
On Open Workflows for Processing of Standardized
Electroencephalography Data
Roman Mou
ˇ
cek
a
and Filip Kupil
´
ık
Department of Computer Science and Engineering, Faculty of Applied Sciences, University of West Bohemia,
Univerzitnı 8, Plzen, Czech Republic
Keywords:
Deep Learning, EEG Data Standards, EEG Workflows, EEG Pipelines, Electroencephalography, Event-related
Potentials, Human Brain, Machine Learning, Reproducibility.
Abstract:
With increasing amounts of experimental data, openness, fairness, and reproducibility of scientific experi-
mental work have become important factors for researchers, journals and funding bodies. However, these
kinds of challenges are not easily and directly achievable. The goal of this paper is to contribute to these
efforts by introducing advances in building more mature lifecycle of electroencephalography/event-related
potential data. The progressive data standardization initiatives, data formats, and trends in using machine
and deep learning methods for processing of domain data are described and discussed. An open process-
ing workflow based on the analysis of current software tools for preprocessing, processing and classification
of electroencephalography/event-related potential data is proposed, implemented and verified on a publicly
available dataset.
1 INTRODUCTION
The electrical activity of the human brain has been
investigated, among other, by the methods and tech-
niques of electroencephalography (EEG) and event-
related potentials (ERPs) when brain data are col-
lected and processed to answer the questions of broad
scientific interest.
The EEG method has many advantages: afford-
ability, non-invasiveness, routine examination proto-
cols, and the opportunity to measure spontaneous ac-
tivity. However, it also has a significant disadvantage,
which is evident in scientific experiments. The result-
ing picture of brain activity (the EEG signal) is very
rough since it represents a huge number of sources of
neuronal activity. It is challenging to derive the cor-
responding neurocognitive processes from the mea-
sured brain activity.
ERPs are changes in brain activity that are time-
locked to particular events (stimuli). Generally, they
have a very small amplitude (up to tens of microvolts)
and can be assessed in small time windows (tens or
hundreds of milliseconds).
When performing EEG/ERP experiments, it is es-
sential to run a laboratory with appropriate equipment
for event (stimuli) presentation, collect EEG/ERP
a
https://orcid.org/0000-0002-4665-8946
data, analyze, and interpret them.
Experimental work, data processing and analy-
sis, interpretation, as well as subsequent publishing
of findings, have been traditional activities that dom-
inated the lifecycle of EEG/ERP data. However, dur-
ing the last decade, these activities were extended
with the requirements for the long-term effectiveness
and efficiency of experimental work. Requirements
for openness, fairness, reproducibility, and citabil-
ity of important data lifecycle artefacts have arisen
and started to be demanded by respected journals and
funding bodies.
It brought an essential change to the culture of
scientific work (not only) in this field. Open, well-
annotated, standardized, and shared EEG/ERP data
and well-described and publicly shared procedures
and workflows have slowly but continuously entered
experimental work and the entire EEG/ERP data life-
cycle. These approaches in a discipline of neu-
roinformatics started to influence traditional scientific
fields related to neural systems investigations. Neu-
roscience, cognitive sciences, or neurolinguistics are
typical examples of them.
The goal of this paper is to contribute to the open-
ness and reproducibility of the experimental work in
EEG/ERP research by making a step in defining open
workflows for processing standardized EEG/ERP
data. More specifically, the achievable goal is to in-
Mou
ˇ
cek, R. and Kupilík, F.
On Open Workflows for Processing of Standardized Electroencephalography Data.
DOI: 10.5220/0010345006670676
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 5: HEALTHINF, pages 667-676
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
667

tegrate a selected set of machine learning (ML) and
deep learning (DL) methods (suitable for EEG/ERP
data processing) with open-source software tools de-
veloped to process standardized EEG/ERP data.
The paper is organized as follows. The state of the
art section presents a traditional EEG/ERP data life-
cycle and its possible improvements, FAIR data prin-
ciples, electrophysiological data standardization ini-
tiatives, and standardized data and metadata formats
used in the EEG/ERP domain. Then current trends in
using ML/DL methods to process the EEG/ERP data
are introduced, and popular ML/DL libraries contain-
ing these classification methods are mentioned.
Subsequently, software tools suitable for the anal-
ysis of standardized EEG/ERP datasets and their abil-
ity to work together with ML/DL libraries are pre-
sented. As the result of the comparison of these
software tools, the feasibility of the selected set of
tools and libraries creating a workflow (pipeline) for
EEG/ERP standardized data processing, is verified by
implementing (reproducing) an analysis based on the
existing and already published EEG/ERP dataset; the
replicated workflow is presented. The last session
concludes the obtained findings.
2 STATE OF THE ART
In this section we introduce a traditional EEG/ERP
experiment, related data lifecycle, FAIR data prin-
ciples, current electrophysiological data standardiza-
tion initiatives, standardized data and metadata for-
mats used in the EEG/ERP domain, and ML/DL
methods suitable to process the EEG/ERP data.
2.1 EEG/ERP Experiment
A traditional EEG/ERP experiment and related data
lifecycle include the following steps and activities:
• research planning - includes experimental design
and its implementation respecting the advantages
and disadvantages of the EEG/ERP method and
its general principles, rules and strategies, ap-
proval of the experiment design by the ethics com-
mittee, the document of informed consent, and lab
setting,
• experimental work - includes preparation of the
participants for an experiment and instructions
given to them, performance of the experiment pro-
tocol, visual inspection of the course of the experi-
ment and its outputs, and collection and storage of
the EEG signal, event markers, and related meta-
data,
• data preprocessing - includes referencing, channel
selection, filtering, ERP epochs extraction, arti-
facts detection, their removal or subtraction, base-
line correction, and epochs averaging,
• data processing and analysis - includes grand av-
eraging, feature extraction, detection (classifica-
tion) of ERP components (power spectral analysis
in case of pure EEG recordings) and their further
detailed investigation,
• data visualization and interpretation - usually in-
clude visualization of channel spectra, topograph-
ical maps, tabular and graphical representation of
averages and grand averages on various electrodes
for selected stimuli, periodograms, classification
results, and statistical interpretation of the find-
ings,
• research publication - includes the publication of
research findings (experimental design, statistics
related to participants, found ERP components or
EEG frequencies), and their interpretation.
2.2 Traditional Lifecycle of EEG/ERP
Data
The traditional EEG/ERP data lifecycle does not at-
tribute too much value to the collected data, quality
of the metadata set, and reproducibility of the entire
experiment. Its typical features are that collected data
and metadata are stored in proprietary formats, tabu-
lar forms, simple files, or even various paper forms.
There is usually little effort to preserve such data and
metadata longer than for the time required to interpret
and publish experiment findings in a scientific jour-
nal. There is also little interest in data and metadata
sharing and transforming data and metadata into well-
organized structures. Specific processing algorithms
and workflows are usually described by referencing
the used software tools. If a non-standard, custom,
or proprietary processing tool is used, the processing
workflow (pipeline) is hardly reproducible.
To cope with the troubles of the experimental
work and drawbacks of the traditional EEG/ERP
data lifecycle and aim for their improvements, op-
portunities to develop open and standardized work-
flows that are based on common EEG/ERP prepro-
cessing/processing methods enriched/integrated with
ML/DL classification methods are addressed in this
paper.
2.3 FAIR Data Principles
The general effort to enhance the openness and re-
producibility of research and reusability and efficient
HEALTHINF 2021 - 14th International Conference on Health Informatics
668

management of raw and derived data led to the design
and joint endorsement of a concise and measurable
set of principles referred to as the FAIR Data Princi-
ples. These principles for scientific data management
and stewardship were first formally published in Sci-
entific Data (Wilkinson et al., 2016). The publication
explained the rationale behind them, gave some initial
implementation, and significantly influenced the view
of scientific data from a long-term point of view.
The FAIR principles, in more detail also explained
in (Wilkinson et al., 2016), are the following ones:
findability, accessibility, interoperability, and reuse of
digital data. They put specific emphasis on enhancing
the ability of machines to automatically find and use
the data, in addition to supporting its reuse by individ-
uals (Wilkinson et al., 2016). Since the interpretation
of the FAIR principles started to be various, several
original authors revisited them in (Mons et al., 2017).
These FAIR principles are highly reflected in the stan-
dardization efforts introduced below.
2.4 Standardization Initiatives
The absence of unified, extensible, open, and widely
accepted descriptive structures, models, and formats
for electrophysiology data has been naturally dis-
cussed in an international forum with the coordinat-
ing role of International Neuroinformatics Coordinat-
ing Facility - INCF (International Neuroinformatics
Coordinating Facility, 2020). Except for proprietary
formats used for simple storage of electrophysiologi-
cal data, there currently exist several competing pro-
posals of flexible data structures, terminologies, and
formats for annotation, organization, and long term
storage of electrophysiological data and metadata.
The common denominator of these proposals is
that they follow the FAIR principles and have been
inspired by open and proprietary data structures and
formats used in electrophysiology, neurophysiology,
electroencephalography, and bioinformatics.
All of them also have to cope with contradictory
requirements: the proposed data/metadata descrip-
tions should be, on the one hand, harmonized and, if
possible, standardized to convey a global descriptive
terminology and support the reproducibility of scien-
tific data, procedures, and results. On the other hand,
they should be flexible enough to meet the require-
ments of individual laboratories. However, even in
the case of the standards endorsed by INCF, these are
still in working progress when concerning the matu-
rity of the whole ecosystem – the entire lifecycle of
electrophysiology data has not been covered so far.
This fact also makes more extensive use of existing
standards difficult.
Three global initiatives are currently worth men-
tioning. The descriptive structures they have pro-
posed usually go beyond the initially intended do-
main boundaries and can be considered to be used
and extended for the description and long-term stor-
age of diverse physiological data. The outcome of
the first initiative, proposed by the German National
Node for Neuroinformatics (G-node), is the Neuro-
science Information Exchange format (NIX) enriched
with odML terminology for the description of meta-
data (Zehl et al., 2016).
Neurodata Without Borders (NWB) (Teeters et al.,
2015) (R
¨
ubel et al., 2019) data structures were in-
troduced by the University of California and fur-
ther elaborated by the scientific community in the
U.S. The BIDS structure (Brain Imaging Data Struc-
ture) (Gorgolewski et al., 2016) introduced at the
McGill University in Montreal has been later ex-
tended with the descriptive structures for EEG
data (Pernet et al., 2019).
In addition to these three global initiatives two
data formats are broadly used for storage of EEG
recordings: the European Data Format (EDF/EDF+)
and the BrainVision Core Data Format. The latter
one defines the structure of data in three files: a text
header file containing metadata, text tag file contain-
ing information about events (stimuli) in data, and
binary data file containing raw EEG data and other
signals recorded together with EEG. All these files
must be stored in one folder; the header and data files
are required. The header and tag files have a fixed
structure (key/value pairs) and their complete descrip-
tion, including a description of all usable keys, can be
found in (Brain Pruducts, 2019).
European data format (EDF) and especially Eu-
ropean data format ’plus’ (EDF+), have been consid-
ered as standard formats for the exchange of physio-
logical data since 2003. EDF+ is a more flexible but
still simple format which is compatible to EDF except
that an EDF+ file may contain interrupted recordings.
When compared to EDF, EDF+ can not only store
annotations but also electromyography, evoked po-
tentials, electroneurography, electrocardiography and
many more types of investigations. (Kemp and Oli-
van, 2003)
Due to the robustness and widespread use of Euro-
pean data format and BrainVision Core Data Format
1.0, the BIDS project recognizes them as the recom-
mended standards for storing EEG/iEEG data (Brain
Products, 2020).
On Open Workflows for Processing of Standardized Electroencephalography Data
669

2.5 Machine and Deep Learning for
EEG/ERP Data Processing
As it has happened in many scientific disciplines,
ML and especially DL methods invaded also the
EEG/ERP domain. The question if DL methods truly
present advantages as compared to more traditional
EEG processing approaches remains an open question
according to a systematic review of DL-based method
for analysis of electroencephalography data that was
published in (Roy et al., 2019).
Effective and efficient EEG/ERP data processing
as part of the whole EEG/ERP data lifecyle suffers
from several limitations. EEG has a low signal-to-
noise ratio (SNR) as the measured brain activity is
often hidden under multiple sources of noise of sim-
ilar or greater amplitude called ’artifacts’. Various
techniques are used to minimize the impact of noise
sources and extract brain activity from the EEG sig-
nal. EEG as a non-stationary signal is also highly
variable across time. Then classifiers trained on
a temporally-limited amount of data might provide
poor generalization to data recorded at a different time
on the same individual. High inter-subject variability
also limits the usefulness of EEG applications. (Roy
et al., 2019)
To cope with the issues mentioned above it seems
to be reasonable to extend traditional and domain-
specific EEG/ERP workflows with DL methods to
clean, extract features and classify EEG/ERP data.
Further automation of these workflows can be done
by utilizing existing data standards, using tools for
EEG/ERP data processing based on these standards
and integrating them with existing ML/DL libraries.
According to (Craik et al., 2019) it is not yet ver-
ified whether DL methods can achieve better results
without use of any pre-processing method. However,
it is surprising that in more than a quarter of the ex-
periments examined, the artifacts were removed man-
ually. This, in addition to being time consuming, also
makes difficult to reproduce the used procedures.
Feature extraction is one of the most challeng-
ing steps in the traditional EEG/ERP data process-
ing. Elimination of the feature extraction procedure
by deep neural networks is the main goal of many
experiments. For example, spectrograms commonly
used to visualize EEG data are used as input to convo-
lutional neural networks (CNNs), whose performance
has proven successful mainly in image data recogni-
tion. Furthermore, it was found that CNNs, which ac-
cept pure EEG signal values for input, performed on
average better than with spectrograms or calculated
features (Craik et al., 2019). This finding contradicts
the idea that feature extraction is an important step
in improving the success of the EEG data classifica-
tion (Craik et al., 2019).
The fundamental decision during the application
of deep-learning methods is the correct selection of
the neural network architecture. For the ERP classi-
fication, CNN is surprisingly the most used architec-
ture, followed by a recurrent neural network (RNN).
Since 2015, there has been a large increase in the use
of CNNs, contrary to expectations that, due to the
temporal nature of the EEG signal, RNNs will be used
more than models that do not naturally work with time
dependencies. The expansion of the use of CNN for
the classification of EEG data can be explained by the
success achieved in computer vision and the use of
a hierarchical structure of the data, as well as recent
discussions and findings concerning the effectiveness
of CNNs for time series processing. However, the fre-
quency of the RNN use is also increasing. (Roy et al.,
2019)
In addition to correct selection of the neural net-
work architecture, researchers must take a decision
about the number of layers. According to (Roy et al.,
2019), most projects used less than five layers for their
models, and it is possible to conclude that shallower
neural networks are currently more suitable for EEG
data. In (Schirrmeister et al., 2017), they directly ad-
dressed the number of CNN layers used for EEG data
and concluded that shallower CNNs successfully out-
performed deeper CNNs.
In the traditional EEG data classification process,
the methods for feature extraction and then traditional
ML methods such as LDA and SVM are often used.
Almost all experiments in (Roy et al., 2019), over
which the successes of DL methods to traditional ML
methods was compared, showed that DL methods im-
proved the classification results.
Currently, the generally well-known and most
popular libraries providing ML/DL methods, scikit-
learn in case of traditional ML methods, and Keras
(based on the TensorFlow library) and PyTorch in
case of DL methods, are also used for EEG/ERP data
processing.
3 SOFTWARE TOOLS FOR
EEG/ERP DATA PROCESSING
The traditional EEG/ERP data lifecycle includes
reading data and pre-/processing them before their
classification. Currently, many various software tools
allow us to do these steps. These tools differ in many
aspects, e.g., in the set of data pre-/processing meth-
ods provided or the data formats they can read or write
to. Some tools provide a user interface; others are
HEALTHINF 2021 - 14th International Conference on Health Informatics
670
offered as libraries. They also differ in their abil-
ity to perform/cooperate/to be integrated with ML/DL
methods and related libraries.
The next subsection describes the tools available
for the pre-/processing of EEG/ERP data and their
current abilities to work/interact with ML/DL meth-
ods and libraries. Since our laboratory primarily
stores EEG/ERP data in the BrainVision format and
the BIDS standard recognizes this format as a do-
main standard, the software tools that can naturally
read/write data in this format are a bit favoured.
3.1 Software Tools Overview
Brainstorm (The Brainstorm team, 2020) is an open-
source tool for the analysis of brain recordings sup-
porting the BrainVision, EDF and NWB formats. It
provides methods for processing EEG data, such as
automated artefact detection, epoch extraction, time-
frequency transformations, and SVM and LDA clas-
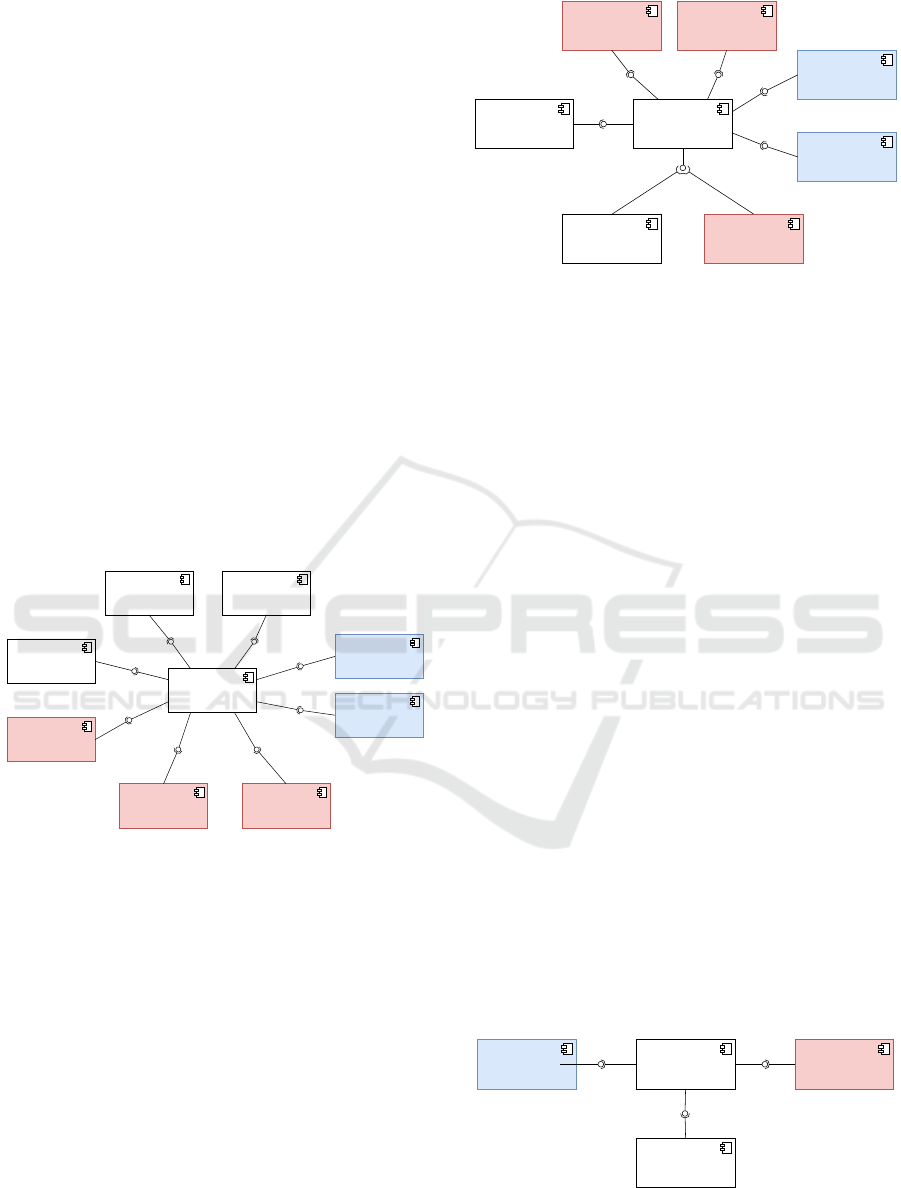
sifiers. Figure 1 shows the UML component diagram
providing the data formats, analytical tools and ML
libraries the Brainstorm can work with. FieldTrip is
Brainstorm
BIDS
export BIDS
MATLAB
Statistics and Machine
Learning Toolbox
LDA a SVM
LibSVM
SVM
EEGLab
read EEGLab sets
FieldTrip
read Fieldtrip structs
NWB
read NWB
MNE
MATLAB - Python
libraries
BrainVision
read Brainvision
read MNE-FIF
Figure 1: UML component diagram - integration of Brain-
storm with data formats, analytical tools and ML libraries.
a MATLAB toolbox for analyzing MEG, EEG, iEEG
and fNIRS data. It contains methods for their prepro-
cessing and more advanced data analysis. It supports
the BrainVision, BIDS, and NWB data formats and
the format used in the EEGLab. The toolbox is free
and can be added to the local MATLAB installation.
FieldTrip does not have any GUI; the methods
provided can be used to create analytical pipelines
in MATLAB. It is integrated with two external ML
MATLAB toolboxes: Donders Machine Learning
Toolbox (DMLT) and MVPA-Light. DMLT offers a
large number of binary and discrete classifiers such as
SVM, Naive Bayesian classifier, or regularized dis-
criminant analysis (van Gerven et al., 2011). Fig-
ure 2 shows the UML component diagram provid-
ing data formats, analytical tools and ML libraries
FieldTripMNE
fieldtrip2fiff
BrainVision
ft_read_data
MVPA-Light
Donders Machine
Learning Toolbox
DML API
cfg.mvpa
EEGLab
BIDS
I/O BIDS
NWB
read NWB
Figure 2: UML component diagram - integration of Field-
Trip with data formats, analytical tools and ML libraries.
FieldTrip can work with. EEGLab is a software tool
written in MATLAB for processing EEG, MEG and
other types of electrophysiological data. It is an open-
source project allowing users to extend the capabili-
ties of the tool by implementing their plugins. It pro-
vides basic preprocessing methods such as filtering,
re-sampling, epoch extraction, baseline correction, or
time-frequency transformation and offers several pro-
cedures for removing artefacts from a noisy signal.
BCILAB (Kothe and Makeig, 2013) (BCILAB,
2020) is an open-source Matlab toolbox for BCI re-
search that allows ML classification algorithms to be
applied to data processed in EEGLab. It provides
algorithms for signal processing and contains imple-
mentations of ML methods including SVM, LDA and
linear regression. Figure 3 shows the UML compo-
nent diagram providing data formats, analytical tools
and ML libraries EEGLab can work with. Neo (Gar-
cia et al., 2014) is a language-independent object
model for handling electrophysiology data in multiple
formats (NIX and BrainVision format are supported).
The motivation for its development was to increase in-
teroperability between Python tools for analysis, visu-
alization and generation of electrophysiological data.
Neo is limited purely to data representation and does
not provide functions for data analysis or visualiza-
tion. Its hierarchical data model is used by several
different tools for data analysis, visualization or sim-
ulation, such as SpykeViewer, Elephant, PyNN, and
EphyViewer.
EEGLab
FieldTrip
fieldtrip2eeglab
BrainVision
read BrainVision
BCILab
read EEGLab
dataset
Figure 3: UML component diagram - integration of
EEGLab with data formats, analytical tools and ML li-
braries.
On Open Workflows for Processing of Standardized Electroencephalography Data
671

SpykeViewer (Pr
¨
opper and Obermayer, 2013),
(The NeuralEnsemble Initiative, 2020b) is a cross-
platform application with a user interface for visual-
izing electrophysiological data.
Elephant (Electrophysiology Analysis Toolkit) is
an open-source library for the analysis of electrophys-
iological data in Python. It focuses on generic an-
alytic functions for action potentials and time-series
records from electrodes, such as local field potentials
(LFP) or intracellular voltage. The aim of the Ele-
phant project is, in addition to a common platform for
analytical codes from different laboratories, to pro-
vide a consistent and homogeneous framework for
analysis built on a modular basis (Elephant authors
and contributors, 2020).
PyNN is a simulator-independent language for
creating spiking neural networks. Its goal is to al-
low users to write code for a simulation model only
once and then run it on any simulator that PyNN sup-
ports. PyNN provides a library of standard models of
neurons, synapses, and synaptic plasticities that have
been proven to work the same on various supported
simulators. It also provides a set of commonly used
connectivity algorithms. (The NeuralEnsemble Initia-
tive, 2020a)
EphyViewer is a Python library for visualizing
electrophysiological data. It supports the display of
possible representations of a given dataset (signal,
epochs, events, action potentials) and provides a stan-
dalone user interface that allows users to view data
from the Neo data format (Garcia and Gill, 2019).
When analyzing the Neo library, no possibility
to use ML classification methods directly was found.
The only way to apply ML methods to EEG/ERP data
stored in the Neo format was by utilizing the Elephant
and Pandas libraries. However, Neo provides func-
tions for manipulating objects in its custom data for-
mat. The data can be extracted in a format that can be
used as an input into DL algorithms within the Keras
library and ML algorithms within the scikit-learn li-
brary. Figure 4 shows the UML component diagram
providing data formats, analytical tools and ML li-
braries can Neo work with. Pandas is an open-source
Python library that provides flexible data structures
designed to work with different types of data; it can
work with the NIX, BIDS and NWB formats. It is
designed as a basic high-level building block for per-
forming practical data analysis. It is useful for differ-
ent types of data such as ordered and unordered time
series or any array data with descriptions of rows and
columns. Pandas is built on the NumPy library; it can
be integrated into the scientific computing environ-
ment with many other libraries such as MNE. (The
pandas development team, 2020)
NEO
BrainVIsion NIX
BrainVisionIO
NIXIO
MNE
MNE from
raw array
SpykeViewer Elephant PyNN EphyViewer
NEO data
model
Pandas
elephnat.pandas_bridge
Figure 4: UML component diagram - integration of Neo
with data formats, analytical tools and ML libraries.
From the internal pandas’ structure, it is easy to
extract data in a format that matches the inputs to
ML classification methods. Although no use cases
for EEG data were found, there are use-cases for pan-
das time series processed using the Keras library. The
sklearn-pandas project was created to integrate pan-
das structures into the machine-learning pipelines of
the scikit-learn library. Another integration of the
pandas library with the library of deep-learning meth-
ods is the keras-pandas project (Herger, 2020). Fig-
ure 5 shows the UML component diagram providing
data formats, analytical tools and ML libraries pan-
das can work with. MNE (Gramfort et al., 2013)
is an open-source Python tool providing algorithms
for data preprocessing, resource localization, statisti-
cal analysis, and estimation of functional connectivity
between distributed areas of the brain. It is integrated
with the basic Python libraries for scientific computa-
tions (NumPy, SciPy) and visualization (Matplotlib).
MNE supports the processing of a number of data
Pandas
NIX NWB
NWBTable.to_dataframe()write_to_pandas()
MNE
to_data_frame()
Elephant
pandas_bridge()
keras-pandas sklearn-pandas
Keras scikit-learn
BIDS
to_df()
Figure 5: UML component diagram - integration of pandas
with data formats, analytical tools and ML libraries.
HEALTHINF 2021 - 14th International Conference on Health Informatics
672

types, such as EEG, MEG, ECG, SEEG, and ECoG.
It supports the BIDS and NIX data formats, BrainVi-
sion and EDF formats, but also data in the formats of
other analytical tools such as FieldTrip, EEGLab and
Brainstorm. MNE cannot work directly with the Neo
data format but provides instructions on how to con-
vert data from Neo to MNE structures.
MNE allows its users to export the processed data
to the structures of the pandas library. Several open-
source projects use the MNE package to process EEG
data, and then apply DL methods from the Keras li-
brary. The integration of these tools can be thus con-
sidered as widespread and proven. The developers of
the MNE library also created and shared examples of
how DL methods from the PyTorch library can be ap-
plied to the data processed in MNE. Figure 6 shows
the UML component diagram providing data formats,
analytical tools and ML libraries MNE can work with.
MNE
NEO
MNE-BIDS NIX-MNE
Brainstorm
MNE from
raw array
read_raw_eeglab()
read_raw_fieldtrip()
bst_raw
EEGLab FieldTrip
BIDS NIX
Braindecode
MNE-torch
deepEEG
arl-EEGmodels
Pandas
to_data_frame()
PyTorch Keras
scikit-learn
mne.decoding
BrainVision
read_raw_brainvision()
Figure 6: UML component diagram - integration of MNE
with data formats, analytical tools and ML libraries.
3.2 Software Tools Comparison
Based on the overview of software tools for EEG/ERP
processing given above, their abilities to work with
machine and DL methods and libraries, and experi-
ence in working with them, we set the criteria and
selected the software tools to form more automated
and open workflows for the processing of standard-
ized EEG/ERP data. The selection criteria are given
and explained below. The evaluation results are sum-
marized in Table 1. Except for licensing, all criteria
are graded in the following three levels:
1. need to get the MATLAB license,
2. level and scope of documentation,
• Low - weak documentation, missing com-
ments in the code, no tutorials,
• Medium - well-commented basic building
blocks of the code, several use cases,
• High - a large number of very detailed instruc-
tions and examples to help users understand the
tool, thoroughly annotated code,
3. current availabilities to use ML/DL learning clas-
sification methods,
• Low - the tool does not provide any possibili-
ties of using ML classification methods, or only
a very limited set of them (up to 3 classifiers),
• Medium - the tool allows to use a more exten-
sive set of ML classification methods, it does
not contain options for the use of DL meth-
ods/neural networks,
• High - the tool provides extensive possibilities
of using machine and DL classification meth-
ods, or is integrated with libraries providing
these methods,
4. the size of the sets of functionalities suitable for
EEG/ERP data processing,
• Low - the tool does not provide any function-
alities for EEG/ERP data processing,
• Medium - the tool provides a limited set of
basic functions for EEG/ERP data processing,
• High - the tool contains extensive possibili-
ties for processing, analysis and visualization
of EEG/ERP data,
5. number of supported formats and tool abilities to
work/interact with other sw tools (the level of in-
tegration),
• Low - the tool supports only a very limited set
of EEG data formats and is not able to interact
with other tools,
• Medium - the tool supports a limited set of
EEG data formats and is integrated with a small
set of tools (up to 3),
• High - the tool supports all or almost all com-
monly used EEG data formats and provides
possibilities of working with other sw tools,
6. community size based on the number of forks of
the project on Github,
• Low - number of forks is < 200,
• Medium - number of forks is between 200 and
1000,
• High - number of forks is > 1000,
7. user friendliness,
On Open Workflows for Processing of Standardized Electroencephalography Data
673

• Low - deep knowledge of the tool is required
for convenient use, some entities are incompre-
hensibly implemented (it is often related to the
lower level of documentation),
• Medium - implementation is difficult to un-
derstand, convenient use of the tool requires
good knowledge of the tool,
• High - implementation is easy to understand,
all functionalities of the tool are easily accessi-
ble.
When evaluating the results given in Table 1, the
MNE library and the related ecosystem (the Brain-
Vision data format, scikit-learn library and Keras li-
brary) were selected as the candidates for an open and
convenient workflow suitable for processing of stan-
dardized EEG/ERP data.
4 USE CASE EXAMPLE
The use case further presented verifies the feasibil-
ity of the selected workflow; integration of appropri-
ate ML/DL classification methods into the chosen tool
for EEG/ERP data pre-/processing is practically eval-
uated. This is demonstrated by replicating a process-
ing workflow of EEG/ERP data described in (Va
ˇ
reka,
2020).
The related experiment called ’Guess the number’
included EEG/ERP data collected from 250 primary
and secondary school children; the underlying data
are also available in (Mou
ˇ
cek et al., 2017). Before the
start of the experiment, each participant was asked to
select a number from 1 to 9 arbitrarily and to concen-
trate on it. Then, the recording of the EEG data was
started (EEG data from the electrodes Fz, Cz, and Pz
and event markers were collected) and the participant
was projected with visual stimuli (numbers between 1
and 9) in random orderNecessary experimental meta-
data from each participant were collected.
The experiment aimed to classify EEG/ERP
epochs into the target (thought number) and non-
target (another number) classes. The division of
epochs into training, validation and testing sets was
random. The success of the classification using a con-
volutional neural network was tested and compared
with the traditional LDA and SVM classifiers. The
MATLAB tools for data loading, epoch extraction and
filtering, and the scikit-learn and Keras libraries for
classification purposes were used in the original pro-
cessing workflow.
The goal of the presented use case is to repli-
cate the original processing workflow described
in (Va
ˇ
reka, 2020) entirely in the Python ecosystem se-
lected above and thus prove that this processing work-
flow can be completed (when achieving the similar re-
sults) only with the use of open resources supporting
current standardization initiatives.
The data in the BrainVision format were read
and preprocessed (filtering, epochs extraction) using
the same methods as described in (Va
ˇ
reka, 2020),
but utilizing the MNE library. The data were fur-
ther classified with the LDA and SVM ML meth-
ods from the scikit-learn library as well as convolu-
tional and recurrent neural networks from the Keras
library (the same convolutional neural network as
in (Va
ˇ
reka, 2020) was used). The success of the clas-
sifiers was evaluated using the Monte-Carlo cross-
validation. The results were compared with the results
obtained in (Va
ˇ
reka, 2020). The resulting implemen-
tation and the detailed manual are publicly available
in (Kupil
´
ık, 2020).
4.1 Testing
Due to the nature of the EEG/ERP data processing
and also because the original experiment is not fully
reproducible (the artefact rejection procedure is not
described in sufficient detail, selection of non-target
epochs is random, and generally, neural networks
are used), the functionalities of the entire processing
workflow and its methods were both inspected visu-
ally by plotting the state of the data before and after
the application of the methods and by comparing the
achieved classification results.
4.2 Results
The entire processing workflow was found functional.
The filtering method was applied to the raw data of all
250 subjects, and the method for removing artefacts
removed approximately 30% of epochs from both tar-
get and non-target sets of epochs. Table 2 shows
the averaged results obtained after ten iterations of
the classification method using standard metrics. The
standard deviations are given in parentheses. The
CNN classifier achieved the best AUC (area under
the ROC curve), and the SVM classifier achieved the
best accuracy and precision. The values closely cor-
respond to the results obtained in (Va
ˇ
reka, 2020) for
the case when the core CNN architecture was used;
a positive change of 1-3 percentage points depending
on the metric is observable. The RNN classifier that
was tested and evaluated only in this processing work-
flow achieved decent results in terms of AUC and pre-
cision compared to other classifiers, but significantly
worse values when recall is considered.
The MNE library was found to be a comprehen-
HEALTHINF 2021 - 14th International Conference on Health Informatics
674

Table 1: Evaluation of software tools for EEG/ERP data processing according to the given criteria.
Sw tool/Criteria Brainstorm FieldTrip EEGLab Neo Pandas MNE
License required N (Y for ML methods) Y (Matlab) Y (Matlab) N N N
Documentation
level
High High High Middle High High
ML/DL methods
utilization
Low Middle Middle Low High High
Number of
EEG/ERP data
processing methods
High High High Low Low High
Level of integration High High Middle High High High
Size of community Low Middle Low Low High Middle
User friendliness Middle Middle Middle Low High High
Table 2: Average cross-validation classification results.
AUC accuracy precision recall
CNN 69.62% (0.79) 64.42% (0.74) 64.89% (1.17) 62.59% (2.52)
RNN 66.2% (0.87) 63.43% (0.83) 65.05% (1.77) 58.58% (4.51)
SVM 65.21% (0.44) 65.22% (0.45) 66.11% (0.64) 62.47% (1.04)
LDA 62.87% (0.38) 62.86% (0.38) 61.94% (0.43) 66.14% (0.8)
sive and convenient software tool for processing and
analyzing EEG/ERP data. Its structure is easy to un-
derstand, and its resources are logically and hierar-
chically well arranged. The level of documentation,
instructions provided, and commented use cases are
very high, and all the above support is continuously
updated. For ERP classification, the great advantage
of the tool is the possibility to export the analyzed
epochs to the format of the NumPy library, which is
used by both libraries (scikit-learn and Keras) provid-
ing classification methods.
5 CONCLUSIONS
The paper introduced an important step in defin-
ing and implementing a processing workflow for
EEG/ERP data when open-source software ecosys-
tem and standardized EEG data format are used. In
parallel, this step contributes to the completion and
maturity of the whole lifecycle of electrophysiology
data. The EEG/ERP data lifecycle, standardization
initiatives, related EEG data formats, and current view
on the use of ML/DL approaches and libraries for
EEG/ERP data processing were introduced. Further-
more, software tools for processing EEG/ERP data
were analyzed (not only) concerning the utilization of
ML/DL methods contained in widespread ML/DL li-
braries.
Based on the results of the analysis, the MNE,
scikit-learn and Keras libraries were chosen for
the processing and classification of standardized
EEG/ERP data. The proposed and implemented pro-
cessing workflow was evaluated over publicly avail-
able EEG/ERP datasets by replicating the processing
workflow described in (Va
ˇ
reka, 2020). The achieved
classification results were compared with similar re-
sults. If we take into account positive user experience,
the proposed workflow was recommended for open
and convenient processing of standardized EEG/ERP
data.
ACKNOWLEDGEMENTS
This work was supported by the University specific
research project SGS-2019-018 Processing of hetero-
geneous data and its specialized applications (project
SGS-2019-018).
REFERENCES
BCILAB (2020). Matlab toolbox for brain-computer inter-
face research. https://github.com/sccn/BCILAB, last
accessed on 2020-09-29.
Brain Products (2020). Bids adopts brainvision core
data format 1.0 as one of the recommended official
eeg/ieeg data formats. https://pressrelease.brainpro
ducts.com/bids/, last accessed on 2020-08-18.
Brain Pruducts (2019). Description of the brainvision
core data format 1.0. https://www.brainproducts.com/
files/public/products/more/BrainVisionCoreDataForm
at 1-0.pdf, last accessed on 2020-08-17.
On Open Workflows for Processing of Standardized Electroencephalography Data
675

Craik, A., He, Y., and Contreras-Vidal, J. L. (2019). Deep
learning for electroencephalogram (EEG) classifica-
tion tasks:a review. Journal of Neural Engineering,
16(3):031001.
Elephant authors and contributors (2020). Ele-
phant - electrophysiology analysis toolkit.
https://elephant.readthedocs.io/en/latest/, last ac-
cessed on 2020-04-19.
Garcia, S. and Gill, J.(2019). ephyviewer 1.3.2.dev.
https://ephyviewer.readthedocs.io/en/latest/, last ac-
cessed on 2020-04-19.
Garcia, S., Guarino, D., Jaillet, F., Jennings, T., Pr
¨
opper, R.,
Rautenberg, P., Rodgers, C., Sobolev, A., Wachtler,
T., Yger, P., and Davison, A. (2014). Neo: an object
model for handling electrophysiology data in multiple
formats. Frontiers in Neuroinformatics, 8:10.
Gorgolewski, K., Auer, T., Calhoun, V., Craddock, R., Das,
S., Duff, E., et al.(2016). The brain imaging data
structure, a format for organizing and describing out-
puts of neuroimaging experiments. Scientific Data, 3.
Gramfort, A., Luessi, M., Larson, E., Engemann,
D., Strohmeier, D., Brodbeck, C., Goj, R., Jas,
M., Brooks, T., Parkkonen, L., and H
¨
am
¨
al
¨
ainen,
M.(2013). Meg and eeg data analysis with mne-
python. Frontiers in Neuroscience, 7:267.
Herger, B. (2020). keras-pandas 3.1.0. https://pypi.org/
project/keras-pandas/, last accessed on 2020-09-30.
International Neuroinformatics Coordinating Facility
(2020). https://www.incf.org/, last accessed on
2020-08-17.
Kemp, B. and Olivan, J. (2003). European data format
‘plus’ (edf+), an edf alike standard format for the ex-
change of physiological data. Clinical Neurophysiol-
ogy, 114(9):1755 – 1761.
Kothe, C. and Makeig, S. (2013). Bcilab: A platform for
brain–computer interface development. Journal of
neural engineering, 10:056014.
Kupil
´
ık, F. (2020). MNE ML - EEG data processing
pipeline using the MNE, Keras and the scikit-learn
libraries. https://github.com/fkupilik/MNE ML, last
accessed on 2020-10-27.
Mons, B., Neylon, C., Velterop, J., Dumontier, M.,
Da Silva Santos, L., and Wilkinson, M. (2017).
Cloudy, increasingly fair; revisiting the fair data guid-
ing principles for the european open science cloud. In-
formation Services and Use, 37(1):49–56.
Mou
ˇ
cek, R., Va
ˇ
reka, L., Prokop, T.,
ˇ
St
ˇ
ebet
´
ak, J., and Br
˚
uha,
P. (2017). Event-related potential data from a guess
the number brain-computer interface experiment on
school children. Scientific Data, 4 cited By 5.
Pernet, C., Appelhoff, S., Gorgolewski, K., Flandin, G.,
Phillips, C., Delorme, A., et al. (2019). Eeg-bids, an
extension to the brain imaging data structure for elec-
troencephalography. Scientific Data, 6(1).
Pr
¨
opper, R. and Obermayer, K. (2013). Spyke viewer:
a flexible and extensible platform for electrophysio-
logical data analysis. Frontiers in Neuroinformatics,
7:26.
Roy, Y., Banville, H., Albuquerque, I., Gramfort, A., Falk,
T. H., and Faubert, J. (2019). Deep learning-based
electroencephalography analysis: a systematic review.
Journal of Neural Engineering, 16(5):051001.
R
¨
ubel, O., Tritt, A., Dichter, B., Braun, T., Cain, N., Clack,
N., et al. (2019). Nwb:n 2.0: An accessible data stan-
dard for neurophysiology. bioRxiv.
Schirrmeister, R., Springenberg, J., Fiederer, L., Glasstet-
ter, M., Eggensperger, K., Tangermann, M., Hutter,
F., Burgard, W., and Ball, T. (2017). Deep learning
with convolutional neural networks for eeg decoding
and visualization: Convolutional neural networks in
eeg analysis. Human Brain Mapping, 38.
Teeters, J., Godfrey, K., Young, R., Dang, C., Friedsam, C.,
Wark, B., et al.(2015). Neurodata without borders:
Creating a common data format for neurophysiology.
Neuron, 88(4):629–634.
The Brainstorm team (2020). Brainstorm. https:// neuroim-
age.usc.edu/brainstorm/, last accessed on 2020-05-02.
The NeuralEnsemble Initiative (2020a). Pynn -
a python package for simulator-independent
specification of neuronal network models.
https://neuralensemble.org/PyNN/, last accessed
on 2020-04-19.
The NeuralEnsemble Initiative (2020b). Spykeviewer.
https://neuralensemble.org/SpykeViewer/, last ac-
cessed on 2020-05-03.
The pandas development team (2020). pandas - data analy-
sis and manipulation tool. https://pandas.pydata.org/,
last accessed on 2020-09-30.
Va
ˇ
reka, L. (2020). Evaluation of convolutional neu-
ral networks using a large multi-subject p300
dataset. Biomedical Signal Processing and Control,
58:101837.
van Gerven, M., Bahramisharif, A., Farquhar, J., and
Heskes, T. (2011). Donders machine learning tool-
box. https://github.com/distrep/DMLT, last accessed
on 2020-05-02.
Wilkinson, M., Dumontier, M., Aalbersberg, I., Appleton,
G., Axton, M., Baak, A., et al.(2016). The fair guiding
principles for scientific data management and steward-
ship. Scientific Data, 3.
Zehl, L., Jaillet, F., Stoewer, A., Grewe, J., Sobolev, A.,
Wachtler, T., et al.(2016). Handling metadata in a neu-
rophysiology laboratory. Frontiers in Neuroinformat-
ics, 10:26.
HEALTHINF 2021 - 14th International Conference on Health Informatics
676
