
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance
Compound Identification
Teodora Gerasimoska
1 a
, Milka Ljoncheva
2,3 b
and Monika Simjanoska
1 c
1
Faculty of Computer Science and Engineering, Ss. Cyril and Methodius University,
Rugjer Boshkovikj 16, 1000 Skopje, N. Macedonia
2
Jo
ˇ
zef Stefan Institute, Jamova cesta 39, 1000 Ljubljana, Slovenia
3
International Postgraduate School Jo
ˇ
zef Stefan, Jamova Cesta 39, 1000 Ljubljana, Slovenia
Keywords:
Mass Spectral Library, Batch Search Tool, Mass Spectrometry.
Abstract:
Identification of new organic compounds through suspect screening (SS) and non-targeted analysis (NTA)
became the most challenging task in environmental and metabolomics research in the recent two decades.
Identification of thousands of organic compounds is performed using the recent technology advancements in
chromatography-mass spectrometry as the core analytical platform, assisted by multitude of cheminformatics-
assisted approaches. As many of those approaches rely on mass spectral libraries (MSLs) search, the avail-
ability of comprehensive MSLs with engines for batch search and export of MS data and batch search engines
for simultaneous search and export of MS data from multiple MSLs is of crucial importance. In lack of such,
analysts perform this step in a laborious, time-consuming manual manner, importing significant risk of com-
pound misidentification.
This paper presents MSL-ST, the first tool for automated batch search and storage of MS spectra that uses
two of the largest publicly available MSLs as data source, the MassBank of North America (MoNa) and the
MassBank of Europe. MSL-ST assembles large amount of MS data in an automated, time- and cost-effective
manner in a format which allows its further processing, especially for the purpose of compound identification.
The tool, accompanied with user manual, is publicly available on GitHub.
1 INTRODUCTION
The typical targeted analysis of organic compounds,
based on the identification of the presence and de-
termination of the concentration of a predetermined
list of organic compounds is the golden standard in
all monitoring analyses of organic compounds. De-
spite it, over the last two decades, SS and NTA
have evolved into effective screening strategies for
“known unknowns” and ”unknown unknowns” com-
pounds in complex environmental samples, respec-
tively. In addition to the challenge of developing sen-
sitive generic analytical methods able to identify as
many analytical signals as possible that correspond
to organic compounds, the next major challenge is
to identify the compounds to which these analytical
signals correspond. Appropriate data processing and
a
https://orcid.org/0000-0001-9923-6483
b
https://orcid.org/0000-0001-6664-0287
c
https://orcid.org/0000-0002-5028-3841
compound identification tools have to be employed
for this task in order to avoid false positive or false
negative identifications, most frequently due to the
presence of interfering singals. Acquisition of sig-
nals of thousands of compounds in a single environ-
mental or biological sample is achieved by employ-
ing two analytical platforms: nuclear magnetic res-
onance (NMR) and gas chromatography (GC) or liq-
uid chromatography (LC) coupled to mass spectrome-
try (MS). While the first analytical platform performs
nondestructive and noninvasive sample analysis with
concentration-dependent sensitivity, GC/LC-MS an-
alytical platforms offer sensitive detection of thou-
sands of analyte signals, accompanied with various
established cheminformatics-assisted approaches for
their annotation and identification. High resolution-
accurate mass MS (HR/AM-MS) became the indis-
pensable analytical platform for accurate and reliable
compound identification. GC and LC are the analyt-
ical techniques used for selective compound separa-
tion, that, when coupled to various HR/AM-MS ana-
Gerasimoska, T., Ljoncheva, M. and Simjanoska, M.
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification.
DOI: 10.5220/0010424101950203
In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2021) - Volume 3: BIOINFORMATICS, pages 195-203
ISBN: 978-989-758-490-9
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
195

lyzers with high mass accuracy, resolving power and
sensitivity and wide linear range ensure reliable com-
pound identification across a wide mass and concen-
tration range.
One of the main applications of cheminformat-
ics is generating methods for storage and retrieval of
chemical structures of compounds. Effective search
through structural information for compounds in-
volves the use of information tools (data search and
machine learning), as well as knowledge in the field
of graph theory (or more precisely, chemical graph
theory). The automated retrieval and storage of chem-
ical, structural compound information is an important
step that would facilitate and speed up the process
that is currently being performed manually. In recent
decade, multitude of cheminformatics approaches are
developed that perform compound identification us-
ing MS data as initial input data. Numerous tools,
such as MOLGEN-MS (Schymanski et al., 2008), the
GMD algorithm (Hummel et al., 2010), FT-BLAST
(Rasche et al., 2012) and NEIMS (Wei et al., 2019)
include MSL(s) search of the MS spectra of the candi-
date compound, followed by comparison of the mea-
sured MS spectra of the unknown compound with
the MS spectra of the candidate compounds con-
tained in the searched MSLs. As NTA often results
with identification of hundreds to thousands of an-
alyte signals, identical is the number of candidate
compounds for which MS data should be searched
across one or multiple MSLs. The automation of
the MSL(s) search, as well as improvements in spec-
trum search tools in terms of user interface, search
algorithms, speed and IT support are the future chal-
lenges addressed in this research. Literature reviews
from the last decade indicate difficulties that persist
to this day. Namely, in order to enable the develop-
ment of data-driven algorithms for identification of
small molecules, it is necessary that all sets of MS
spectra are available for public use through appro-
priate MSLs. The fact that most MSLs do not al-
low data download, ie. batch data download suggests
that for optimal use of MSL data, research commu-
nities should embrace new technologies and mecha-
nisms that support interoperability, and promote the
advancement of an ”open data sharing” culture. They
would find even more frequent use as input data on
several ML models (Scheubert et al., 2013). The en-
tries of spectral data in MSLs is often accompanied by
rich metadata annotations, including compound struc-
ture, name, molecular descriptors (InChI, InChIKey,
SMILES), instrument type, ionization mode, opera-
tion conditions (e.g. GC oven temperature program,
LC method, collision energy etc.), adduct ion type and
product ion annotations. An extensive overview of the
literature on MSLs and the identification of peptides
and small molecules using cheminformatics-assisted
approaches indicates that the lack of ability to search
and retrieve batch mass spectra from publicly avail-
able MSLs significantly impedes the development of
new ML models for predicting molecular mass, MF
and / or the structure of small organic compounds,
thus preventing the identification of new organic com-
pounds. Hence the main impetus for the development
of an MSLs search tool, used for the identification
of organic compounds, which has the option of batch
search of mass spectra for more data and their auto-
mated retrieval.
The paper first discusses some of the notable tools
related to the problem at hand (section 2), moves for-
ward to the presentation of the main data sources and
methods used for the tool development (Section 3)
and of the comprehensive functionality of the tool
(Section 4) and concludes the directions of further de-
velopment (section 5).
2 RELATED WORK
Searching and providing high quality chemical infor-
mation from multiple data sources is an extreme chal-
lenge, but also essential before performing SS and
NTA, given that the direct approach to identifying
compounds is to search for one or more MSLs. Chal-
lenges for chemical DBs of compounds include at-
taching high quality data and thus providing access to
chemical structures and related metadata (experimen-
tal/predicted properties, toxicity data, literature data,
etc.). The publication of chemical metadata with an
appropriate molecular descriptor (eg SMILES, InChI,
InChI Key, PubChem ID, ChemSpider ID, Dashboard
DTXSID, CAS numbers or MOL files) is extremely
important for unique self-identification display, but
also for in the database.
Most MSLs are publicly available, including
METLIN (Guijas et al., 2018), MassBank, GMD
(Hummel et al., 2007), although many do not allow
batch download. Some MSLs are specific to a specific
research area, such as Human Metabolome Database
(HMDB) (Wishart et al., 2007), which includes the
MS / MS spectrum of human metabolites, and Re-
Spect (Sawada et al., 2012) contains mass spectra
of plant metabolites. Also, depending on the type
of chromatography, for GH-MS large MSLs are rou-
tinely used, while for LC-MS MSLs are used which
usually contain a smaller number of mass spectra for
a smaller number of compounds.
MSLs can be classified according to various crite-
ria, such as resolution (low/high), number and type of
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
196

mass analyzers used to generate mass spectra (MS
1
,
MS
2
, ...., MS
n
) (method type of ionization, type of
chromatography (GH, LH), number and type of com-
pounds with mass spectra included in MSLs, tak-
ing into account their physicochemical properties and
source (human, animal, plant, synthetic compound,
environmental pollutant).
Statistical processing of the information extracted
from the relevant scientific literature in 2015 shows
that EI-MS libraries for volatile compounds (such
as NIST (NIST: National Institue of Standard and
Technology, 2020) / EPA / NIH, Wiley (McLafferty
and Stauffer, 2009), GMD (Hummel et al., 2007),
MassBank (Horai et al., 2020) ) are most commonly
cited and used MSLs (82%), followed by ESI-MS /
MS MSLs (such as METLIN (Guijas et al., 2018)
and HMDB (Wishart et al., 2007) ) for non-volatile
compounds (16%) (Ljoncheva, Milka and Stepi
ˇ
snik,
Toma
ˇ
z and D
ˇ
zeroski, Sa
ˇ
so and Kosjek, Tina, 2020).
GC-MS spectra have a great advantage over LC-
MS/MS spectra due to their higher reproducibility,
which makes the generated mass spectra comparable,
regardless of whether they are generated using mass
analyzers of different configuration and/or manufac-
turer, which in turn allows the application of com-
prehensive MBSs. The limited use of the GC-MS
analytical platform for the analysis of volatile com-
pounds leads to a reduced number of GH-MS spectra
in MSLs. Namely, Mass Bank of Europe (Horai et al.,
2020) contains mass spectra of 15000 non-volatile
organic compounds out of a total of 41,092 com-
pounds, METLIN (Guijas et al., 2018) only 12,057
out of 242,032 compounds, compared to libraries with
volatile compounds such as NIST (NIST: National
Institue of Standard and Technology, 2020), Wiley
(McLafferty and Stauffer, 2009), MassBank of Eu-
rope (Horai et al., 2020), Fiehn Lib (Kind et al.,
2009), and the Golm Metabolome Database (Hummel
et al., 2007), containing a significantly larger number
of compounds.
3 MATERIALS AND METHODS
3.1 Materials
3.1.1 MassBank of Europe
MassBank of Europe (Horai et al., 2020) is the
first publicly available MSL containing comprehen-
sive corpus of MS data generated using versatile
analytical platforms, such as LC-ESI-QTOF, LC-
ESI-ITFT, LC-ESI-QFT, EI-B, LC-ESI-QQ, LC-ESI-
Q, LC-ESI-QQQ and GC-EI-TOF. As of December
2020, electrospray ionization (ESI) (60.0%), elec-
tron ionization (EI) (30.0%), chemical ionization
(CI) (2.0%), atmospheric-pressure chemical ioniza-
tion (APCI) (1.6%) and matrix-assisted laser desorp-
tion/ionization (MALDI) (1.5%) mass spectra rep-
resent the most valuable MS data from the 88 168
unique spectra (19 981 MS
1
, 67 188 MS
2
, 929 MS
3
and 70 MS
4
spectra) of 14 838 unique compounds this
MSL includes, making it one of the most comprehen-
sive MS data repository available MassBank (Horai
et al., 2020). MassBank of Europe’s unique feature
is the inclusion of merged spectra, that are spectra
containing MS data from numerous spectra of a sin-
gle compound generated under different instrumental
conditions (usually at different ionization energies).
MassBank of Europe is also repository for trial and
unknown MS data, that are of great importance for SS
and NTA. Despite being especially user-friendly and
with versatile search options, including name, exact
mass, molecular formula, peak list, peak differences,
InChIKey and SPLASH search, the search engine of
this MSL does not offer batch data search and export.
3.1.2 MoNa
The MassBank’s version of North America,MoNa
(Mehta, 2020) is the most exhaustive readily-
downloadable MS data repository. As of December
2020, it contains 672 751 MS spectra of more than
90 300 organic compounds (20 756 MS
1
, 635 129
MS
2
, 929 MS
3
and 70 MS
4
spectra), 490 087 of which
in silico and 179 535 experimental spectra. MoNa’s
search engine apart from offering quick spectra search
by compound name, compound class, molecular for-
mula and/or exact mass includes the unique feature of
searching for spectra similar to the one of the user, but
lacks to offer batch search and download of spectral
data and accompanying metadata.
3.2 Methods
MSL-ST relies on both Python and Django frame-
work, and the concept of simulating human approach
for data assessment. This approach saves significant
amount of time otherwise used for manual spectral
data download. The process itself involves fetch-
ing the web page and downloading data by utilising
Python libraries, which is automating the scenario
when the web page would have been displayed to
the user through a web browser. Once data is down-
loaded, the extraction begins. The content of the page
can be parsed, searched, and reformatted.
Python is often described as the ”batteries in-
cluded” language because of its comprehensive stan-
dard library and modules for variety of tasks (DeBill,
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification
197

2010). Besides being the main tool for the automated
data extraction functionality, it was as well selected
as the most appropriate programming language for
developing MSL-ST by using the Django free open
source framework, which follows the model-view-
template (MVT) architectural pattern (Holovaty and
Kaplan-Moss, 2009), including usage of Python even
for the configuration files and data models. Django
was selected as web framework due to the emphasiz-
ing reusable components, shorter codes, rapid devel-
opment and principle of non-repetition, which lead to
facilitated creation of complex data-driven web pages.
The source code of MSL-ST is available at GitHub
htt ps : //github.com/gerasimoska/mass spectra,
and is also hosted and can be accessed at the
following link htt ps : //massspectra.dev/.
4 MSL-ST FUNCTIONALITY
In this section, the functionality of MSL-ST is pre-
sented by depicturing two experimental scenarios of
obtaining data from the two possible sources: the first
one represents batch search and export from MoNa
and the second one from MassBank of Europe.
The web interface of MSL-ST (Fig. 1) consists of:
(1) Mass spectral library section, in which the user
selects the MSL to be searched against. Selecting one
among the two MSLs (one-at-a-time) enables parts of
the interface that correspond with the selected library,
that are searching parameters valid for the library at
hand. Selecting MoNa as source MSL enables the Li-
brary filter section, where the user is allowed to select
one or more of the sub-MSLs included in MoNa (Fig.
2). Selecting MassBank of Europe as source MSL, on
the other hand, enables the mass Spectra Library fil-
ter section, the Other Instrument Types filter, which is
only available when searching the Mass Bank, is no
longer blurred and disabled (Fig. 3);
(2) Search parameter filter sections, used to cus-
tom the search parameters according to the user’s re-
quirements. Apart from the library-specific parame-
ters made available after MSL selection, other filters
are available for both MSLs, including:
• The ion mode filter, that allows the selection of
the type of ionization in MS, positive or negative;
• The MS Type filter which indicates the number
and type of mass spectrometers used to generate
MS and
• The Source Introduction filter that allows the se-
lection of the type of chromatography, whether it
is gas chromatography, liquid chromatography, or
capillary electrophoresis.
offering selection of ion mode (positive/negative), MS
type (MS
1
, MS
2
, MS
3
, MS
4
) and source introduction
(LC, GC, capillary electrophoresis (CE)). The search
filters are represented as tabs where the header is the
name of the filter, and the available options that can
be selected are radio buttons (only one of the offered
options is allowed), or, checkboxes (one or more of
the offered options are allowed). Since we already
described the different contents the .csv file might
contain, the user has to select the type of input data:
InChiKey, molecular formula or exact mass. Selec-
tion of exact mass as input data triggers another input
number field requiring the user to define the mass dif-
ference tolerance level of the exact mass variation (in
Da);
(3) Upload file section, which allows attachment of
a .csv file with a list of compound names, molecu-
lar formulae, InChIKeys or molecular masses of com-
pounds for which MS data is required. The file upload
is performed by clicking the Browse button, which al-
lows attachment of files locally stored on the device.
After selecting the file, the window closes automati-
cally and the name of the selected file is written in the
Upload File tab and thus is visible for the user.
Furthermore, MSL-ST implements validation of
the attached file, approving attachment only of files
with .csv or .txt extension. This feature warns the user
fo attaching inappropriate file format by displaying an
error message in a warning window.
After uploading and selecting the desired filter op-
tions, by clicking the Search button, the request is
submitted and the data retrieval process begins, af-
ter which a file is automatically generated and down-
loaded (Fig. 8). In case the file is not in the appro-
priate format, and the source is not selected from the
Mass Spectral Library filter, this button remains dis-
abled.
After clicking Search, a POST request is sent and
a message for successful processing is displayed in
a warning window. This means that an HttpRequest
object is passed from the displayed template as an ar-
gument in the function of the view pointed from the
URL. The view expects an HttpResponse that will be
returned after executing the function for data retrieval.
As soon as the OK button is clicked, all filters are
disabled and a spinner is displayed, while the request
is processed and an output file is generated.
The data retrieval process, as shown in Fig. 5
starts by scrolling through all the input items, and
invoking the corresponding function from additional
helpers file in the system. Input arguments are the
values of selected filters from the application (ION
MODE, MS TYPE, SOURCE INTRODUCTION, LI-
BRARY), while the other arguments are initialized
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
198
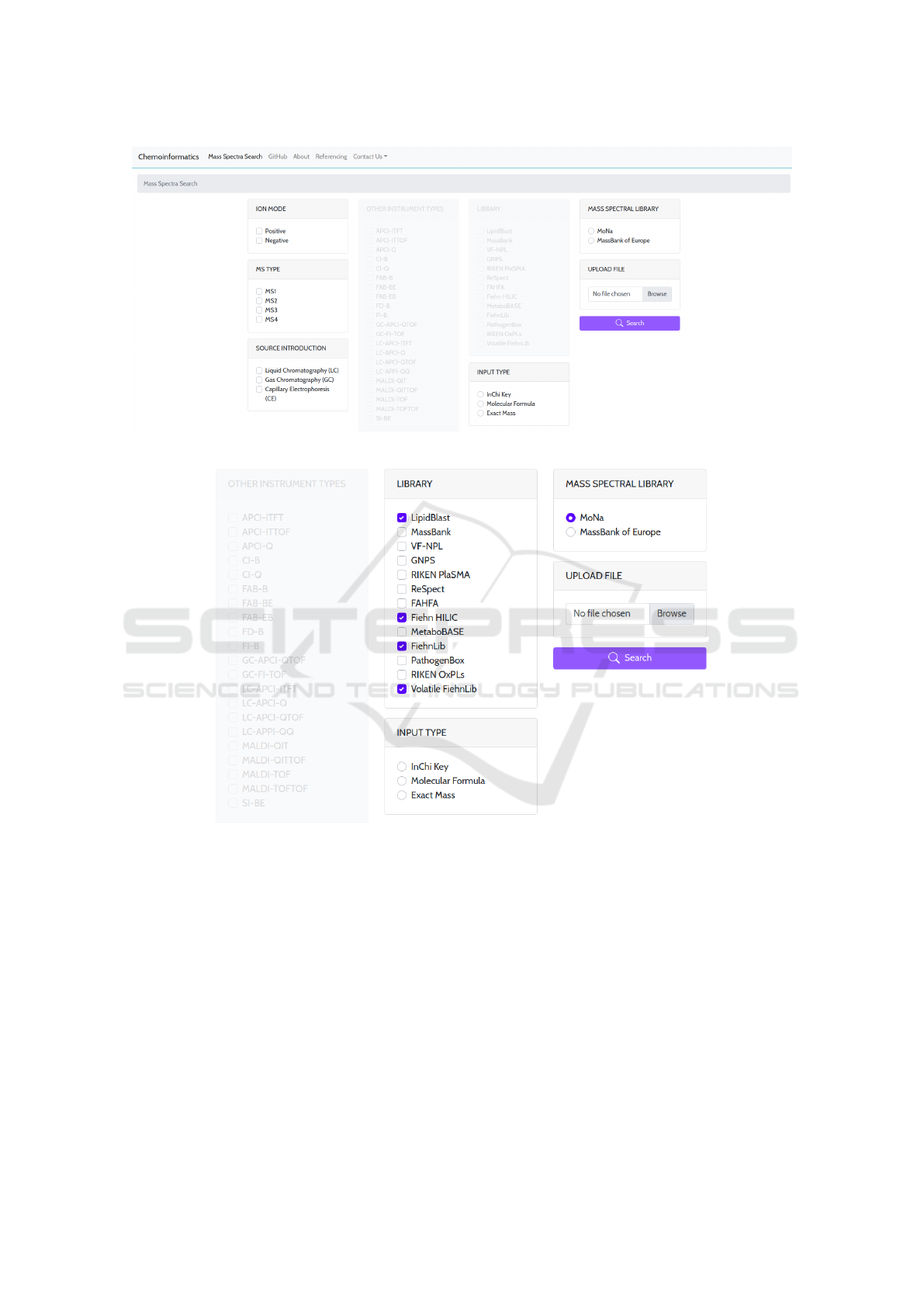
Figure 1: MSL-ST web interface.
Figure 2: MSL-ST with MoNa selected as source MSL.
to an empty list. Using the selected filters, a URL
where querystring passes the values, is generated and
a GET request is sent to it. The response of the request
contains list of ids from all of the compounds whose
metadata is contained in the selected MSL, that fulfil
the selected parameters.
Using the output arguments of the function, it is
iterated through each of the received IDs, and a GET
request is sent to each of the generated URLs from
the ID. The response contains all the data for the
given compound (name, exact mass, molecular for-
mula, molecular descriptors etc.) and the metadata
corresponding to instrument part and conditions, as
shown in Fig. 5. Since the response is in JSON for-
mat, using build-in functions of data structure objects
eases the process of parsing the data. Additionally,
formatting of the retrieved data in an easy-to-use out-
put .csv format is done before download (Fig. 8).
The output .csv file, where the header contains
the metadata/parameters name, molecular formula,
ions, etc.). The described data retrieval procedure is
repeated for all compounds from the input file and the
results are added to the output file accordingly. As
soon as the data retrieval process is finished for all
input items that have been iterated, the Django view
that has received the POST HttpRequest as an input
argument, returns the response as an output argument
of the function. After receiving the response from
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification
199

Figure 3: MSL-ST with MassBank of Europe selected as source MSL.
Figure 4: Obtained metadata for a given compound.
the POST request, when the output file is ready to be
downloaded, this option becomes available and easily
visible to the user as shown in Fig. 6. In real time,
the MS data for all MS entries of the selected MSL
will be downloaded and written in the output file. As
shown in Fig.7, it contains all the metadata available
in both MSLs.
Figure 5: UML diagram for the POST request.
Apart from the MSLs search engine, the navi-
gation menu of MSL-ST’s interface provides about
page with introduction to the project aim, mass spec-
trometry and organic compound identification, a con-
tent aimed to aid the user’s experience while utilizing
MSL-ST. A link to the GitHub repository and author’s
contact information are also provided (Fig. 8).
In this way, MSL-ST allows time-effective re-
trieval of large data quantities, replacing multiple
weeks of manual MS data search and export with few
minutes of automated gathering of MS data for thou-
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
200
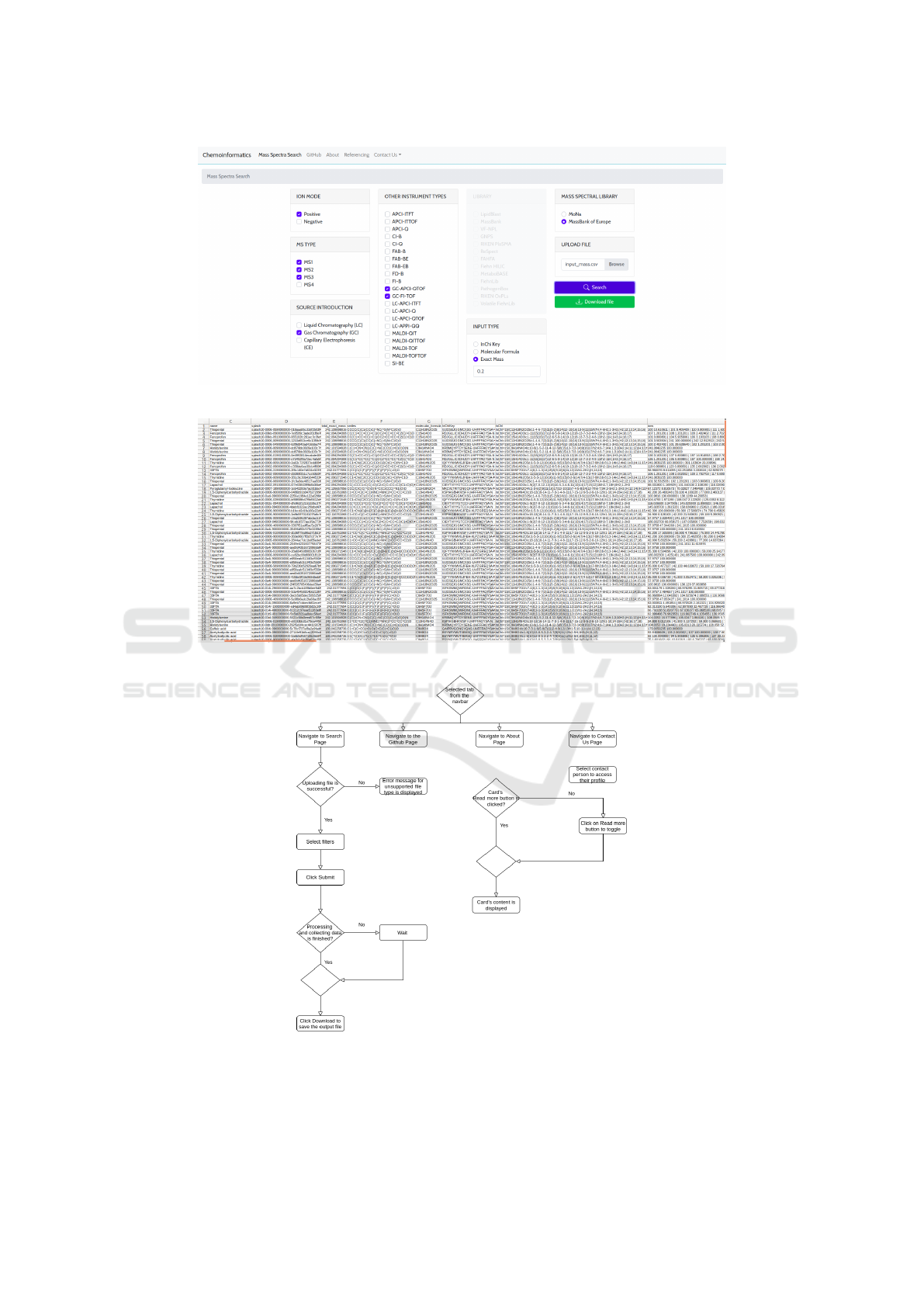
Figure 6: Data ready to be downloaded.
Figure 7: Several columns from the generated output file containing retrieved metadata for the retrieved spectra.
Figure 8: Path diagram of MSL-ST.
sands of compounds. As MSL-ST is the first batch
MSLs search and export engine available, no com-
parative analysis with similar tools can be performed.
Due to the lack of batch search option in MassBank
of Europe and MoNa, comparison with single MSL
batch search and export is also not possible.
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification
201

5 CONCLUSION
The development of MSL-ST, the first publicly avail-
able MS data batch search and export engine using
two of the most comprehensive MSLs is pivotal step
in MSL-chemoinformatics-based compound identifi-
cation. By offering a time-, cost- and labour-effective
solution for data extraction that can be easily im-
plemented in custom-made workflows, MSL-ST is
clearly addressing three of the current challenges of
chemoinformatics, that are: (1) centralization of mul-
tiple MSLs and uniformation of searching through the
empirical data they contain; (2) enabling search and
retrieval of batch data, instead of manually repeat-
ing the process, and (3) automated download of com-
pound data in a structured tabular format, instead of
time-consuming manual storage.
Since MSL-ST is the first MSLs batch search
and export engine, it can be easily assumed that
the idea of developing such tools in order to aim
chemoinformatics-assisted compound identification
is in its earliest infancy and thus, many more advance-
ments are to be added to the basic functionalities of
MSL-ST available in its first version, presented in this
paper. Consequently, its further upgrades would in-
clude:
• Addition of more publicly available MSLs, that
would allow access to larger amount of exepri-
mental and metadata, thus spreading the capabili-
ties of the MSL(s)-based chemoinformatics tools;
• The MS Type filter which indicates the number
and type of mass spectrometers used to generate
MS, and
• The Source Introduction filter that allows the se-
lection of the type of chromatography, whether it
is gas chromatography, liquid chromatography, or
capillary electrophoresis.
This would allow access to a large number of ex-
perimental data on compounds of different species
(metabolites, peptides, etc.). The process of real-
time data extraction would be expanded by searching
through other structured databases, using them as a
”living resource” that is updated daily.
The web application provides an easy way to se-
lect the characteristics of the mass spectrometry of the
whole pile of input compounds, which will be applied
in the search in the selected MSL. There is still room
for future work in improving the interface of the web
application, which would result in a better user expe-
rience and easier use of the application.
REFERENCES
DeBill, E. (2010). Module counts. WWW], http://www.
modulecounts. com/.[Haettu 1.11. 2016.].
Guijas, C., Montenegro-Burke, J. R., Domingo-Almenara,
X., Palermo, A., Warth, B., Hermann, G., Koel-
lensperger, G., Huan, T., Uritboonthai, W., Aisporna,
A. E., et al. (2018). Metlin: a technology platform for
identifying knowns and unknowns. Analytical chem-
istry, 90(5):3156–3164.
Holovaty, A. and Kaplan-Moss, J. (2009). The defini-
tive guide to Django: Web development done right.
Apress.
Horai, H., Suwa, K., Arita, M., Nihei, Y., and Nishioka,
T. (2020 (accessed November 16, 2020)). Massbank:
Mass spectral database for metabolome analysis. In
The 56th ASMS Conference on Mass Spectrometry
and Allied Topics, Denver, CO.
Hummel, J., Selbig, J., Walther, D., and Kopka, J. (2007).
The golm metabolome database: a database for gc-ms
based metabolite profiling. In Metabolomics, pages
75–95. Springer.
Hummel, J., Strehmel, N., Selbig, J., Walther, D., and
Kopka, J. (2010). Decision tree supported substruc-
ture prediction of metabolites from gc-ms profiles.
Metabolomics, 6(2):322–333.
Kind, T., Wohlgemuth, G., Lee, D. Y., Lu, Y., Pala-
zoglu, M., Shahbaz, S., and Fiehn, O. (2009). Fiehn-
lib: mass spectral and retention index libraries for
metabolomics based on quadrupole and time-of-flight
gas chromatography/mass spectrometry. Analytical
chemistry, 81(24):10038–10048.
Ljoncheva, Milka and Stepi
ˇ
snik, Toma
ˇ
z and D
ˇ
zeroski, Sa
ˇ
so
and Kosjek, Tina (2020). Cheminformatics in MS-
based environmental exposomics: Current achieve-
ments and future directions. Trends in Environmental
Analytical Chemistry, page e00099.
McLafferty, F. W. and Stauffer, D. (2009). Wiley registry of
mass spectral data, volume 662. John Wiley Hobo-
ken, NJ.
Mehta, S. (2020 (accessed November 16, 2020)). Mass-
bank of north america (mona): An open-access, auto-
curating mass spectral database for compound identi-
fication in metabolomics presentation.
NIST: National Institue of Standard and Technology (2020
(accessed November 16, 2020)). The nist mass spec-
trometry data center.
Rasche, F., Scheubert, K., Hufsky, F., Zichner, T., Kai, M.,
Svato
ˇ
s, A., and B
¨
ocker, S. (2012). Identifying the un-
knowns by aligning fragmentation trees. Analytical
chemistry, 84(7):3417–3426.
Sawada, Y., Nakabayashi, R., Yamada, Y., Suzuki, M.,
Sato, M., Sakata, A., Akiyama, K., Sakurai, T.,
Matsuda, F., Aoki, T., et al. (2012). Riken tan-
dem mass spectral database (respect) for phytochem-
icals: a plant-specific ms/ms-based data resource and
database. Phytochemistry, 82:38–45.
Scheubert, K., Hufsky, F., and B
¨
ocker, S. (2013). Computa-
tional mass spectrometry for small molecules. Journal
of cheminformatics, 5(1):12.
BIOINFORMATICS 2021 - 12th International Conference on Bioinformatics Models, Methods and Algorithms
202

Schymanski, E. L., Meinert, C., Meringer, M., and Brack,
W. (2008). The use of ms classifiers and structure
generation to assist in the identification of unknowns
in effect-directed analysis. Analytica chimica acta,
615(2):136–147.
Wei, J. N., Belanger, D., Adams, R. P., and Sculley, D.
(2019). Rapid prediction of electron–ionization mass
spectrometry using neural networks. ACS central sci-
ence, 5(4):700–708.
Wishart, D. S., Tzur, D., Knox, C., Eisner, R., Guo, A. C.,
Young, N., Cheng, D., Jewell, K., Arndt, D., Sawh-
ney, S., et al. (2007). Hmdb: the human metabolome
database. Nucleic acids research, 35(suppl 1):D521–
D526.
MSL-ST: Development of Mass Spectral Library Search Tool to Enhance Compound Identification
203
