern
´
andez, S., Graves, A., and Schmidhuber, J. (2007). An
application of recurrent neural networks to discrim-
inative keyword spotting. In International Confer-
ence on Artificial Neural Networks, pages 220–229.
Springer.
Fojt
´
ıkov
´
a, L.,
ˇ
Sul
´
akov
´
a, A., Bla
ˇ
zkov
´
a, M., Holubov
´
a, B.,
Kucha
ˇ
r, M., Mik
ˇ
s
´
atkov
´
a, P., Lap
ˇ
c
´
ık, O., and Fukal, L.
(2018). Lateral flow immunoassay and enzyme linked
immunosorbent assay as effective immunomethods
for the detection of synthetic cannabinoid jwh-200
based on the newly synthesized hapten. Toxicology
reports, 5:65–75.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Hu, J., Wang, L., Li, F., Han, Y. L., Lin, M., Lu, T. J., and
Xu, F. (2013). Oligonucleotide-linked gold nanoparti-
cle aggregates for enhanced sensitivity in lateral flow
assays. Lab on a Chip, 13(22):4352–4357.
Hu, J., Wang, S., Wang, L., Li, F., Pingguan-Murphy, B.,
Lu, T. J., and Xu, F. (2014). Advances in paper-based
point-of-care diagnostics. Biosensors and Bioelec-
tronics, 54:585–597.
Jozefowicz, R., Vinyals, O., Schuster, M., Shazeer, N., and
Wu, Y. (2016). Exploring the limits of language mod-
eling. arXiv preprint arXiv:1602.02410.
Mak, W. C., Beni, V., and Turner, A. P. (2016). Lateral-flow
technology: From visual to instrumental. TrAC Trends
in Analytical Chemistry, 79:297–305.
Moghadam, B. Y., Connelly, K. T., and Posner, J. D. (2015).
Two orders of magnitude improvement in detection
limit of lateral flow assays using isotachophoresis. An-
alytical chemistry, 87(2):1009–1017.
Picon, A., Irusta, U.,
´
Alvarez-Gila, A., Aramendi, E.,
Alonso-Atienza, F., Figuera, C., Ayala, U., Garrote,
E., Wik, L., Kramer-Johansen, J., et al. (2019). Mixed
convolutional and long short-term memory network
for the detection of lethal ventricular arrhythmia. PloS
one, 14(5):e0216756.
Quesada-Gonz
´
alez, D. and Merkoc¸i, A. (2017). Mobile
phone-based biosensing: An emerging ”diagnostic
and communication” technology. Biosensors and Bio-
electronics, 92:549–562.
Qureshi, A., Gurbuz, Y., and Niazi, J. H. (2012). Biosensors
for cardiac biomarkers detection: A review. Sensors
and Actuators B: Chemical, 171:62–76.
Ridker, P. M. (2003). Clinical application of c-reactive pro-
tein for cardiovascular disease detection and preven-
tion. Circulation, 107(3):363–369.
Ridker, P. M., Hennekens, C. H., Buring, J. E., and Rifai,
N. (2000). C-reactive protein and other markers of
inflammation in the prediction of cardiovascular dis-
ease in women. New England Journal of Medicine,
342(12):836–843.
Rivas, L., Medina-S
´
anchez, M., de la Escosura-Mu
˜
niz, A.,
and Merkoc¸i, A. (2014). Improving sensitivity of gold
nanoparticle-based lateral flow assays by using wax-
printed pillars as delay barriers of microfluidics. Lab
on a Chip, 14(22):4406–4414.
Torres, J. L. and Ridker, P. M. (2003). High sensitivity c-
reactive protein in clinical practice. American Heart
Hospital Journal, 1(3):207–211.
Xiong, Z., Nash, M. P., Cheng, E., Fedorov, V. V., Stiles,
M. K., and Zhao, J. (2018). Ecg signal classification
for the detection of cardiac arrhythmias using a convo-
lutional recurrent neural network. Physiological mea-
surement, 39(9):094006.
Zeng, N., Wang, Z., Zhang, H., Liu, W., and Alsaadi, F. E.
(2016). Deep belief networks for quantitative analysis
of a gold immunochromatographic strip. Cognitive
Computation, 8(4):684–692.
Detection and Categorisation of Multilevel High-sensitivity Cardiovascular Biomarkers from Lateral Flow Immunoassay Images via
Recurrent Neural Networks
183
