
A Meta-ontology Framework for Parameter Concepts of Disease Spread
Simulation Models
Le Nguyen and Deborah Stacey
a
School of Computer Science, University of Guelph, Guelph, Ontario, Canada
Keywords:
Meta-ontology, Parameters, Animal Disease Spread, Simulation Models, Transformation, Parameters
Assessment.
Abstract:
This work reports on an ontological organization (framework) that separates domain knowledge from knowl-
edge of specific views and formalizes conceptual relationships by linking to the meta-ontology structure. We
use parameters of animal disease spread simulation models as an example, although all concepts presented
could apply to human disease spread simulation as well. A meta-ontology is created to document parameter
concepts in different comparable simulation models. It formalizes relationships between parameter concepts.
This offers several advantages such as allowing explicit domain knowledge representation and provenance,
allowing for the assessment of parameters with respect to domain knowledge, and assisting in usage and eval-
uation of the models. The meta-ontology allows views about parameter concepts to be captured. This is
important because it establishes a neutral view point which allows the assessment of parameter semantics in
respect to documented domain knowledge. While this work uses the domain of animal disease spread, the
principles of ontological representation of model parameters is applicable to a wide range of domains.
1 INTRODUCTION
Today, we live in an era of global markets. Products
and livestock are shipped from one part of the globe
to another. While globalization might have benefits to
the world economy, the world is facing greater risks
of transmission of infectious (including zoonotic) dis-
eases than ever before. It is imperative and impor-
tant that a country is well prepared to deal with these
risks. Simulation models for the spread of diseases
are popular tools to study the spread of diseases and to
evaluate the effectiveness of control strategies. Over
the last decades, several simulation models for ani-
mal disease spread have been developed and achieved
several objectives such as to mimic the outbreak of
animal diseases, to study the aspects of animal dis-
ease transmission, to develop support decision sys-
tems, to support preparedness planning, and to assess
economic impacts, etc. These agent-based simulation
models are characterized by large numbers of parame-
ters. Because of the large numbers of parameters, it is
challenging to make these models work together and
to share the knowledge of a model (as expressed in its
parameters) to others or to compare them. This often
a
https://orcid.org/0000-0002-2019-9905
contributes to high costs and is time consuming. One
of the reasons for this problem is that the semantics
of these parameters are often overlooked by the mod-
els. The semantics of these parameters are determined
by the reality that the modeller wish to emulate. This
has a great implication because emulation of the same
reality can be different based on the views of the mod-
ellers, e.g. views with different semantics, views with
different granularity, and views in different contexts,
etc. It is often done implicitly. Because of this im-
plicit representation of the parameters, there are sev-
eral disadvantages of the current models. This paper
explores the use of a meta-ontology framework to rep-
resent explicitly the semantics of agent based simula-
tion model parameters to address some shortcomings
of the current models such as knowledge representa-
tion, knowledge sharing, and assessment of domain
knowledge (as expressed in the parameters) allowing
a means to share information across simulation mod-
els and to assist usage and comparison of these mod-
els.
Nguyen, L. and Stacey, D.
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models.
DOI: 10.5220/0010299302230233
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 223-233
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
223
2 RESEARCH CONTEXT
Parameter-based simulation models are characterized
by large numbers of parameters and processes that
model the behaviours of phenomena. In this con-
text, we examine parameters of the animal disease
spread models for Foot and Mouth Disease (FMD)
such as the North American Animal Disease Spread
(NAADSM) and InterSpread Plus models. The dis-
ease spread simulation models’ parameters are used
to describe animal units, farm locations, movement
of animal units, spread mechanisms and courses of
infection. Without disease control mechanisms, a dis-
ease spread in an animal population is a result of com-
bination of the farm network, the movement of herds,
herd infectiousness (the course of an infection), and
disease spread mechanisms. Regardless of how well
a model is built, it always is an approximate version
of reality. There always exist uncertainties associated
with simulation models. There are two types of un-
certainty associated with simulation models:
• Model structural uncertainty: “the imperfect rep-
resentation of processes within a model” (MA
et al., 2013)(Kennedy and Hagan, 2001)(Arendt
et al., 2012).
• Model parameter uncertainty: “the imperfect
knowledge of the values of parameters” (bi-
ological parameters, model parameters, and
model artifacts) associated with modelling pro-
cesses (Kennedy and Hagan, 2001)(MA et al.,
2013)(Arendt et al., 2012).
For users, the ease of use and interpretation of pa-
rameters are key requirements for simulation model
builders. In order to evaluate simulation models, we
first must agree on parameters and their semantics be-
fore an evaluation can take place. They should reflect
the intended meaning with respect to the simulation
models and the related domain knowledge. It is chal-
lenging because there exist many implicit facts that
relate to simulation models. It is a result of the mod-
ellers’ views on how they wish simulation models to
be perceived. The progression of the simulation mod-
els through time (life cycle) also contributes to the
changing of parameters (and especially the changing
of their semantics) that makes them harder to use, to
maintain and to evaluate.
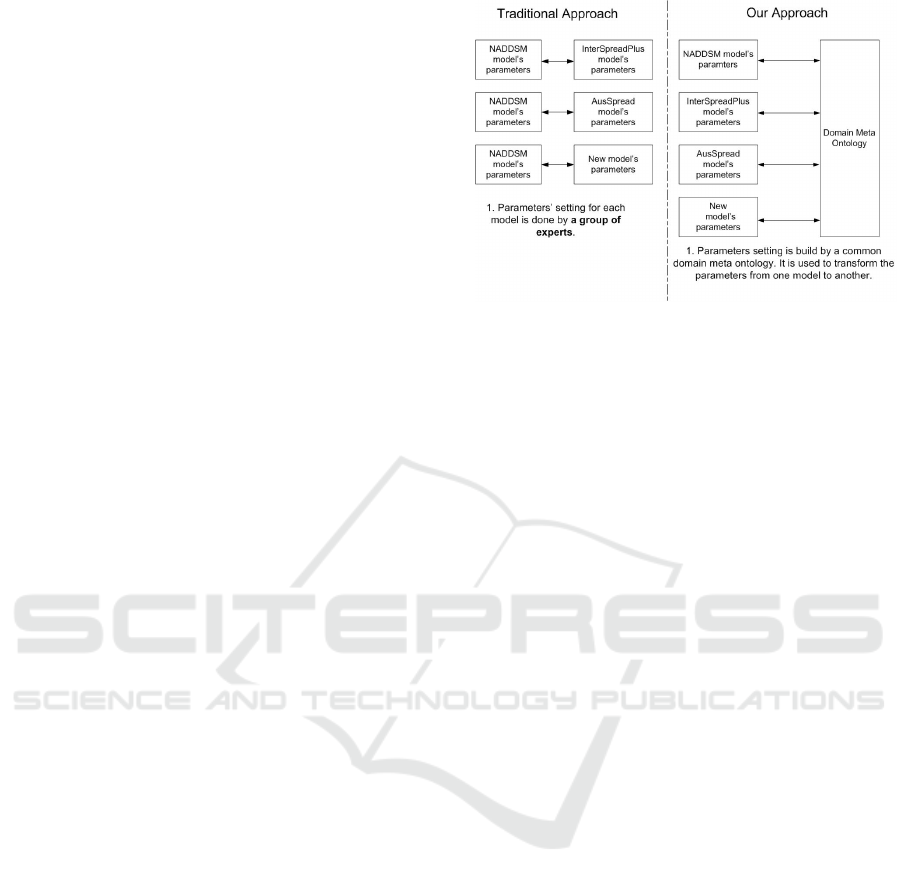
Figure 1: Traditional disease spread parameters setting and
ontology approach for comparing models.
3 OUR APPROACH
3.1 Design Scope and Restrictions
The scope of our ontology is to capture core concepts
of simulation models’ parameters and related domain
knowledge. Our study uses:
• Concepts related to the parameters of the
NAADSM, InterSpread Plus models and the re-
lated FMD domain knowledge for an FMD course
of infection, i.e., we use animal disease spread do-
main as our example. Restrictions within this do-
main include:
– For the duration of a state in the FMD infection,
the distribution is assumed to be a normal dis-
tribution and a Poisson distribution is used as
an example
– A single production type (type of animal) is
used for both models
– Parameters for farm/herd information in
NAADSM and InterSpread Plus models are
used
– The farm network is not included in this study
– Spread mechanism is not included in this study
3.2 Overview of Our System
An overview of our system is depicted in Figure 2.
There are two (2) basis components in our approach:
• Knowledge Representation
– A domain meta-ontology is used to capture
core concepts of parameters related to the FMD
course of infection as reflected in the FMD do-
main literature and simulation models. It pro-
vides vocabularies to describe parameter con-
cepts of simulation models and related FMD
HEALTHINF 2021 - 14th International Conference on Health Informatics
224

domain knowledge for an FMD course of in-
fection.
– Conceptual descriptions of model parameters.
They are descriptions of simulation model pa-
rameter settings and the related FMD domain
knowledge.
• Semantic Engine
– A semantic engine is used to examine and to
assess the parameter concepts and perform pa-
rameter transformation from one model to an-
other.
Figure 2: Our system overview.
3.3 Knowledge Representation
3.3.1 Ontology Architecture
A two-layered architecture for our ontology is pro-
posed. It is shown in Figure 3. There are two lay-
ers associated with our ontology. In the first layer,
the domain meta-ontology has concepts shared by
the simulation models and the related FMD domain
knowledge. It provides shared vocabularies to de-
scribe parameter concepts and the related FMD do-
main knowledge. The second layer describes the con-
ceptual descriptions of models’ parameter settings for
the FMD simulation models and related FMD domain
knowledge. The shared vocabularies permit parame-
ter descriptions of new models since new model con-
cepts are generally taken from the domain knowledge.
Thus, it provides a scalable way to describe the con-
ceptual descriptions of simulation models’ parame-
ters.
Figure 3: Ontology architecture.
3.4 Knowledge Acquisition
We adopted the method from Uschold and Gruninger
(Fox and Gruninger., 1995) by capturing the do-
main in natural language. Other automatic and semi-
automatic knowledge extraction techniques may be
used, however, most of these methods are fairly prim-
itive and do not work well on a large and complex do-
main. We acquired domain knowledge by examining
a number of literature works as follows:
• Foot and mouth disease papers (G., 2001)(Mar-
dones et al., 2010)(C. et al., 2016)(Sanson,
1993)(S. et al., 2003)(P. et al., 2006)(R. et al.,
2009)(K. et al., 2007)(JM. et al., 2012)(M. et al.,
2009)(van Roermund H. et al., 2010)
• NAADSM papers (Harvey et al., 2007) and re-
lated papers (Harvey and Reeves, 2012)
• InterSpread Plus papers (MA et al., 2013) and re-
lated papers (team, 2018)
We used the above literature to acquire knowledge
and case study scenarios to aid in building our on-
tology.
3.5 Ontology Specification
Our ontology specification provides the core vocabu-
laries or concepts to describe the parameter concep-
tual model of animal disease spread simulation mod-
els such as NAADSM and InterSpread Plus models,
and related FMD domain knowledge for an FMD
course of infection. There are two components of
this specification. The first component is the do-
main meta-ontology component. It provides the core
concepts to describe the parameter conceptual mod-
els. The second component is the conceptual de-
scriptions of FMD simulation models’ parameters.
These are the descriptions of the parameter models for
NAADSM, Interspread Plus, and knowledge domain.
Among these models, they share the same fundamen-
tal parameter concepts related to an FMD course of
infection. However, with respect to each concept, the
parameters of each model might be set differently.
This reflects the complexity and the differences in
semantics in choosing the parameter settings for the
models. In this section, we construct a series of ques-
tions that the domain meta-ontology must be able to
answers. These serve as a basis of our specification. It
is important to note that our competency questions are
to check whether the domain meta-ontology answers
to these questions at the terminological level.
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models
225

3.5.1 Domain Meta-ontology
Our domain meta-ontology is only about the con-
ceptual description of an FMD course of infection
(i.e. we do not consider the containment or control
of the spread). The following questions and answers
are to address core concepts that are captured in our
ontology (only a small selection are presented here).
Who are the Users of Ontology?
• The users of the ontology are FMD experts and
FMD simulation modellers.
What Does the Ontology need to Describe?
• The ontology is to describe parameters and their
semantics related to an FMD course of infection
for NAADSM and InterSpread Plus simulation
models and related FMD domain knowledge.
What is a State?
• A state is a basic unit of a state transition models.
It indicates a disease state of an animal unit. A
state has following properties:
– State name
– State order
– State duration
What is a State Duration?
• A state duration is the amount of time (usually,
days) that an animal unit is in a state.
• It can be specified by the users, e.g. modelled as a
probability density function.
A Course of FMD Infection might have following
States:
• Infected State
– Latent State
– Subclinical Infectious State
– Clinical Infectious State
– Clinical Non-infectious State
– Immune State or Naturally Immune State
– Incubation State
– Infectious State
• Noninfected State
– Susceptible State
In the domain meta-ontology, we built the core con-
cepts that are needed to describe the semantics of pa-
rameter settings of simulation models and the related
FMD domain knowledge. Our emphasis is on the
states’ concepts and other concepts related to an FMD
course of infection. We create and use the ontology
structure of the states to find and to reason about the
incubation state, infected states, non-infected states,
infectious states, and non-infectious states related to
the simulation models’ parameter settings. It can be
further developed to work with the states of many
other diseases.
3.5.2 Conceptual Descriptions of Simulation
Models’ Parameter Settings
In this section, we discuss the usage of the domain
meta-ontology for the description of FMD simulation
models’ parameters settings and highlight differences
between the simulation model concepts. First, we ex-
amined some examples of FMD courses of infection.
We show the states of an FMD course infection and
their definitions which aims to clarify the semantics
of the states of simulation models. Second, from the
examples, we construct a list of competency questions
that the ontology needs to answer. Normally, an an-
imal or an animal unit that associated with an FMD
course of infection goes through a number of states
as the disease progresses. In essence, we want to de-
scribe these concepts as reflected by a model’s param-
eter settings. To show how domain meta-ontology
can be used to describe the FMD course of infec-
tion, we examine the following cases: the descrip-
tion of an FMD course of infection for FMD domain,
NAADSM model, and InterSpread Plus model. The
concepts of parameter settings are depicted in Figure
4, 5, and 6.
Figure 4: An example of FMD domain course of infection.
It is taken and modified from (C. et al., 2016).
In Figure 4, 5, and 6, we provide the state con-
cepts, and their descriptions related to the models’
parameter settings. We note that there are differ-
ences in the description of the states and state names
as shown in the figures. For example, in NAADSM
model, clinical state means clinical infectious state
whereas in FMD domain, clinical state might have
different meaning. The figures show that there are dif-
HEALTHINF 2021 - 14th International Conference on Health Informatics
226

Figure 5: An example of NAADSM model’s FMD course
of infection. It is modified from (Harvey et al., 2007).
Figure 6: An example of InterSpread Plus FMD course of
infection.
ferent states if we compare the FMD course of infec-
tion between the NAADSM model, the InterSpread
Plus model and the FMD domain knowledge. In
the NAADSM model, states are modelled in differ-
ent granularity as compared to the InterSpread Plus
model. Given the conceptual models of the parameter
presented by the figures, we want to use our domain
meta-ontology to describe the parameters setting. We
want to answer the following questions:
• What are states that associated with an FMD sim-
ulation model parameter settings?
• What are concepts related to an FMD model pa-
rameter setting?
• What is a duration of a state?
• What types of infectious states are associated with
an FMD simulation model?
• Which models have an incubation state?
• Which models have a clinical non-infectious
state?
In section 3.6, we formally provide a discussion
on FMD state relations and descriptions of an FMD
course of infection.
3.6 Formal Knowledge Representation
In this section, we present the core part of the formal
knowledge representation of our ontology. A com-
plete ontology can be accessed via the link provided
in (Nguyen, 2020). There are two parts of the on-
tology: the terminological components, and the as-
sertion components. We present only core termino-
logical components. The assertion component can be
accessed via the previous link. We use Manchester
OWL syntax and Prot
´
eg
´
e (Musen, 2015) for our for-
mal knowledge representation.
3.6.1 Terminological Components
They are used to describe an FMD course of infection
in the FMD domain, and NAADSM and InterSpread
Plus models. In this section, we discuss core compo-
nents of our ontology. The generic components are
vocabularies that can be used to construct an FMD
course of infection. State concepts are key compo-
nents of our ontology. The domain meta-ontology
state relations are depicted in Figure 7. In this fig-
ure, it shows the relationship between the primitive
classes and defined classes.
Figure 7: Domain meta-ontology state relation.
In Figure 7, primitive classes are depicted in light
yellow ovals. The defined classes or named classes
are depicted in orange ovals. We can use the named
classes for reasoning purpose. We can use primitive
classes to define more named classes to fit the ontol-
ogy requirements.
A Description of an FMD Course of Infection.
The description of an FMD course of infection is gen-
erally defined in the ProductionType subclass. It oc-
curs here since a course of infection is specific to the
animal in which it occurs, i.e. the production type
since these models are restricted to agricultural an-
imal species. It uses concepts in the Generic class
component to describe the concepts related to an
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models
227

FMD course of infection. We will examine the FMD-
ContextSingleProductionType, NAADSMSinglePro-
ductionType, and InterSpreadPlusSingleProduction-
Type classes.
1. FMD Course of Infection: This is the FMD do-
main course of infection. It is reflected in the
definition of the FMDContextSingleProduction-
Type class. In this class, a cover axiom is used
to ensure only necessary concepts are used to de-
scribe FMDContext. The description of FMD-
ContextSingleProductionType has an object prop-
erty hasModelState. It is used to establish a re-
lation between the production type and a state.
In this class, each production type has several
states with exactly 1 LatentState, Subclinical-
InfectiousState,ClinicalInfectiousState, Clinical-
NonInfectiousState, and NaturallyImmuneState.
Class: PO:FMDContextSingleProductionType
SubClassOf:
PO:ProductionType,
PO:hasModelState only
(PO:ClinicalInfectiousState
or PO:ClinicalNonInfectiousState
or PO:LatentState
or PO:NaturallyImmuneState
or PO:SubclinicalInfectiousState),
PO:hasModelState exactly
1 PO:ClinicalInfectiousState,
PO:hasModelState exactly
1 PO:ClinicalNonInfectiousState,
PO:hasModelState exactly
1 PO:LatentState,
PO:hasModelState exactly
1 PO:NaturallyImmuneState,
PO:hasModelState exactly
1 PO:SubclinicalInfectiousState
2. An FMD course of infection in NAADSM model:
It is defined in the NAADSMSingleProduction-
Type class.
Class: PO:NAADSMSingleProductionType
SubClassOf:
PO:ProductionType,
PO:isProductionTypeOf some PO:UnitOfNAADSM,
PO:hasModelState only
(PO:ClinicalInfectiousState
or PO:LatentState
or PO:NaturallyImmuneState
or PO:SubclinicalInfectiousState
or PO:SusceptibleState),
PO:hasModelState exactly
1 PO:ClinicalInfectiousState,
PO:hasModelState exactly
1 PO:LatentState,
PO:hasModelState exactly
1 PO:NaturallyImmuneState,
PO:hasModelState exactly
1 PO:SubclinicalInfectiousState,
PO:hasModelState exactly
1 PO:SusceptibleState
3. An FMD course of infection in InterSpread Plus
model: It is defined in the InterSpreadPlusSin-
gleProductionType class.
Class: PO:InterSpreadPlusSingleProduction-
Type
SubClassOf:
PO:ProductionType,
PO:hasModelState only
(PO:IncubationState
or PO:InfectiousState
or PO:NaturallyImmuneState
or PO:SusceptibleState),
PO:hasModelState exactly
1 PO:IncubationState,
PO:hasModelState exactly
1 PO:InfectiousState,
PO:hasModelState exactly
1 PO:NaturallyImmuneState,
PO:hasModelState exactly
1 PO:SusceptibleState
Similar to the FMD course of infection for
the FMD domain knowledge, descriptions of FMD
courses of infection for NAADSM and InterSpread
Plus are formally presented. They use the same vo-
cabularies to describe the states related to the FMD
course of infection. These models are different in the
way that the FMD course of infection is defined as
discussed previously. The concepts of these models
are reflected via parameter settings aligned with the
FMD domain knowledge. Thus, we can leverage the
ontology structure to share and infer new knowledge
associated with the models.
3.7 Domain Meta-ontology Queries
There are nine competency question (CQ) queries
which are used to ask about classes associated with
animal unit states, production type, animal unit and
duration that are related to the states. They are pre-
sented in the section 3.7.1. In this table, we present
the CQs and the corresponding queries for domain
meta-ontology. The translations for the CQs are very
straight forward. Most of the time, they are self-
explanatory. In query 7, we filter other subclass
based on the super class DiscreteProbabilityDistribu-
tion since we use Poisson distribution as an example
for all duration of the states. We use the filter clause to
remove owl:Nothing from the set of answers because
owl:Nothing is a subclass of any class expression.
3.7.1 Domain Meta-ontology CQ’s and Queries
1. What all states do an FMD course of infection
have?
SELECT ?x WHERE {
?x rdfs:subClassOf po:ModelState .
FILTER(?x !=owl:Nothing)}
HEALTHINF 2021 - 14th International Conference on Health Informatics
228

2. What are subclasses of Incubation State?
SELECT ?x WHERE {
?x rdfs:subClassOf po:IncubationState .
FILTER(?x !=owl:Nothing)}
3. What are subclasses of InfectiousState?
SELECT ?x WHERE {
?x rdfs:subClassOf po:InfectiousState .
FILTER(?x !=owl:Nothing)}
4. What are subclasses of NonInfectiousState?
SELECT ?x WHERE {
?x rdfs:subClassOf po:NonInfectiousState .
FILTER(?x !=owl:Nothing)}
5. What are substates of InfectedState?
SELECT ?x WHERE {
?x rdfs:subClassOf po:InfectedState .
FILTER(?x !=owl:Nothing)}
6. What production types are captured in the ontol-
ogy?
SELECT DISTINCT ?x WHERE {
?x rdfs:subClassOf po:ProductionType .
FILTER(?x !=owl:Nothing)}
7. What durations are associated with a state?
SELECT DISTINCT ?t WHERE{
?x rdf:type po:ModelState .
?x po:hasMathematicalFunction ?y .
?y rdf:type ?t .
?t rdfs:subClassOf ?super .
?otherSub rdfs:subClassOf ?super .
?t rdfs:subClassOf ?otherSub .
FILTER (?otherSub != ?t)
FILTER (?super =
po:DiscreteProbabilityDistribution)}
8. What animal species concepts are captured?
SELECT DISTINCT ?x WHERE {
?x rdfs:subClassOf po:AnimalSpecies .
FILTER(?x !=owl:Nothing)}
9. What animal units are captured?
SELECT DISTINCT ?x WHERE {
?x rdfs:subClassOf po:Unit .
FILTER(?x !=owl:Nothing)}
3.8 Application of Meta-ontology
Queries
The queries related to the application of the meta-
ontology are to further test the meta-ontology, and its
objectives. We would like to be able to answer the
questions shown in section 3.8.1. We show queries
related to the NAADSM model. Similar queries work
with the InterSpread Plus model and the FMD domain
knowledge conceptual model. We include the com-
plete queries in the link previously provided.
3.8.1 NAADSM Application of Meta-ontology:
Competency Questions and Queries
1. What are individual states of NAADSM model?
PO:NAADSMModel(?m) ˆ PO:hasUnit(?m, ?u)
ˆ PO:hasProductionType(?u, ?pt)
ˆ PO:hasModelState(?pt, ?s)->
sqwrl:select(?s)
2. What is incubation state of NAADSM model?
PO:NAADSMModel(?m)
ˆ ParameterOntology:hasUnit(?m, ?u)
ˆ PO:hasProductionType(?u, ?pt)
ˆ PO:hasModelState(?pt, ?s)
ˆ PO:IncubationState(?s)->sqwrl:select(?s)
3. What is incubation durations means and variance
of a NAADSM model?
PO:NAADSMModel(?m1)
ˆ PO:hasUnit(?m1, ?u1)
ˆ PO:hasProductionType(?u1, ?pt1)
ˆ PO:hasModelState(?pt1, ?s1)
ˆ PO:IncubationState(?s1)
ˆ PO:hasMathematicalFunction(?s1, ?pd1)
ˆ PO:hasMeanValue(?pd1, ?mean1)
ˆ PO:hasVarianceValue(?pd1, ?variance1)
-> sqwrl:sum(?mean1)ˆsqwrl:sum(?variance1)
4. What are state concept differences between
NAADSM and Interspread Plus models?
PO:NAADSMModel(?m1)
ˆ PO:hasUnit(?m1, ?u1)
ˆ PO:hasProductionType(?u1, ?pt1)
ˆ PO:hasModelState(?pt1, ?ms1)
ˆ abox:caa(?class1, ?ms1)
. sqwrl:makeSet(?set1, ?class1)
. sqwrl:size(?size1, ?set1)
ˆ PO:InterSpreadPlusModel(?m2)
ˆ PO:hasUnit(?m2, ?u2)
ˆ PO:hasProductionType(?u2, ?pt2)
ˆ PO:hasModelState(?pt2, ?ms2)
ˆ abox:caa(?class2, ?ms2)
ˆ sqwrl:makeSet(?set2, ?class2)
ˆ sqwrl:size(?size2, ?set2)
ˆ sqwrl:difference(?set3, ?set1, ?set2)
ˆ sqwrl:size(?size3, ?set3)
ˆ sqwrl:element(?e3, ?set3)
-> sqwrl:select(?class1, ?size1,
?class2, ?size2, ?size3, ?e3)
3.9 Semantic Engine
The semantic engine is responsible for performing the
following two tasks:
• Parameter assessment
• Transformation of parameters from one model to
another
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models
229

The parameter assessment and transformation can
be performed by leveraging the meta-ontology struc-
ture. Queries can be used to extract and evaluate con-
cepts with evidence in domain knowledge and other
models. These tasks can be machine driven. How-
ever, in the complex scenario, we can use an external
framework to assist with these tasks. For example,
we can use statistical framework to analyse the statis-
tical distribution related to the duration of a state. We
have constructed a small framework to illustrate these
tasks but will concentrate here in showing how we
use queries and rules to perform required tasks. Fur-
thermore, to demonstrate these tasks, we restrict our-
selves to state concepts of an FMD course of infection
to show the parameter assessment and transformation
tasks.
3.9.1 Parameters Assessment
To perform the parameter assessment, we need to per-
form the assessment of the state concepts of the sim-
ulation models with respect to the concepts of the
FMD domain knowledge model. We use the follow-
ing steps:
• Get state concepts that are aligned between pa-
rameters of the simulation models and the FMD
domain knowledge.
• Get the instances of the state concepts. Compare
the state instances or individuals of FMD domain
knowledge to those of the simulation model.
• Do the same for other aligned concepts.
The parameter assessment allows us to know the dif-
ference in parameter settings between the parameters
of the simulation models and the FMD domain knowl-
edge. We examined two cases:
• Case 1: NAADSM parameter assessment
– Assessment of latent state
– Assessment of sub-clinical infectious state
– Assessment of clinical infectious state
– Assessment of naturally immune state
• Case 2: InterSpread Plus parameter assessment
– Assessment of incubation state
– Assessment of infectious state
– Assessment of naturally immune state.
Given the knowledge base, we want to answer the
queries that are related to the assessment of param-
eters concepts related to simulation models.
• Simulation model parameter assessment queries
– Given the asserted state concepts individuals of
a simulation model and FMD domain knowl-
edge, can we find the aligned concepts of the
two models?
– Can we infer and assess the duration of the state
concepts with respect to FMD domain knowl-
edge given the asserted individuals of the two
models?
3.9.2 Transformation of Parameters from One
Model to Another
To show how the transformation works for an FMD
course of infection, we need to discuss some concepts
in FMD domain knowledge that are used to describe
an FMD course of infection. An FMD course of infec-
tion description is based on primitive classes, defined
classes and attributes. The primitive classes are used
to construct the defined class. The defined classes,
incubation, and infectious states, are defined as:
• Incubation state is a state from infection to onset
infectiousness. It is a union of latent state and sub-
clinical infectious state.
• Infectious state is a state of infectiousness. It is a
union of sub-clinical infectious state and clinical
infectious state.
Without a disease control mechanism, an FMD
course of infection is related to the disease state con-
cepts. Because the NAADSM and InterSpread Plus
models are designed differently, the incubation, in-
fectious, and infected states are set differently. In
the NAADSM simulation model, there are explicit
states such as latent state, subclinical infectious state,
clinical infectious state and naturally immune state as
compared to the InterSpread Plus model’s states such
as susceptible and infected states (incubation state, in-
fectious state, immune state are explicit, and latent
state, subclinical infectious state, clinical infectious
state, and immune states may be implicit states). To
show how the domain meta-ontology can be used in
the parameter transformation of simulation models,
we examine how a parameter setting can be expressed
or transformed in terms of another model.
We examine the following case:
• Given a parameter set for the NAADSM model,
can we generate a semantically equivalent param-
eter set for the InterSpread Plus model or vice
versa?
In order to perform the transformation, we need to
define the transformation criteria and transformation
procedure.
Definition of Transformation Criteria.
Typically, a transformation criterion for parameters is
a number of concepts that are reflected in the simula-
tion models’ parameters. These concepts must exist
HEALTHINF 2021 - 14th International Conference on Health Informatics
230

in both source and destination simulation models for
a transformation to take place. If there are missing
concepts in the simulation models, the transformation
is not possible or might be possible with high uncer-
tainty due to the conceptual heterogeneity existing in
the simulation models.
Transformation Procedure.
There are two transformation procedures: Concepts-
based transformation procedure and missing concept
procedure.
1. Concepts-based transformation procedure:
• Get the concepts of source models.
• From the concepts, we can generate required
correspondent criteria concept parameters for
the destination simulation model’s parameters
with respect to source concepts.
2. Concepts-based transformation procedure with
missing concepts:
• Perform the concepts-based transformation for
aligned concepts of models as described above.
• With missing concepts, we can estimate or infer
missing concepts based on the concepts from
the source model’s parameters if it is possi-
ble to infer or estimate the destination settings.
With random estimation, these parameter set-
tings may have high uncertainty because of our
lack of knowledge, and it must then be left
for the users to decide if they wish to pro-
ceed with this level of uncertainty. We can use
FMD course of infection domain knowledge
that aligned with the source model to assist in
the transformation.
We examine the following cases for parameter
transformation:
Case 1: Parameter Transformation from
NAADSM Model to InterSpread Plus Model.
Let us set the criteria for the transformation as:
• Farm unit related concepts
• State concepts: Incubation state, infectious state,
immune state.
The NAADSM model can infer the incubation state,
infectious state, and naturally immune state from its
basic states.
• Incubation state can be obtained from latent state
and subclinical infectious state.
• Infectious state can be obtained from subclinical
infectious state and clinical infectious state.
• Naturally immune state can be obtained from its
state.
The incubation, infectiousness state, and naturally
immune state exist in InterSpread Plus. Thus, we can
transform from NAADSM to InterSpread Plus in this
case.
Case 2: Parameter Transformation from
InterSpread Plus to NAADSM. Using the same
criterion as in the previous case, however, the state
concepts are latent, subclinical infectious, clinical
infectious, and naturally immune states. In the In-
terSpread Plus model we can set the incubation state
and the infectiousness state of the FMD course of
infection by specifying these states in the following
forms:
• With infectiousness state, incubation state, im-
mune state.
• With user defined latent state, infectious state, in-
cubation state, immune state and implicit clinical
infectious state and subclinical infectious state.
Figure 8: Disease state transition models for NAADSM and
InterSpread Plus without control measures.
The challenge in transformation from InterSpread
Plus to NAADSM is the missing concepts (latent,
sub-clinical, and clinical states) that are required in
the NAADSM model. This is depicted in Figure 8.
We examine the following cases:
• Case 2a: With latent state, and incubation state,
infectious state and naturally immune state it is
possible to transform into NAADSM parameter
setting because we can obtain the needed concepts
to construct NAADM’s parameters:
– Subclinical infectious state can be estimated
from incubation and latent states
– Clinical infectious state state can be estimated
with subclinical state and infectious state.
– Naturally immune state can be obtained from
the immune state
• Case 2b: With only infectiousness state and in-
cubation state, a transformation is not possible
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models
231

because we cannot obtain the latent, subclinical
infectious, and clinical infectious states from in-
cubation and infectious states. Although we can
randomly generate the latent and sub-clinical du-
rations to match the incubation duration and the
sub-clinical and clinical durations to match the
infectious duration in InterSpread Plus, this will
generate high uncertainty due to lack of knowl-
edge.
In summary, the definition of criteria for parame-
ter transformation is dependent on a number of con-
cepts that are reflected in the parameters’ settings.
• Concepts must exist in both models. It is a condi-
tion for a transformation to take place.
• With missing concepts, we can perform the pa-
rameter transformation as described in a transfor-
mation procedure with uncertainty.
4 CONCLUSIONS
In this paper, we propose the use of a meta-ontology
framework to capture the semantics of parameters and
related domain knowledge associated with an FMD
course of infection in animal disease spread simula-
tion models. It permits parameter knowledge sharing,
parameter assessment and parameter transformation
between models. Our motivation for this approach
is to minimize the ambiguity that exists in parame-
ter settings and allow a standard way to describe pa-
rameter settings and the related domain knowledge.
It promotes the interoperability between simulation
models, and the ability to assess domain knowledge.
By explicitly describing parameter knowledge and es-
tablishing the linkage between parameters and doc-
umented domain knowledge, this allows us to have
an understanding of the differences between different
models’ parameters and views of the domain knowl-
edge. It strives to provide a basis for a new way to
understand and assess parameter and related views of
domain knowledge. The central piece of this work is
the focus on the meta-ontology framework construc-
tion in capturing the semantics of the parameters, the
related FMD course of infection domain concepts and
assisting in the assessment and the transformation of
parameters between models. This work reports on a
novel ontological organization that separates domain
knowledge from the knowledge about the parameters
in different comparable simulation models and for-
malizes a relationship between parameters by linking
to the domain knowledge part of the ontological struc-
ture. It allows explicit knowledge representation, a
means to compare animal disease spread simulation
models and a means to evaluate views (as expressed
in parameters) related to simulation models and do-
main knowledge. This work also acknowledges the
limitations in ontology creation. It is a time consum-
ing process that requires great effort and collaboration
of a number of experts in different domains. In gen-
eral, without experts’ assistance, parameter settings
alone are not sufficient to account for the differences
between the models’ parameters due to differences in
parameter representation of the models and their as-
sumptions. The introduction of an ontology provides
a standard means to document and describe the views
of simulation models and views of the domain knowl-
edge. These views are built from ontological concepts
that are reflected by the parameters, their semantics
and related domain knowledge. The ability to cap-
ture conceptual relations, properties and the ability
to verify the consistency of the ontology allows facts
related to parameter settings to be assessed not only
with other simulation models but also to the related
domain knowledge.
In future work, we hope to extend our ontological
concepts to other domains and to extend the number
and types of tasks that our semantic engine can per-
form including validation of requirements, compari-
son of concepts between related ontologies, and the
transformation of concepts and values between on-
tologies. We anticipate that these extensions will find
use in many domains where there is a need to compare
and reconcile competing ontologies.
REFERENCES
Arendt, P. D., Apley, D. W., and Chen, W. (2012). Quantifi-
cation of model uncertainty: Calibration, model dis-
crepancy, and identifiability. Journal of Mechanical
Design, Transactions of the ASME.
C., K. A., Patterson, G., L., V. K., E., C. M., and M., P. A.
(2016). Parameter values for epidemiological models
of foot-and-mouth disease in swine. Frontiers in Vet-
erinary Science, 3:44.
Fox, M. S. and Gruninger., M. (1995). Methodology for the
design and evaluation of ontologies. In Proc. of the
Workshop on Basic Ontological Issues in Knowledge
Sharing, 1995.
G., D. (2001). The foot and mouth disease (fmd) epidemic
in the united kingdom. Comp imm, Microb and Inf Dis
2002, 1(25):331–343.
Harvey, N. and Reeves, A. (2012). Model Description for
the North American Animal Disease Spread Model
4.0. The NAADSM Development team.
Harvey, N., Reeves, A., Schoenbaum, M., et. al. (2007).
The North American Animal Disease Spread Model:
A simulation model to assist decision making in eval-
HEALTHINF 2021 - 14th International Conference on Health Informatics
232

uating animal disease incursions. Preventive Veteri-
nary Medicine, 82(34):176–197.
JM., P., M., T., E., H., E., B., J., A., and L., R. (2012). Direct
contact transmission of three different foot-and-mouth
disease virus strains in swine demonstrates important
strain-specific differences. Veterinary Journal.
K., O., de Jong MC., A., B., JA., S., and A., D. (2007).
Foot and mouth disease virus transmission among
vaccinated pigs after exposure to virus shedding pigs.
Vacine.
Kennedy, M. C. and Hagan, A. O. (2001). Bayesian cal-
ibration of computer models. Journal of the Royal
Statistical Society.
M., Q., C., M., Z., Z., S., D., I., E., C., D., and S., A.
(2009). Influence of exposure intensity on the effi-
ciency and speed of transmission of foot-and-mouth
disease. Journal of comparative pathology.
MA, S., RL, S., MW, S., BD, O., M, S., and et al. (2013).
Interspread plus: A spatial and stochastic simulation
model of disease in animal. Prev Vet Med, 1(109):10–
24.
Mardones, F., Perez, A., Sanchez, J., Alkhamis, M., and
Carpenter, T. (2010). Parameterization of the dura-
tion of infection stages of serotype o foot-and-mouth
disease virus: an analytical review and meta-analysis
with application to simulation models. Veterinary Re-
search, 41(4).
Musen, M. A. (2015). The prot
´
eg
´
e project: a look back and
a look forward. AI Matters, 1(4):4–12.
Nguyen, L. (2020). http://doi.org/10.5683/SP2/VSHYAA.
P., E., de Koeijer A., A., B., A., S., and A., D. (2006). Quan-
tification of within- and between-pen transmission of
foot-and-mouth disease virus in pigs. Veterinary re-
search.
R., H., M., Q., NJ., S., L., M., S., A., and M., W. (2009). Ef-
fect of the initial dose of foot-and-mouth disease virus
on the early viral dynamics within pigs. Journal of
royal society interface.
S., A., M., Q., C., M., J., K., and Z., Z. (2003). Studies of
quantitative parameters of virus excretion and trans-
mission in pigs and cattle experimentally infected with
foot-and-mouth disease virus. Journal of comparative
pathology.
Sanson, R. L. (1993). The development of a decision sup-
port system for an animal disease emergency. PhD
thesis, Massey University.
team, I. P. (2018). InterSpread Plus manual. EpiSoft.
van Roermund H., P., E., de Jong M., and A., D. (2010). No
between-pen transmission of foot-and-mouth disease
virus in vaccinated pigs. Vacine.
A Meta-ontology Framework for Parameter Concepts of Disease Spread Simulation Models
233
