
Enhancing Cross-lingual Semantic Annotations using Deep Network
Sentence Embeddings
Ying-Chi Lin
a
, Phillip Hoffmann
b
and Erhard Rahm
c
Department of Computer Science, Leipzig University, Germany
Keywords:
Semantic Annotation, UMLS, Sentence Embedding, BERT, Medical Forms.
Abstract:
Annotating documents using concepts of ontologies enhances data quality and interoperability. Such semantic
annotations also facilitate the comparison of multiple studies and even cross-lingual results. The FDA therefore
requires that all submitted medical forms have to be annotated. In this work we aim at annotating medical
forms in German. These standardized forms are used in health care practice and biomedical research and are
translated/adapted to various languages. We focus on annotations that cover the whole question in the form
as required by the FDA. We need to map these non-English questions to English concepts as many of these
concepts do not exist in other languages. Due to the process of translation and adaptation, the corresponding
non-English forms deviate from the original forms syntactically. This causes the conventional string matching
methods to produce low annotation quality results. Consequently, we propose a new approach that incorporates
semantics into the mapping procedure. By utilizing sentence embeddings generated by deep networks in the
cross-lingual annotation process, we achieve a recall of 84.62%. This is an improvement of 134% compared
to conventional string matching. Likewise, we also achieve an improvement of 51% in precision and 65% in
F-measure.
1 INTRODUCTION
Semantic annotation using ontology concepts plays
an important role in data integration. The US
Food and Drug Administration (FDA) and the Clini-
cal Data Interchange Standards Consortium (CDISC)
have jointly developed a series of study data stan-
dards. Since 2016, the submissions to FDA, such
as new drug applications or biologics license appli-
cations have to comply with CDISC standards (FAD,
2017). Among them, the semantic annotation of any
submitted medical form is also compulsory. These
study data standards help the FDA to receive, pro-
cess and review submissions more efficiently. Fur-
ther, they also enable the FDA to explore many re-
search questions by combining data from multiple
studies. The submitted forms shall be annotated using
concepts in the Study Data Tabulation Model Con-
trolled Terminology (SDTM-CT). The SDTM-CT is
part of the CDISC standards and is maintained and
distributed as part of the NCI Thesaurus. This ter-
minology covers a large set of medical forms used
a
https://orcid.org/0000-0003-4921-5064
b
https://orcid.org/0000-0002-9699-1376
c
https://orcid.org/0000-0002-2665-1114
in clinical studies, for instance, the Epworth Sleepi-
ness Scale (ESS) Questionnaire and the St. George’s
Respiratory Questionnaire (SGRQ). In SDTM-CT, a
unique NCI concept is assigned to each question of
these forms. We refer to this type of annotations
as Question-as-Concept (QaC) annotations, i.e., the
whole question is mapped to corresponding concepts
in the ontology.
In this study, we are aiming at identifying
QaC-annotations of medical forms but in a cross-
lingual setting. We use the term cross-lingual se-
mantic annotation to indicate the process of anno-
tating non-English documents using English ontol-
ogy concepts. This is especially a necessity for
QaC-annotations as to the best of our knowledge,
there exists no such concepts in other languages. Not
only the submissions to the FDA but also clinical
or epidemiological studies might use such medical
forms in different languages. Annotating these forms
using the same English concepts provides a stepping
stone for cross-country comparisons. Figure 1 shows
examples of such cross-lingual semantic annotations.
To ensure that multilingual versions of such stan-
dardized forms can obtain conceptually equivalent re-
sults, it is generally necessary to apply some cul-
188
Lin, Y., Hoffmann, P. and Rahm, E.
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings.
DOI: 10.5220/0010256801880199
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 188-199
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Sheet1
Page 1
Associated UMLS concepts
Question 1 CUI Concept Name Form
OE Poor appetite or overeating C2706943 Poor appetite or overeating in last 2W.presence:^Patient:Ord:Observed PHQ-9
DE
C2706945 Poor appetite or overeating in last 2W.frequency:Patient:Ord:Observed PHQ-9
C2707461 Poor appetite or overeating in last 2W.presence:^Patient:Ord:Reported PHQ-9
GO Decreased appetite or excessive need to eat C2707462 Poor appetite or overeating in last 2W.frequency:^Patient:Ord:Reported PHQ-9
Question 2
OE Tension C3639361 Tension BPRS-A
DE Anspannung C4086709 Tension PANSS
GO Tension C3640479 Tension HAMA
Verminderter Appetit oder übermäßiges
Bedürfnis zu essen
Figure 1: Cross-lingual annotation examples of two questions of medical forms. On the left, the original English questions
(OE), their German version (DE) and their translations using Google Translate (GO) are listed. On the right, the mapped
UMLS concepts are shown. In UMLS, each concept is assigned with a CUI (Concept Unique Identifier).
tural adaptation and validation for a certain language
and/or a specific population. For instance, the Gener-
alized Anxiety Disorder-7 (GAD-7) is a GAD screen-
ing form consisting of seven questions. Since the
publishing of GAD-7 (Spitzer et al., 2006), it has
been translated and adapted/validated into Portuguese
(Sousa et al., 2015) for the European Portuguese pop-
ulation, into German for the German general popula-
tion (L
¨
owe et al., 2008), into Spanish for the people
in Spain (Garc
´
ıa-Campayo et al., 2010) and into Chi-
nese for people with epilepsy (Tong et al., 2016). The
adaptation might result in further modifications on
the translated versions that can complicate the cross-
lingual annotation process.
Our previous study (Lin et al., 2020) shows that
using conventional string matching approaches to find
QaC-annotations for non-English medical forms is a
challenging task. The unsatisfactory matching results
can source from two factors. Firstly, due to the trans-
lation from English to target language and the cultural
adaptation, the English and non-English versions can
vary in formulation and wording. Secondly, to be able
to annotate the German forms using concepts in En-
glish, we apply machine translators to translate these
forms into English. This translation also increases
the deviation from the original questions. As a re-
sult, the translated English questions were not identi-
cal but rather paraphrases of their original questions.
This can lead to low annotation quality when using
string matching methods such as N-grams, TF/IDF or
LCS (longest common substring). One intuitive solu-
tion towards this problem is to use semantic match-
ing instead of conventional string matching as either
cultural adaptation of the forms or the translation be-
tween different language versions shall ideally still re-
tain the semantics of the original question.
In this study, we propose the use of deep network
language models to encode the questions in the med-
ical forms and use these sentence encodings to gen-
erate semantic annotations. The deep network ex-
tracts the semantic features of the input sequence and
embeds it into a vector, a so-called sentence embed-
ding. The current state-of-the-art deep network in nat-
ural language processing (NLP) is the language model
BERT (Bidirectional Encoder Representations from
Transformers, Devlin et al., 2018). Using the con-
textual embeddings generated by BERT and its vari-
ants such as RoBERTa (Liu et al., 2019) and ALBERT
(Lan et al., 2019), many models have achieved best
results in NLP tasks (examples see GLUE
1
and Su-
perGLUE
2
benchmarks). This includes the Seman-
tic Textual Similarity benchmark (STSb, Cer et al.,
2017) task, the most related task to this study. The
goal of STSb is to assign scores for a pair of sen-
tences based on their similarity. The training and
fine-tuning of such models is computationally expen-
sive and requires large dataset. For instance, Con-
neau et al. (2019) suggests that a few hundred MiB of
text data is usually a minimum for learning a BERT
model. Since our dataset size is limited, we selected
existing pretrained and already fine-tuned models to
apply to our task. This study has the following main
contributions:
1) We manually build a parallel corpus in both
English and German of medical forms.
2) We manually build a Gold Standard Corpus
(GSC) to enable annotation quality evaluation.
3) We propose two workflows using deep network
sentence embedding generation models for cross-
lingual annotation.
4) We analyze the main factors in the proposed
workflows and give recommendations on best
practice.
5) We further improve the annotation quality by
combining results using intersect and union.
1
General Language Understanding Evaluation https://
gluebenchmark.com
2
SuperGLUE https://super.gluebenchmark.com/
leaderboard
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
189

2 BACKGROUND AND
RELATED WORK
Biomedical Cross-lingual Semantic Annotation.
The Conference and Labs of the Evaluation Forum
(CLEF) has hosted cross-lingual annotation chal-
lenges on biomedical named entities in 2013, 2015
and 2016. The CLEF-ER 2013 evaluation lab
(Rebholz-Schuhmann et al., 2013) resulted in two
multilingual gold standard corpora (GSC): the Mantra
GSC (Kors et al., 2015) containing 5530 annotations
in five languages and the QUAERO corpus (N
´
ev
´
eol
et al., 2014), a larger GSC with 26,281 annotations in
French. However, since these corpora do not contain
QaC-annotations, we have to build our own GSC in
this study. The QUAERO corpus was used in the fol-
lowing two cross-lingual annotation challenges: the
CLEF eHealth 2015 Task 1b (N
´
ev
´
eol et al., 2015) and
the 2016 Task 2 (N
´
ev
´
eol et al., 2016). The winning
team in 2015 (Afzal et al., 2015) uses the intersec-
tion of two translators to expand the UMLS terminol-
ogy into French. They apply a rule-based dictionary
lookup system to generate the annotation candidates
that are further post-processed to reduce false posi-
tives. The best team in 2016 (Cabot et al., 2016) in-
corporates bag-of-words and pattern-matching to ex-
tract concepts. In 2018, Roller et al. (2018) propose a
sequential concept normalization system which uses
Solr to lookup French and English concepts sequen-
tially and use the same post-processing as in (Afzal
et al., 2015). Their system outperform the winning
teams in the previous CLEF challenges. The above
mentioned studies all focus on annotating biomedical
name entities but not questions in the medical forms.
Lin et al. (2020) propose two workflows for
cross-lingual annotation of non-English medical
forms, in their case, in German. The first workflow
annotates the German forms using all available Ger-
man ontologies in the UMLS. The second workflow
uses machine translators to translate the German
forms into English and use three conventional string
matching annotators: MetaMap (Aronson and Lang,
2010), cTAKES (Savova et al., 2010) and AnnoMap
(Christen et al., 2015) to identify annotations. Com-
pared to the second workflow, the first workflow
produced very limited amount of annotations mainly
due to the scarcity of German concepts in the UMLS
compared to the English ones. By combining the
three annotator results using union, they achieved
a recall of 68.3% of their silver standard corpus.
They also investigated the annotation quality of
QaC-annotations using AnnoMap. On annotating
original English forms, AnnoMap obtains 82.7% on
precision, 82.3% on recall and 82.5% on F-measure.
The annotation quality dropped significantly when
annotating translated forms: 49.1%, 26% and 34% on
precision, recall and F-measure, respectively. Hence,
we seek to improve these results in this current study
by incorporating deep networks such as BERT and
its variants. In the following, we briefly describe the
related models used in this study.
BERT and Its Variants. BERT is composed of
multi-layer bidirectional Transformer encoders based
on the Transformer implementation in Vaswani et al.
(2017). It is pretrained using two methods: 1) masked
language model (MLM) objective and 2) next sen-
tence prediction (NSP). During MLM, some percent-
age of the input tokens are masked at random and
the model learns to predict those masked tokens. The
NSP task helps BERT to understand the relationship
between sentences such as in Question Answering
(QA) and Natural Language Inference (NLI) tasks.
The corpora used for pretraining are the BooksCor-
pus (800M words) (Zhu et al., 2015) and the English
Wikipedia (2,500M words). BERT was released as
two sizes: the BERT
base
consists of 12 Transformer
layers and the BERT
large
has 24 layers.
The authors of RoBERTa (Robustly optimized
BERT approach, Liu et al., 2019) propose an al-
tered pretraining on BERT resulting in a better per-
forming model. The new approach comprises mod-
ifications such as using dynamic masking, remov-
ing the NSP objective and using larger mini-batch
size. However, the largest gain in performance of
RoBERTa is due to an extensive training data expan-
sion (use 160GB instead of 13GB) and training for
more steps. RoBERTa achieves a Pearson-Spearman
correlation of 92.4 on STSb, an improvement of 2.4
over BERT
large
. Based on BERT, RoBERTa has also
two model sizes: RoBERTa
base
and RoBERTa
large
.
Opposite to RoBERTa that aims to gain better
performance through more pretraining, DistilBERT
(Sanh et al., 2019) focuses on producing a light-
weighted version of BERT without losing too much
performance. As in RoBERTa, DistilBERT also uses
dynamic masking, a large batch size and removes the
NSP task during the pretraining. The key feature of
DistilBERT is a compression technique, the so-called
knowledge distillation (Bucilu
ˇ
a et al., 2006; Hinton
et al., 2015), where a compact model - the student
- is trained to reproduce the behavior of a more com-
plex model - the teacher. The DistilBERT model com-
prises only 6 Transformer layers and 40% fewer pa-
rameters but is 60% faster and still retains roughly
97% of BERT’s performance on the GLUE bench-
mark.
HEALTHINF 2021 - 14th International Conference on Health Informatics
190

Sentence-BERT (SBERT). In this study we mainly
utilize the pretrained SBERT models (Reimers
and Gurevych, 2019). SBERT was developed to
overcome the inefficiency of BERT for finding the
most similar pair of sentences in a large dataset.
Using BERT for the comparison, each pair has to be
input into the network separately and consequently
a comparison of 10,000 sentences takes 50 million
inference computations (∼65 hours). This is too
expensive in terms of time and resources in our case
as the ontologies we use contain over one million
entries. SBERT uses different above-mentioned
BERT variants as backbone and adds a pooling
operation to generate a fixed sized sentence em-
bedding. The authors applied siamese and triplet
networks to fine-tune the models to generate better
sentence embeddings for similarity comparison. The
pretrained models of SBERT are fine-tuned either
on the NLI datasets
3
with classification objective
alone or additionally also on the STSb dataset with
regression objective. The testing result on STSb
(Spearman rank correlation coefficient = 79.23 using
BERT
large
trained on NLI) outperforms the average
BERT embeddings, InferSent (Conneau et al., 2017)
and the Universal Sentence Encoder (Cer et al., 2018).
SBERT-WK. The SBERT-WK
4
(Wang and Kuo,
2020) aims to refine the sentence embeddings gener-
ated by SBERT. The word embeddings generated by
SBERT are further modified based on how informa-
tive / important the word is. The word importance is
derived from its neighboring words of the same layer
and its cosine similarity changes through layers. The
idea is if a word aligns well with its neighboring
word vectors, it is less informative. Similarity, if a
word evolves faster across layers (larger variance of
the pair-wise cosine similarity), it is more important.
Since it works on already generated embeddings, no
further training is needed. Adding SBERT-WK can
improve the SBERT results on STSb for approxi-
mately 5 scores on the Spearman rank correlation
coefficient.
Multilingual Language Models. In addition to the
English sentence encoders mentioned above, we also
applied multilingual language models to generate sen-
tence embeddings. Multilingual alignment of sen-
tence embeddings aims to achieve that the different
embeddings of different languages of a sentence shall
be mapped to the same vector space. This is the basic
3
Containing the Stanford Natural Language Inference
dataset (Bowman et al., 2015) and the Multi-Genre NLI
dataset (Williams et al., 2017)
4
WK stands for the initials of the two authors
hypothesis when using similarity or distances mea-
sures of the embeddings to estimate the closeness of
the multilingual sentences.
The multilingual Universal Sentence Encoder
(mUSE) is such a multilingual language model (Yang
et al., 2019). It is pretrained on 16 different lan-
guages simultaneously. The term ”Multi-task Dual-
Encoder Model” denotes mUSE’s framework, com-
prising a single encoder handling a variety of down-
stream task. The encoder consists of either Trans-
formers, guaranteeing a better accuracy, or of Con-
volutional Neural Networks (CNN) that ensure an ef-
ficient computation. The mBERT
5
, the multilingual
version of BERT, is based on BERT
base
model and
is trained on 104 languages using Wikipedia. Since
mBERT was not trained on parallel corpus, the sen-
tence embeddings do not align well.
Lample and Conneau (2019) introduced the cross-
lingual language models (XLMs) and utilise two
methods to improve multilingual alignment. First,
they use so-called translation language modeling
(TLM) to learn XLM. Instead of using sentences of
same language as in BERT MLM, TLM uses two
parallel sentences of different languages as input se-
quence. The second method is using Byte Pair En-
coding (BPE, Sennrich et al., 2015) on the same
shared vocabulary instead of word or characters as in-
put. Conneau et al. (2019) introduced a further cross-
lingual pretrained model called XLM-R. R stands for
RoBERTa as it is trained similar as RoBERTa. XLM-
R uses only MLM as training objective as RoBERTa
(i.e. no TLM) but with multilingual corpus. It is
trained on even larger dataset (the CommonCrawl
corpus (Wenzek et al., 2019), 2.5 TB) and in 100 lan-
guages. XLM-R sets a new stat of the art on XNLI of
an 83.6% average accuracy.
In this study we used pretrained multilingual lan-
guage models developed by the authors of SBERT.
They proposed a new approach called multilingual
knowledge distillation that aims to reinforce better
multilingual alignment of the generated embeddings
(Reimers and Gurevych, 2020). The student model
ˆ
M, generally (but not restricted to) a multilingual
pretrained model, learns the behavior of the teacher
model M, generally an intensively trained mono-
lingual (English) model, so that the sentence em-
beddings of different languages shall be mapped to
the same vector space. The training requires a set
of parallel (translated) sentences ((s
1
, t
1
), ··· , (s
n
, t
n
))
where t
i
is the translation of s
i
. The objective is
to minimize the mean squared loss so that
ˆ
M(s
i
) ≈
M(s
i
) and
ˆ
M(t
i
) ≈ M(s
i
). The training dataset in-
5
Multilingual BERT https://github.com/
google-research/bert/blob/master/multilingual.md
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
191

(a) Workflow-Multi
Output
Annotation
mappings
𝒜ℳ
!
!
,#$%&
…
𝒜ℳ
!
"
,#$%&
Set of German
questions
𝑞
1
… 𝑞
𝑛
Multilingual
Sentence
Embedding
Generation
UMLS English
concepts 𝑐
1
… 𝑐
𝑚
Cosine Similarity
𝑆𝑖𝑚(𝑞
𝑖
, 𝑐
𝑗
)
𝑞
𝑖
∈ 𝑞
1
… 𝑞
𝑛
𝑐
𝑗
∈ (𝑐
1
… 𝑐
𝑚
)
ℳ𝑎𝑥
(b) Workflow-Eng
Output
English
Sentence
Embedding
Generation
UMLS English
concepts 𝑐
1
… 𝑐
𝑚
Cosine
Similarity
𝑆𝑖𝑚(𝑞
𝑖
, 𝑐
𝑗
)
ℳ𝑎𝑥
Set of
German
questions
𝑞
1
… 𝑞
𝑛
–DeepL
–Microsoft
–Google
Machine
translator
Set of
English
questions
𝑞
1
… 𝑞
𝑛
Annotation
mappings
𝒜ℳ
!
!
,#$%&
…
𝒜ℳ
!
"
,#$%&
SBERT-
WK
Figure 2: Two workflows to generate cross-lingual annotations using sentence embeddings.
cludes bilingual dictionaries and also several parallel
corpora from the OPUS website (Tiedemann, 2012)
such as translated subtitles of TED talks and paral-
lel sentences extracted from the European Parliament
website. The experiments on multilingual STS 2017
dataset (Cer et al., 2017) shows these multilingual
SBERT models outperform mBERT and XLM-R sig-
nificantly.
3 METHODS
3.1 Corpus and Ontology
We selected 21 German medical forms that contain
in total 497 questions as our corpus. Since we need
to build a Gold Standard Corpus (GSC) for evalua-
tion, the selection criteria of our corpus include 1) the
forms must have corresponding English forms and 2)
the questions in the English forms must have corre-
sponding QaC-annotations in the UMLS. Many of the
forms included in our GSC were used in the LIFE
6
-
Adult-Study (Loeffler et al., 2015). The study inves-
tigates prevalences, early onset markers, genetic pre-
dispositions, and the role of lifestyle factors of major
civilization diseases, such as metabolic and vascular
diseases, heart function, depression and allergies.
We choose the Unified Medical Language System
(UMLS) Metathesaurus as our ontology source.
The UMLS integrates a large set of biomedical
ontologies so that we can maximize the semantic
6
LIFE stands for Leipzig Research Center for Civ-
ilization Diseases https://life.uni-leipzig.de/en/life health
study.html
interoperability for our corpus. We use the 2019AB
version which contains approximately 4.26 million
concepts from 211 source vocabularies. Since using
deep networks to generate sentence embeddings is
a time consuming and resource intensive task, we
conducted a selection process to reduce the number
of ontologies. Henceforth, the resulting UMLS
subset contains only 428,000 concepts (1,03 million
entries) from three source ontologies (instead of 211)
but still covers 99.1% of the GSC annotations. The
subset consists of 1) NCI Thesaurus, 2) LOINC, and
3) Consumer Health Vocabulary. As mentioned in
Section 1, the CDISC Controlled Terminology is
included as part of the NCI Thesaurus.
Gold Standard Corpus (GSC). A manually built
GSC enables us to compare the annotation quality
of different sentence embedding models. The ques-
tions of the original English forms are entered into
the UMLS UTS Metathesaurus Browser
7
and the con-
cepts having exactly the same text are considered as a
correct annotation in our GSC.
3.2 Annotation Workflows
We applied two workflows to annotate the German
medical forms: the Workflow-Multi uses multilingual
sentence encoders (Figure 2(a)) and the Workflow-
Eng integrates machine translators and English sen-
tence encoders (Figure 2(b)). For the Workflow-Multi,
we input the German questions directly into the mul-
tilingual encoders. In Workflow-Eng, the German
questions are firstly translated into English using ma-
7
UMLS Terminology Services https://uts.nlm.nih.gov/
metathesaurus.html
HEALTHINF 2021 - 14th International Conference on Health Informatics
192

Table 1: Models and configurations used in Workflow-Eng. GO: Google Translate, DL: DeepL, MS: Microsoft Translator.
Model Size Training SBERT-WK Corpus Result size
BERT large / base
NLI
NLI + STSb
with
w/o
(only on base-models)
GO
DL
MS
k ∈ {1, 2, 3, 5}
DistilBERT base
RoBERTa large / base
Table 2: The three multilingual embedding generation mod-
els used in Workflow-Multi. To simplify the description in
the text, we introduce codes in the first column to refer to
these models.
Code Teacher model Student model
M1 mUSE DistilmBERT
M2 SBERT (BERT
base
-NLI) XLM-R
M3 SBERT (BERT
base
-NLI+STSb) XLM-R
chine translators before input into English encoders.
We use three machine translators, namely DeepL
8
,
Microsoft Translator
9
and Google Translate
10
and
hence obtained three translated corpora. The optional
SBERT-WK is applied on the embeddings generated
by English encoders (i.e. in Workflow-Eng).
A preliminary experiment concludes that using
cosine similarity produced better results than using
Euclidean or Manhattan distances (data not shown).
Hence, for both workflows, the cosine similarities
are calculated between the generated sentence em-
beddings of each question and that of each UMLS
concept. The resulting mappings of each question
are ranked based on their cosine similarities. We
then retain various Top k results for evaluation where
k ∈ {1, 2, 3, 5}. For instance, a Top3 result set con-
tains mappings having the highest three cosine sim-
ilarities of that question. We use precision, recall
and F-measure to present our results. We also use
Workflow-Eng to annotate the original English corpus
as the reference comparison.
3.3 Baseline: AnnoMap
Our baseline annotator is AnnoMap (Christen et al.,
2015, 2016). AnnoMap generates annotation can-
didates based on three string similarity functions:
TF/IDF, Trigram and LCS. After candidate genera-
tion, an optional group-based selection can be ap-
plied to improve precision (Christen et al., 2015).
AnnoMap retains only candidates whose similarity
scores are above a given threshold δ. For our exper-
iment, we set two thresholds δ ∈ {0.6, 0.7} and set
the same result sizes as in approaches using deep net-
works, i.e. k ∈ {1, 2, 3, 5}. We do not use group-based
selection so that we can expand the result to meet the
8
https://www.deepl.com/translator
9
https://www.microsoft.com/en-us/translator/
10
https://translate.google.com
larger result set size setting. We annotate the three
translated corpora used in the Workflow-Eng and also
the original English corpus as reference.
3.4 Deep Network Language Models
Multilingual Embedding Generation Models.
We selected three pretrained models of Reimers
and Gurevych (2020) for the multilingual sentence
embedding generation in Workflow-Multi. These
pretrained models are obtained from the SBERT
repository
11
. Table 2 lists their corresponding teacher
and student models. The first model (M1) uses
multilingual models as teacher and student models.
The models M2 and M3 both use English models as
the teacher models (two SBERT models trained on
different pretraining dataset) and the same multilin-
gual model (XLM-R) as the student model. These
models are introduced in Section 2.
English Embedding Generation Models. Table 1
details the SBERT models we used in this study with
their further configurations for the Workflow-Eng.
There are three pooling strategies used in SBERT:
1) use the CLS token 2) max pooling and 3) mean
pooling. We choose the models using mean pool-
ing as they are the best performing models proven
by a preliminary test using our dataset. In con-
trast to BERT and RoBERTa, DistilBERT has only
the base-model. These models are fine-tuned us-
ing two labeled training datasets: 1) NLI only or
2) NLI + STSb. We utilize the implementation of
SBERT-WK in the SBERT repository on the base-
models. Since the SBERT-WK is computationally
expensive and primary results of large-models with
SBERT-WK show no significant benefit, we do not
apply SBERT-WK on large-models. We used three
translated corpora and at the end retain result sets of
4 different sizes. Consequently, we have 192 configu-
ration settings (configs): 144 configs for the base-
models
12
and 48 configs for the large-models.
11
https://public.ukp.informatik.tu-darmstadt.de/reimers/
sentence-transformers/v0.2/
12
3 models × 2 training datasets × 2 SBERT-WK-setting
× 3 corpora × 4 result sizes = 144 configs
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
193

121
293
28
27
8
6
5
1
3
2
1
2
0
100
200
300
1 2 3 4 5 6 7 8 9 11 12 13
Number of GSC annotations of a question
Frequency
Figure 3: Frequency distribution of number of annotations
of a question in the GSC.
4 EVALUATION
4.1 Gold Standard Corpus
We manually identified 1105 GSC annotations for the
497 questions. Figure 3 shows the frequency distribu-
tion of number of annotations of a question. Most
of the questions have 1 or 2 GSC annotations and
about 10% of the questions have 3 or 4 annotations.
There are only few questions having more than 5
mapped UMLS concepts. From our observations, this
is mainly due to 1) same question of a form might
be given multiple CUIs in the UMLS or 2) the same
question occurs in different forms and hence has dif-
ferent CUIs. Figure 1 shows such examples.
4.2 Cross-lingual Annotation
Baseline. When annotating the original English
corpus, our baseline, AnnoMap, achieves the best
precision of 93.61%, the best recall of 86.52% and
the best F-measure of 80.08% using different settings
(Table 3). The best results on the translated corpora,
however, drop significantly. The best precision on
the Google Translate corpus decreases to 61.69%
and best recall is rather low (36.20%). Overall, the
best F-measure AnnoMap achieves on the translated
corpora is 38.47%. The results using the DeepL and
Microsoft Translator corpora are slightly worse than
Google Translate results. This confirms our previous
study (Lin et al., 2020), concluding that conventional
string matching methods deliver unsatisfactory
results for such a cross-lingual annotation problem.
Workflow-multi. In this part we compare the an-
notation quality of the three multilingual models in
Workflow-Multi. Table 4 shows the mean precision,
recall and F-measure, averaged over different result
sizes. The best performing model is M1. It also
obtained the best precision (57.14% with Top1), best
recall (61% with Top5) and best F-measure (48.69%
with Top2). The sole difference between M2 and
M3 is that the teacher models are fine-tuned on dif-
Table 3: The best precisions, recalls and F-measures ob-
tained by AnnoMap with thresholds δ ∈ {0.6, 0.7} and re-
sult sizes k ∈ {1, 2, 3, 5}. The last row of each metric in grey
is the best corresponding result obtained using the original
English corpus (OE). GO: Google Translate.
Corpus δ Result size Precision Recall F-measure
Precision
GO 0.7 Top1 61.69 13.85 22.62
OE 0.7 Top1 93.61 41.09 57.11
Recall
GO 0.6 Top5 35.65 36.20 35.92
OE 0.6 Top5 51.43 86.52 64.51
F-measure
GO 0.6 Top2 53.19 30.14 38.47
OE 0.7 Top2 89.31 72.58 80.08
Table 4: Averaged annotation quality using multilingual en-
coders. For the model codes refer to Table 2.
Model code M
Precision
M
Recall
M
F−measure
M1 43.66 46.38 41.61
M3 39.48 42.02 37.63
M2 38.90 41.06 36.96
ferent datasets (NLI and NLI + STSb, respectively).
However, this does not affect much on the annotation
quality as the results from M2 and M3 are very
similar.
Workflow-eng. We took the means of the precision,
recall and F-measure values across different configs
to see how various factors such as model type or cor-
pus influence annotation quality. Tables 5 and 6 show
the results. Firstly, we compare the annotation qual-
ity produced by the three models BERT, RoBERTa
and DistilBERT. Since DistilBERT has only base-
model, we excluded the results of the large-models
of BERT and RoBERTa from comparison. The aver-
aged performance of BERT and RoBERTa are almost
identical with BERT producing slightly better results
(Table 5(a)). Considering that DistilBERT is signif-
icantly smaller in size, the annotation quality is only
marginally worse than the other two models. In terms
of fine-tuning data used to train the SBERT models,
the configs fine-tuned on both NLI and STSB per-
form slightly better than the models solely tuned on
NLI (Table 5(b)). This result consents to the findings
of the original paper (Reimers and Gurevych, 2019).
Among the corpora generated by three machine trans-
lators, using Google Translate results in the best per-
formance with a 1% lead over DeepL in all metrics
and outperforms Microsoft Translator by more than
2.5% (Table 5(c)). Table 5(d) confirms that the best
precision is achieved when taking only Top1 results,
while, as expected, the Top5 result sets obtain the best
recalls. However, the highest F-measure scores are
HEALTHINF 2021 - 14th International Conference on Health Informatics
194

generated by the Top2 result sets. This is conceivable
as most of the questions have two or less GSC anno-
tations (see Figure 3).
Table 5: Averaged annotation quality based on different
factors. n denotes the number of configs used for aver-
aging. Note that large-models are excluded from Model
comparison as DistilBERT has only base-models. GO:
Google Translate, DL: DeepL, MS: Microsoft Translator.
M
Precision
M
Recall
M
F−measure
(a) Model (n = 48)
BERT 48.04 50.75 45.68
RoBERTa 47.64 50.53 45.39
DistilBERT 46.41 49.09 44.15
(b) Training data (n = 96)
NLI + STSb 47.83 50.60 45.51
NLI 47.34 50.10 45.06
(c) Corpus (n = 64)
GO 48.78 51.72 46.47
DL 47.68 50.40 45.36
MS 46.28 48.93 44.03
(d) Result size (n = 48)
Top1 62.36 28.05 38.69
Top2 56.50 50.83 53.52
Top3 42.44 57.26 48.75
Top5 29.02 65.26 40.18
Large-models generate better results but not al-
ways. Among the 48 comparisons between configs,
in 12 cases the large-models do not perform better
than base-models. The refinement of SBERT-WK
on the SBERT-generated embeddings improves the
annotation quality on base-models and even exceeds
the large-models (Table 6). All 72 comparisons
between configs of base-models show that the
application of SBERT-WK yields better results than
without using SBERT-WK. The largest improvement
of using SBERT-WK on base-models is 6.84% in
precision, 6.42% in recall and 5.62% in F-measure.
Table 6: Averaged annotation quality of configs using dif-
ferent SBERT-WK settings and model sizes. The results
of DistilBERT are excluded because it does not have large-
models. Hence, the results of each row are averaged over
n = 48 configs.
SBERT-WK Size M
Precision
M
Recall
M
F−measure
with base 49.36 52.21 46.97
w/o large 48.23 51.04 45.91
w/o base 46.33 49.06 44.11
Best Performing configs. Table 7 presents the
best performing configs in precision, recall and
F-measure using Workflow-Eng. The best perform-
ing configs using the original English corpus
(listed as the last column of each metric) indicates
the best achievable results. The best performing
configs in each metric all applied SBERT-WK and
Google Translate in their workflows. The highest
precision, 66.6%, is attained by DistilBERT
base
trained on NLI + STSb. The best three recalls are
generated by RoBERTa. The best recall achieved
is 70.86% by RoBERTa
base
. Interestingly, the best
F-measure (57.17%) is yield by RoBERTa
base
which
is only trained on the NLI corpus. As the table shows,
the differences in performance between models
trained on NLI + STSb or NLI only are marginal.
Notably, the best DistilBERT model (trained on
NLI + STSb and with SBERT-WK) accomplished the
best precision, rank 6 in recall (c.a. 2% less than the
best recall) and rank 3 in F-measure (0.38% less).
Computation Efficiency. Table 8 shows the com-
puting time per embedding for different model
settings. While BERT and RoBERTa have similar
efficiency, DistilBERT is almost 2 times faster.
Adding SBERT-WK on the base models of BERT
and RoBERTa prolongs the embedding generation
by a factor of 2.6. In our case with a dataset of
more than 1,000,000 concepts, the total run time
increases from approximately 4 hours to 10.5 hours.
Interestingly, the time increase of adding SBERT-WK
on DistilBERT is ”only” doubled. This is probably
due to DistilBERT’s architecture, as it only has
half the amount of Transformer layers as BERT or
RoBERTa, resulting in reduced computation steps
of SBERT-WK. Using the large-models requires
on average another 17ms per embedding than using
base-models with SBERT-WK.
Combination of Results. In our previous studies
(Lin et al., 2017, 2020) we showed that combining
using union or intersect on the result sets generated
by different annotation tools and corpora can further
improve annotation quality. Therefore, we apply two
combinations in this study. In Combi-1, we combine
the results generated from SBERT with those from
AnnoMap. The combinations are carried out on re-
sults having same result size and same corpora. With
Combi-2, we combine the results of SBERT configs
that are generated using all three different translated
corpora. With respect to Combi-1, Table 9 shows
that the best precision of combined result sets reaches
85.63%. This is a significant improvement of the
precision of the best AnnoMap and SBERT results
(61.69% and 66.6%, respectively). Using Combi-2
we accomplish a precision of 93.27%, a further im-
provement of 7.64%. The best recall generated by
Combi-1 is 73.94%, that is an increase of 37.64%
on AnnoMap (36.20%) and merely 3.08% on SBERT
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
195

Table 7: The configs of the best precisions, recalls and F-measures. The last row of each metric in grey is the best cor-
responding result obtained using the original English corpus (OE). Equally performing configs are either presented in the
same row or with the same ranking number. For example, with respect to precision, three configs rank third, two with the
same model setting (RoBERTa) but different corpora (presented as the same row) and one config (BERT) is noted as rank 3.
GO: Google Translate, DL: DeepL, MS: Microsoft Translator.
Ranking Model Size Training SBERT-WK Corpus Result size Precision Recall F-measure
Precision
1 DistilBERT base NLI + STSb with GO Top1 66.60 29.95 41.32
2 BERT large NLI + STSb w/o GO Top1 66.40 29.86 41.20
3 RoBERTa large NLI + STSb w/o GO/DL Top1 66.20 29.77 41.07
3 BERT base NLI with GO Top1 66.20 29.77 41.07
4 RoBERTa base NLI with GO Top1 66.00 29.68 40.95
5 BERT base NLI with MS Top1 65.39 29.41 40.57
BERT large NLI + STSb w/o OE Top1 96.18 43.26 59.68
Recall
1 RoBERTa base NLI + STSb with GO Top5 31.51 70.86 43.62
2 RoBERTa large NLI + STSb w/o GO Top5 31.43 70.68 43.51
3 RoBERTa base NLI with GO Top5 31.11 69.95 43.06
4 BERT base NLI with GO Top5 30.95 69.59 42.84
5 RoBERTa large NLI + STSb w/o DL Top5 30.74 69.14 42.56
6 DistilBERT base NLI + STSb with GO Top5 30.58 68.78 42.34
BERT base NLI + STSb with OE Top5 40.97 92.13 56.71
F-measure
1 RoBERTa base NLI with GO Top2 60.36 54.30 57.17
2 BERT base NLI with GO Top2 60.16 54.12 56.98
2 RoBERTa base NLI + STSb with GO Top2 60.16 54.12 56.98
3 DistilBERT base NLI + STSb with GO Top2 59.96 53.94 56.79
3 RoBERTa large NLI + STSb w/o GO/DL Top2 59.96 53.94 56.79
4 RoBERTa large NLI w/o DL Top2 59.66 53.67 56.50
5 BERT base NLI with MS Top2 59.56 53.57 56.41
RoBERTa base NLI + STSb with OE Top2 85.31 76.74 80.80
Table 8: Averaged computation time per embedding of dif-
ferent models, sizes and with or w/o SBERT-WK.
Model Size SBERT-WK Time (ms)
BERT base w/o 25.1
BERT base with 65.3
BERT large w/o 81.5
RoBERTa base w/o 23.5
RoBERTa base with 62.4
RoBERTa large w/o 80.0
DistilBERT base w/o 13.6
DistilBERT base with 27.3
(70.86%). This indicates that combining AnnoMap
and SBERT does not add many more correct annota-
tions to the SBERT result sets. Encouragingly, with
Combi-2 we achieve the best recall of 84.62%, that is
another gain of 10.68% compared to Combi-1. The
best F-measure using Combi-1 (56.96%) is slightly
lower than SBERT’s best F-measure (57.17%) but
Combi-2 achieves 63.45% by 2-vote-agreement (an
annotation is considered as correct by at least two of
the three configs).
4.3 Result Summary
In this section we summarize our findings. Con-
sidering the settings in Workflow-Eng, the differ-
ences in annotation quality between different mod-
els and training data are neglectable. However, since
DistilBERT is much more efficient, it would be a bet-
ter choice if computational resources are restricted
and/or the annotation task is time-critical. The
best performing machine translator for our dataset is
Google Translate. Instead of using the large-models,
adding SBERT-WK to the base-models can improve
the results more significantly whilst being more effi-
cient. In order to find such QaC-annotations using the
UMLS as concept source, we recommend at least to
retain the Top2 results.
Figure 4 presents the best results of all approaches
we investigated in this study. Notably, if the task
is not cross-lingual but to annotate the original En-
glish forms, the performance of the newly proposed
system and the baseline is rather similar (80.80%
using SBERT and 80.08% using AnnoMap in F-
HEALTHINF 2021 - 14th International Conference on Health Informatics
196
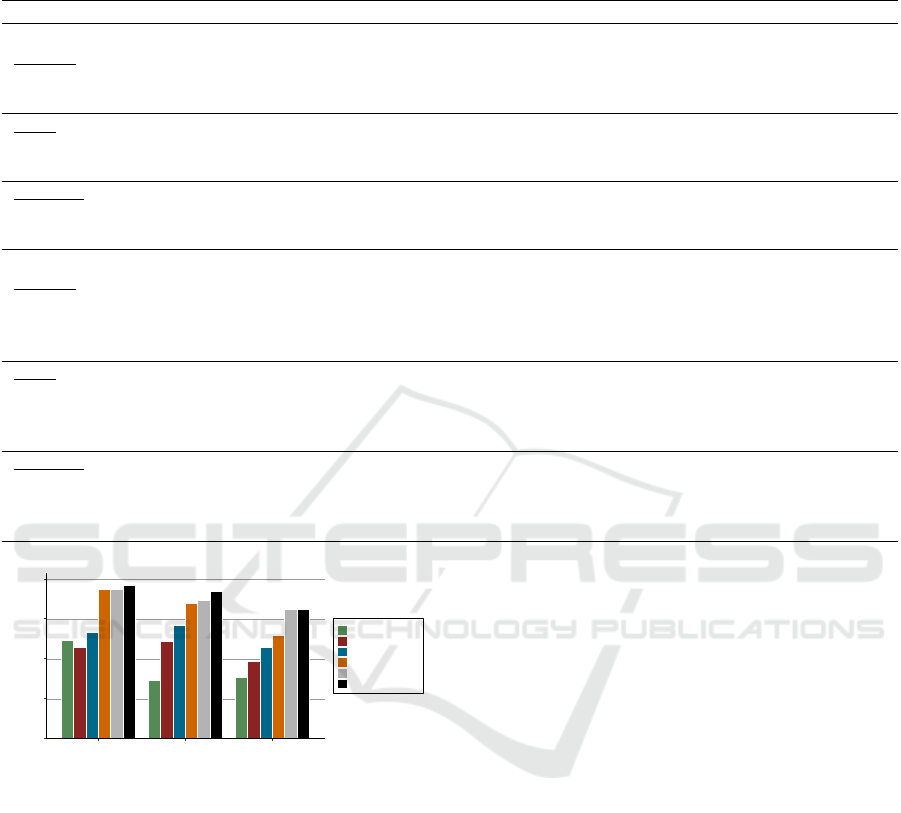
Table 9: The best combination results. The rows in gray are the best corresponding results obtained using the original English
corpus (OE). GO: Google Translate, DL: DeepL, MS: Microsoft Translator. The threshold (δ) of AnnoMap used in the
combination is indicated in the Corpus column.
Method Model Size Training SBERT-WK Corpus (δ) result size Precision Recall F-measure
SBERT + AnnoMap
Precision
intersect BERT base NLI w/o GO (0.6) Top2 85.63 25.34 39.11
intersect RoBERTa large NLI w/o OE (0.6) Top1 99.62 23.89 38.54
Recall
union RoBERTa large NLI + STSb w/o GO (0.6) Top5 27.35 73.94 39.93
union RoBERTa large NLI w/o OE (0.6) Top5 34.12 95.75 50.31
F-measure
union RoBERTa base NLI + STSb with GO (0.7) Top2 56.38 57.56 56.96
union BERT base NLI + STSb with OE (0.7) Top2 83.11 83.26 83.18
3 Corpora
Precision
intersect
BERT large NLI w/o GO
Top2 93.27 36.38 52.34
RoBERTa base NLI + STSb w/o DL
BERT base NLI with MS
Recall
union
RoBERTa base NLI + STSb with GO
Top5 20.10 84.62 32.49
RoBERTa large NLI + STSb w/o DL
RoBERTa large NLI w/o MS
F-measure
2-vote-
agreement
DistilBERT base NLI + STSb with GO
Top2 77.79 53.57 63.45
RoBERTa large NLI w/o DL
BERT base NLI with MS
0
25
50
75
100
Precision Recall F−measure
%
AnnoMap
Workflow−Multi
Workflow−Eng
Combination
AnnoMap_OE
SBERT_OE
Figure 4: The best results of each approach. GO:
Google Translate, OE: original English corpus.
measure). However, in the cross-lingual scenario,
the newly proposed deep network annotation system
exceeds conventional string matching methods sig-
nificantly in all three measures. We achieved an
improvement of 134% in recall (from 36.20% of
AnnoMap to 84.62% of combination), 51% in preci-
sion (AnnoMap: 61.69%, combination: 93.27%) and
65% improvement in F-measure (AnnoMap: 38.47%,
combination: 63.45%). Secondly, the best perform-
ing configs using Workflow-Eng outperform the best
model in Workflow-Multi. This indicates that it is
still inevitable to apply the more complex workflow
(i.e. Workflow-Eng) as long as the multilingual en-
coders do not generate better aligned sentence em-
beddings. Instead of using a single SBERT model,
our study conveys that combining SBERT result sets
of all three language corpora can further improve the
annotation quality. Hence, having different translated
versions of the questions is beneficial. Overall, if
we can apply manual verification on the Top5 results
(semi-automatic annotation) and achieve a precision
of 100%, with the best recall of 84.62%, a F-measure
of 91.67% is feasible.
5 CONCLUSIONS
In this study we propose to use deep network gener-
ated sentence embeddings to tackle the cross-lingual
annotation task. The results are very promising for
questions of medical forms. For future work, we seek
to investigate the application of such semantic an-
notation technique on other type of annotations (e.g.
biomedical name entities) or in other domains.
We used current state of the art models for the em-
bedding generation in this study. Our result shows
that the annotation quality using multilingual en-
coders still fall behind that of using English encoders.
A further improvement of the multilingual encoders
to produce better aligned sentence embeddings is ben-
eficial for our task. As this simplifies the annota-
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
197

tion procedure significantly by excluding the machine
translation.
ACKNOWLEDGEMENTS
This work is funded by the German Research Foun-
dation (DFG) (grant RA 497/22-1, ”ELISA - Evolu-
tion of Semantic Annotations”). Computations for
this work were done using resources of the Leipzig
University Computing Centre.
REFERENCES
Afzal, Z., Akhondi, S. A., van Haagen, H., van Mulligen,
E. M., and Kors, J. A. (2015). Biomedical concept
recognition in French text using automatic translation
of English terms. In CLEF (Working Notes).
Aronson, A. R. and Lang, F.-M. (2010). An overview of
MetaMap: historical perspective and recent advances.
Journal of the American Medical Informatics Associ-
ation, 17(3):229–236.
Bowman, S. R., Angeli, G., Potts, C., and Manning, C. D.
(2015). A large annotated corpus for learning natural
language inference. arXiv preprint arXiv:1508.05326.
Bucilu
ˇ
a, C., Caruana, R., and Niculescu-Mizil, A. (2006).
Model compression. In Proceedings of the 12th
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining, pages 535–541.
Cabot, C., Soualmia, L. F., Dahamna, B., and Darmoni, S. J.
(2016). SIBM at CLEF eHealth Evaluation Lab 2016:
Extracting concepts in French medical texts with ecmt
and cimind. In CLEF (Working Notes), pages 47–60.
Cer, D., Diab, M., Agirre, E., Lopez-Gazpio, I., and Spe-
cia, L. (2017). Semeval-2017 task 1: Semantic tex-
tual similarity-multilingual and cross-lingual focused
evaluation. arXiv preprint arXiv:1708.00055.
Cer, D., Yang, Y., Kong, S.-y., Hua, N., Limtiaco, N., John,
R. S., Constant, N., Guajardo-Cespedes, M., Yuan,
S., Tar, C., et al. (2018). Universal sentence encoder.
arXiv preprint arXiv:1803.11175.
Christen, V., Groß, A., and Rahm, E. (2016). A reuse-based
annotation approach for medical documents. In Inter-
national Semantic Web Conference, pages 135–150.
Springer.
Christen, V., Groß, A., Varghese, J., Dugas, M., and Rahm,
E. (2015). Annotating medical forms using UMLS. In
International Conference on Data Integration in the
Life Sciences, pages 55–69. Springer.
Conneau, A., Khandelwal, K., Goyal, N., Chaudhary, V.,
Wenzek, G., Guzm
´
an, F., Grave, E., Ott, M., Zettle-
moyer, L., and Stoyanov, V. (2019). Unsupervised
cross-lingual representation learning at scale. arXiv
preprint arXiv:1911.02116.
Conneau, A., Kiela, D., Schwenk, H., Barrault, L., and Bor-
des, A. (2017). Supervised learning of universal sen-
tence representations from natural language inference
data. arXiv preprint arXiv:1705.02364.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). BERT: Pre-training of deep bidirectional
transformers for language understanding. arXiv
preprint arXiv:1810.04805.
FAD (2017). Stady data standards: What you need to know.
https://www.fda.gov/media/98907/download.
Garc
´
ıa-Campayo, J., Zamorano, E., Ruiz, M. A., Pardo, A.,
P
´
erez-P
´
aramo, M., L
´
opez-G
´
omez, V., Freire, O., and
Rejas, J. (2010). Cultural adaptation into Spanish of
the generalized anxiety disorder-7 (GAD-7) scale as a
screening tool. Health and Quality of Life Outcomes,
8(1):8.
Hinton, G., Vinyals, O., and Dean, J. (2015). Distilling
the knowledge in a neural network. arXiv preprint
arXiv:1503.02531.
Kors, J. A., Clematide, S., Akhondi, S. A., Van Mulligen,
E. M., and Rebholz-Schuhmann, D. (2015). A mul-
tilingual gold-standard corpus for biomedical concept
recognition: the Mantra GSC. Journal of the Ameri-
can Medical Informatics Association, 22(5):948–956.
Lample, G. and Conneau, A. (2019). Cross-lingual
language model pretraining. arXiv preprint
arXiv:1901.07291.
Lan, Z., Chen, M., Goodman, S., Gimpel, K., Sharma, P.,
and Soricut, R. (2019). ALBERT: A lite BERT for
self-supervised learning of language representations.
arXiv preprint arXiv:1909.11942.
Lin, Y.-C., Christen, V., Groß, A., Cardoso, S. D., Pruski,
C., Da Silveira, M., and Rahm, E. (2017). Evaluat-
ing and improving annotation tools for medical forms.
In Proc. Data Integration in the Life Science (DILS
2017), pages 1–16. Springer.
Lin, Y.-C., Christen, V., Groß, A., Kirsten, T., Cardoso,
S. D., Pruski, C., Da Silveira, M., and Rahm, E.
(2020). Evaluating cross-lingual semantic annotation
for medical forms. In Proceedings of the 13th Inter-
national Joint Conference on Biomedical Engineering
Systems and Technologies - Volume 5 HEALTHINF,
pages 145–155.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen,
D., Levy, O., Lewis, M., Zettlemoyer, L., and Stoy-
anov, V. (2019). RoBERTa: A Robustly Opti-
mized BERT Pretraining Approach. arXiv preprint
arXiv:1907.11692.
Loeffler, M., Engel, C., Ahnert, P., Alfermann, D., Are-
lin, K., Baber, R., Beutner, F., Binder, H., Br
¨
ahler,
E., Burkhardt, R., et al. (2015). The LIFE-Adult-
Study: objectives and design of a population-based
cohort study with 10,000 deeply phenotyped adults in
Germany. BMC Public Health, 15(1):691.
L
¨
owe, B., Decker, O., M
¨
uller, S., Br
¨
ahler, E., Schellberg,
D., Herzog, W., and Herzberg, P. Y. (2008). Valida-
tion and standardization of the Generalized Anxiety
Disorder Screener (GAD-7) in the general population.
Medical Care, 46(3):266–274.
N
´
ev
´
eol, A., Cohen, K. B., Grouin, C., Hamon, T., Lavergne,
T., Kelly, L., Goeuriot, L., Rey, G., Robert, A., Tan-
nier, X., and Zweigenbaum, P. (2016). Clinical infor-
HEALTHINF 2021 - 14th International Conference on Health Informatics
198

mation extraction at the CLEF eHealth Evaluation lab
2016. CEUR Workshop Proceedings, 1609:28–42.
N
´
ev
´
eol, A., Grouin, C., Leixa, J., Rosset, S., and Zweigen-
baum, P. (2014). The QUAERO French medical cor-
pus: A ressource for medical entity recognition and
normalization. In In Proc BioTextM, Reykjavik. Cite-
seer.
N
´
ev
´
eol, A., Grouin, C., Tannier, X., Hamon, T., Kelly, L.,
Goeuriot, L., and Zweigenbaum, P. (2015). CLEF
eHealth Evaluation Lab 2015 Task 1b: clinical named
entity recognition. In CLEF (Working Notes).
Rebholz-Schuhmann, D., Clematide, S., Rinaldi, F.,
Kafkas, S., van Mulligen, E. M., Bui, C., Hellrich,
J., Lewin, I., Milward, D., Poprat, M., et al. (2013).
Entity recognition in parallel multi-lingual biomedical
corpora: The CLEF-ER laboratory overview. In Inter-
national Conference of the Cross-Language Evalua-
tion Forum for European Languages, pages 353–367.
Reimers, N. and Gurevych, I. (2019). Sentence-BERT:
Sentence embeddings using siamese BERT-networks.
arXiv preprint arXiv:1908.10084.
Reimers, N. and Gurevych, I. (2020). Making monolin-
gual sentence embeddings multilingual using knowl-
edge distillation. arXiv preprint arXiv:2004.09813.
Roller, R., Kittner, M., Weissenborn, D., and Leser, U.
(2018). Cross-lingual candidate search for biomedi-
cal concept normalization. CoRR, abs/1805.01646.
Sanh, V., Debut, L., Chaumond, J., and Wolf, T.
(2019). DistilBERT, a distilled version of BERT:
smaller, faster, cheaper and lighter. arXiv preprint
arXiv:1910.01108.
Savova, G. K., Masanz, J. J., Ogren, P. V., Zheng, J., Sohn,
S., Kipper-Schuler, K. C., and Chute, C. G. (2010).
Mayo clinical Text Analysis and Knowledge Extrac-
tion System (cTAKES): architecture, component eval-
uation and applications. Journal of the American Med-
ical Informatics Association, 17(5):507–513.
Sennrich, R., Haddow, B., and Birch, A. (2015). Neural
machine translation of rare words with subword units.
arXiv preprint arXiv:1508.07909.
Sousa, T. V., Viveiros, V., Chai, M. V., Vicente, F. L., Je-
sus, G., Carnot, M. J., Gordo, A. C., and Ferreira,
P. L. (2015). Reliability and validity of the Portuguese
version of the Generalized Anxiety Disorder (GAD-7)
scale. Health and Quality of Life Outcomes, 13(1):50.
Spitzer, R. L., Kroenke, K., Williams, J. B., and L
¨
owe, B.
(2006). A brief measure for assessing generalized
anxiety disorder: the GAD-7. Archives of internal
medicine, 166(10):1092–1097.
Tiedemann, J. (2012). Parallel data, tools and interfaces in
OPUS. In LREC, volume 2012, pages 2214–2218.
Tong, X., An, D., McGonigal, A., Park, S.-P., and Zhou,
D. (2016). Validation of the Generalized Anxi-
ety Disorder-7 (GAD-7) among Chinese people with
epilepsy. Epilepsy Research, 120:31–36.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. In Advances in
neural information processing systems, pages 5998–
6008.
Wang, B. and Kuo, C.-C. J. (2020). SBERT-WK: A sen-
tence embedding method by dissecting BERT-based
word models. arXiv preprint arXiv:2002.06652.
Wenzek, G., Lachaux, M.-A., Conneau, A., Chaudhary, V.,
Guzm
´
an, F., Joulin, A., and Grave, E. (2019). CCNet:
Extracting high quality monolingual datasets from
web crawl data. arXiv preprint arXiv:1911.00359.
Williams, A., Nangia, N., and Bowman, S. R. (2017).
A broad-coverage challenge corpus for sentence
understanding through inference. arXiv preprint
arXiv:1704.05426.
Yang, Y., Cer, D., Ahmad, A., Guo, M., Law, J., Constant,
N., Abrego, G. H., Yuan, S., Tar, C., Sung, Y.-H., et al.
(2019). Multilingual universal sentence encoder for
semantic retrieval. arXiv preprint arXiv:1907.04307.
Zhu, Y., Kiros, R., Zemel, R., Salakhutdinov, R., Urtasun,
R., Torralba, A., and Fidler, S. (2015). Aligning books
and movies: Towards story-like visual explanations by
watching movies and reading books. In Proceedings
of the IEEE international conference on computer vi-
sion, pages 19–27.
Enhancing Cross-lingual Semantic Annotations using Deep Network Sentence Embeddings
199
