
An Effective Resistance based Genetic Algorithm for Community
Detection
Clara Pizzuti
a
and Annalisa Socievole
b
National Research Council of Italy (CNR), Institute for High Performance Computing and Networking (ICAR),
Via Pietro Bucci, 8-9C, 87036 Rende (CS), Italy
Keywords:
Community Detection, Genetic Algorithm, Effective Resistance, Moore-Penrose Pseudoinverse.
Abstract:
This work presents a new approach based on genetic algorithms (GAs) and the concept of effective resistance
for detecting communities within an undirected graph. The method considers the equivalent electric network
of the input graph, where edges are weighted with their effective resistance, a measure of electrical resistance
between nodes, whose square root has been shown to be a Euclidean metric. The algorithm computes the
similarity between nodes by using the effective resistance values and generates a weighted and sparse graph
by adopting a thresholding sparsification strategy based on the nearest neighbors of each node. Experiments
over synthetic and real-world networks demonstrate the effectiveness of our approach when compared to other
benchmark methods.
1 INTRODUCTION
The community detection problem in complex net-
works plays a fundamental role in several fields, in-
cluding telecommunications, sociology, physics, and
biology. Many real-world systems, in fact, can be rep-
resented through networks where nodes correspond to
the objects of the system and edges model the rela-
tionships between such objects. The task of finding
communities in networks has attracted a lot of atten-
tion in the last years since the presence of commu-
nities indicates the existence of group organization
which could be interesting to uncover for a better un-
derstanding of the system.
The problem of finding a community structure
can be generally formalized as an optimization prob-
lem where a criterion function, catching the intu-
itive concept of group, is defined and optimized. To
date, many community detection algorithms, based
on different approaches, such as consensus clus-
tering, spectral methods, statistical inference-based,
optimization-based, dynamics-based methods, have
been proposed (Fortunato and Hric, 2016).
Real-world networks generally have a very high
density of edges and, often, their edges are weighted
(Barrat et al., 2004). As observed in (Yan et al., 2018),
a
https://orcid.org/0000-0001-7297-7126
b
https://orcid.org/0000-0001-5420-9959
analyzing these networks with computational meth-
ods is very hard, thus preprocessing techniques need
to be applied in order to reduce the number of con-
nections and make the problem tractable.
An approach to eliminate edges is graph sparsifi-
cation (Tumminello et al., 2005; Radicchi et al., 2011;
Spielman and Srivastava, 1996). In particular, (Spiel-
man and Srivastava, 1996) proposed to build a sparse
graph H from the original one G by including in H
edges of G with a probability proportional to their ef-
fective resistance.
The effective resistance (Klein and Randi
´
c, 1993;
Ghosh et al., 2008) of an edge is equal to the proba-
bility that the edge appears in a random spanning tree
of G (Doyle and Snell, 1989), and it was proven to
be proportional to the commute-time between its end-
points (Chandra et al., 1996). Moreover, (Klein and
Randi
´
c, 1993) showed that the square root of the ef-
fective resistance between any couple of nodes (i, j)
is a Euclidean metric, in particular it measures the dis-
tance between nodes i and j. Thus, the computation
of the effective resistance for each edge of G provides
a distance matrix between each couple of nodes of G.
In this paper, given a network G = (V, E), where
V is the set of nodes and E of edges, we propose an
evolutionary algorithm to detect communities by ex-
ploiting the concept of effective resistance. The main
idea is to apply a Genetic Algorithm (GA) (Goldberg,
1989) to find communities on the weighted graph
28
Pizzuti, C. and Socievole, A.
An Effective Resistance based Genetic Algorithm for Community Detection.
DOI: 10.5220/0010644300003063
In Proceedings of the 13th International Joint Conference on Computational Intelligence (IJCCI 2021), pages 28-36
ISBN: 978-989-758-534-0; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

G
0
= (V,W ) obtained from G by computing the ef-
fective resistance of all the node pairs of G. G
0
has
the same set V of nodes of G, while the set W of
edges consists of the effective resistance between any
couple of nodes (i, j) of V . However, the adjacency
matrix Ω corresponding to G
0
is a full matrix, thus
a sparsification procedure is necessary to reduce the
number of edges and make clear the original network
structure.
The simplest way to obtain a sparse graph is the
weight thresholding (Yan et al., 2018), i.e. removing
the edges whose weight is above a fixed threshold.
Deleting as many edges as possible without altering
the original system is a key point.
Our approach considers for each node i only a
fixed number nn of the most similar nodes and re-
moves from Ω all the edges between i and the nodes
not included in this set of nn nearest neighbors to ob-
tain a sparse weighted adjacency matrix
e
Ω. We there-
fore run the GA over the sparsified weighted adja-
cency matrix
e
Ω by evolving a population of individ-
uals and minimizing the concept of modularity of a
partition, the most popular quality function of com-
munity structure introduced by Girvan and Newman
(Girvan and Newman, 2002).
A comparison with a baseline genetic algorithm
optimizing modularity and running on the original
graph G, along with two of the best-known commu-
nity detection methods Louvain (Blondel et al., 2008)
and Infomap (Rosvall and Bergstrom, 2008) shows
that the proposed algorithm obtains results better than
the contestant methods both over synthetically gener-
ated and real-world networks.
The paper is organized as follows. The next sec-
tion describes some measures proposed for comput-
ing node similarity and how these measures are used
for the community detection task. Section 3 recalls
the concept of effective resistance, defines the com-
munity detection problem we tackle and describes our
method. Section 4 describes the datasets used and the
experiments performed to validate the proposed algo-
rithm. Finally, Section 5 concludes the paper and dis-
cusses the future directions.
2 RELATED WORK
Distances and similarity measures between the nodes
of a graph are widely used in data analysis and es-
pecially in clustering tasks. Many measures have
been proposed so far including the widely investi-
gated Shortest Path distance (Dijkstra et al., 1959),
the Resistance (Klein and Randi
´
c, 1993), the loga-
rithmic Walk measure, the Forest measure related to
Resistance, and many others (Deza and Deza, 2009)
(Avrachenkov et al., 2017).
In their pioneering work, Klein and Randic (Klein
and Randi
´
c, 1993) proposed the use of the effective
resistance, also named resistance distance, between
two nodes as a meaningful distance measure. Indeed,
it has been shown that this measure is a Euclidean
distance.
The close link between the effective resistance and
the commute-time of a random walker on a graph
has been studied in (Chandra et al., 1996). More-
over, the relationship between the Laplacian matrix
of the graph and the commute-time was investigated
in (Saerens et al., 2004). For this reason, the effective
resistance is also named commute-time distance.
In (Avrachenkov et al., 2019), a set of similar-
ity measures on graphs based on three fundamental
graph matrices, the adjacency matrix, the Laplacian
matrix, and the stochastic Markov matrix are analyt-
ically studied and compared. For each measure, the
work investigates if it is (a) a kernel, (b) a proximity
measure, and (c) a transitional measure.
In (Yen et al., 2007), the commute-time kernel
is used for clustering the nodes of a weighted undi-
rected graph. The method is based on a two-step pro-
cedure that initially computes the sigmoid commute-
time kernel matrix from the adjacency matrix of the
graph, providing a similarity measure between nodes,
and then, clusters the nodes by exploiting a kernel-
based k-means or fuzzy k-means on the obtained ker-
nel matrix. The proposed methodology combining
commute-time kernel and kernel clustering outper-
forms standard k-means, as well as spectral cluster-
ing, on a difficult graph clustering problem.
A comprehensive study on graph nodes clustering
with the sigmoid commute-time kernel can be found
in (Yen et al., 2009). In (Sommer et al., 2016), six dif-
ferent distance measures are transformed into kernels
and tested on kernel k-means and a weighted version
of it. A comparison with the Louvain method shows
the effectiveness of the distance-based algorithms.
The impact of network topology on the efficiency
of proximity measures for community detection is in-
vestigated in (Aynulin, 2019). Specifically, the work
checks wether the advantage of using one measure
is kept for different network topologies. The au-
thors showed that there are measures behaving well
for most topologies.
In the next section the concept of effective resis-
tance metric is recalled and how it is exploited for de-
tecting effective community structure is described.
An Effective Resistance based Genetic Algorithm for Community Detection
29

3 COMMUNITY DETECTION
BASED ON EFFECTIVE
RESISTANCE
In this section, we first introduce the concept of ef-
fective resistance as distance metric and then describe
the proposed method.
3.1 Effective Resistance
Klein and Randic, in their seminal work (Klein and
Randi
´
c, 1993), proposed the theory of resistive elec-
trical networks to define a new distance function be-
tween vertices. They suggested that if fixed resis-
tors are assigned to the edge of a connected graph,
the effective resistance between couples of nodes is a
graphical distance.
Given an undirected and connected graph G =
(V,E), an equivalent electric network can be associ-
ated with G by weighting each edge (i, j) ∈ E with
positive weights w
i j
representing the conductance, i.e.
the inverse of the electrical resistance ω
i j
of a resistor,
so that ω
i j
=
1
w
i j
ohm (Klein and Randi
´
c, 1993). As
described in (Klein and Randi
´
c, 1993; Van Mieghem
et al., 2017), for any edge of the graph G, a distance
function can be defined as follows.
The effective resistance ω
i j
between any pair of
nodes i and j is defined as the voltage developed be-
tween i and j when a unit current is injected at node i
and is withdrawn at node j. The corresponding N ×N
matrix including all the ω
i j
between each node pair i
and j is denoted Ω.
The interesting feature is that ω
i j
is upper
bounded by the shortest path distance in a graph
(Van Mieghem, 2010). Moreover, the communte-time
distance C
i j
between two nodes i and j, i.e. the ex-
pected number of steps needed during a random walk
from i to j, is C
i j
= u
T
e
Au ω
i j
, where u is the all
one vector and u
T
e
Au is the double of the sum of all
the edge weights in the weighted adjacency matrix
e
A
(Chandra et al., 1996).
In (Klein and Randi
´
c, 1993), it is shown that the
square root
√
ω
i j
of the effective resistance is a Eu-
clidean metric. More precisely, the effective resis-
tance matrix Ω is a distance matrix, in which for any
triple of non-negative elements, ω
ii
= 0, and the tri-
angle inequality, ω
i j
≤ ω
ik
+ ω
k j
is satisfied.
Moreover, Ω can be defined (Doyle and Snell,
1989; Klein and Randi
´
c, 1993; Van Mieghem, 2010)
as
Ω = ζu
T
+ uζ
T
−2L
+
(1)
where the vector
ζ =
L
+
11
,L
+
22
,.. .,L
+
NN
(2)
contains the diagonal elements of the Moore-Penrose
pseudoinverse matrix L
+
of the weighted Laplacian
matrix
e
L of the graph G.
Recall that, given ∆ = diag(d
i
) the N ×N diago-
nal degree matrix, where d
i
=
∑
N
j=1
a
i j
, the Laplacian
matrix L of the graph G is defined as the N ×N sym-
metric matrix L = ∆ −A, with elements
l
i j
=
d
i
if i = j
−1 if the edge (i, j) ∈ E
0 otherwise
(3)
The effective resistance between two nodes x and
y equals
ω
xy
= (e
x
−e
y
)
T
L
+
(e
x
−e
y
) = l
+
xx
+ l
+
yy
−2l
+
xy
(4)
where e
k
is the basic vector with the m-th component
equal to (e
k
)
m
= δ
mk
and δ
mk
is the Kronecker-delta:
δ
mk
= 1 if m = k, otherwise δ
mk
= 0.
3.2 Problem Definition
Given a graph G, let Ω be the resistance matrix as-
sociated with G, nn the number of nearest neighbors
to consider, and
e
Ω the matrix obtained from Ω with
elements
e
ω
xy
=
ω
xy
if y is among the nn
nearest neighbors of x
0 otherwise
(5)
The community detection problem is defined as:
find a partition C = {C
1
,...,C
k
}of the nodes of G such
that the weighted modularity of C is maximized.
The weighted modularity Q is computed as
Q =
1
2m
∑
i j
e
ω
i j
−
k
i
k
j
2m
δ(C
i
,C
j
) (6)
where m is the sum of the edge weights, k
i
and k
j
the sum of the weights of the edges attached to nodes
i and j respectively, and δ is the Kronecker function
which yields one if i and j are in the same community,
i.e. C
i
= C
j
, zero otherwise. Modularity measures the
expected number of edges within the communities of
a random graph with the same degree distribution.
Basically, we look for a community structure
where both intra-community weighted modularity
and similarity between nodes is high (thus the overall
distance between the nodes of the same community is
low).
3.3 Method Description
In this section, a detailed description of the method is
given. OmeGAnet is a method based on Genetic Algo-
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
30

rithms (GA) (Goldberg, 1989), an evolutionary com-
putation technique which revealed very efficacious
for the task of community detection (Pizzuti, 2018).
The algorithm creates a population of individuals
(i.e. a network division in communities) that are ini-
tially randomly generated, and then evolves the pop-
ulation through variation and selection operators by
optimizing the value of the objective function while
exploring the search space.
Each individual is represented with the locus-
based adjacency representation (Park and Song,
1998) for which an individual I is a vector of N genes
(i.e. nodes). Each gene can assume a value k from 1
to N. When a value k is assigned to the i-th gene, it
means that nodes i and k are connected. A decod-
ing step identifies all the connected components of
the graph which correspond to the network division
in communities.
As crossover operator, OmeGAnet exploits the
uniform crossover which generates a random binary
mask of length equal to the number of nodes. The
offspring is then generated by selecting from the first
parent the genes where the mask is 0, and from the
second parent the genes where the mask is 1.
Finally, the mutation operator randomly assigns
the value of a i-th gene to one of its neighbors.
The steps performed by OmeGAnet are the fol-
lowing. It receives in input the graph G = (V,E), the
number of nearest neighbors nn to consider, and per-
forms the following steps:
1. compute the Laplacian L of G;
2. compute the Moore-Penrose pseudoinverse ma-
trix L
+
of L;
3. compute the effective resistance matrix Ω from
L
+
as ω
xy
= l
+
xx
+ l
+
yy
−2l
+
xy
;
4. make Ω a distance matrix by substituting each el-
ement as ω
xy
=
√
ω
xy
;
5. normalize the elements of Ω and make Ω a sym-
metric matrix Ω = 0.5[
Ω
max(Ω)
+ (
Ω
max(Ω)
)
T
];
6. generate the sparse similarity weighted matrix
e
Ω
from Ω by maintaining for each node x only the nn
entries ω
xy
having the minimum distance value,
i.e the connections with the nodes y having the
highest similarity values with x;
7. run the Genetic Algorithm on
e
Ω for a number of
iterations by using modularity as fitness function
to maximize, uniform crossover and neighbor mu-
tation as variation operators;
8. obtain the partition C = {C
1
,.. .,C
k
} correspond-
ing to the solution with the highest fitness value.
Table 1: LFR-128 parameters setting.
Parameter Value
Number of nodes 128
Node average degree 8
Node maximal degree 9
Exponent for power law creating degree sequence 2
Exponent for power law creating community sizes 1
Mixing parameter µ [0.1; 0.6]
Maximal community size 40
Minimal community size 20
Average density 0.062
In the next section, we compare OmeGAnet with
three community detection methods and compare
the results they obtain on synthetic and real-world
datasets.
4 EXPERIMENTAL EVALUATION
For validating OmeGAnet, we performed several sim-
ulations using Matlab 2020a and the Global Opti-
mization Toolbox.
Regarding the input parameters to our approach,
we experimentally set them by performing a trial and
error procedure.
In the following subsections we describe the
datasets, the evaluation measures used, the algorithms
in comparison and finally the results obtained.
4.1 Datasets
4.1.1 Synthetic Networks
To create synthetic networks with realistic commu-
nity structures, the Lancichinetti-Fortunato-Radicchi
(LFR) benchmark (Lancichinetti et al., 2008) has
been used.
This well-known and widely used network gen-
erator is able to control the structure of the commu-
nities by properly setting the mixing parameter µ.
The lower the µ, the clearer the resulting community
structure with much more intra-community links than
inter-cluster links. On the contrary, when µ has high
values, the community structure is not clear.
The parameters used for generating the LFR net-
works are shown in Table 1. In particular, for each µ
value we generated 10 network instances.
4.1.2 Real-world Networks
We considered four real-world networks for which the
ground-truth division is known.
• Zachary Karate Club. This well-known dataset
contains the data of the friendship social network
An Effective Resistance based Genetic Algorithm for Community Detection
31

(a) (b)
Figure 1: (a) A 128-nodes LFR network generated with µ = 0.2 and (b) its reduced graph with nn = 5.
of 34 members of a karate club, collected by
Wayne Zachary in 1977. The group has been ob-
served for two years, splitting in two communities
almost of the same size due to disagreements be-
tween the members.
• Amazon US Politics Books. The second dataset
contains the data of books co-purchased on Ama-
zon.com by customers during the US political
elections in 2004. The network was collected by
Krebs and contains 105 nodes and 374 edges. The
books were later classified into the three political
groups liberal, neutral and conservative by New-
man.
• American College Football. This dataset con-
tains the network of 115 American football teams
extracted from the Fall 2000 regular season
games. The teams were linked during a game with
a resulting number of 616 edges/games and parti-
tioned in 12 communities.
• Bottlenose Dolphins. The last dataset con-
tains the network of 62 Bottlenose dolphins from
Doubtful Sound in New Zealand. Linked through
159 edges, the edges represent frequent associa-
tions observed between dolphins. Here, the dol-
phins are partitioned into two communities.
4.2 Evaluation Measure
We evaluate the quality of the solutions by using the
Normalized Mutual Information (NMI).
Given two divisions A and B of a network, and C
the confusion matrix whose element C
i j
is the number
of nodes of community i of the partition A that are
also in the community j of the partition B, the NMI
between A and B is defined as:
NMI(A,B) =
−2
∑
c
A
i=1
∑
c
B
j=1
C
i j
log(C
i j
n/C
i.
C
. j
)
∑
c
A
i=1
C
i.
log(C
i.
/n) +
∑
c
B
j=1
C
. j
log(C
. j
/n)
(7)
where c
A
(c
B
) is the number of groups in the par-
tition A (B), C
i.
(C
. j
) is the sum of the elements of C
in row i (column j), and n is the number of nodes.
If A = B, NMI(A,B) = 1. If A and B are completely
different, NMI(A,B) = 0.
4.3 Algorithms in Comparison
We first compare OmeGAnet with a baseline GA-
based algorithm, denoted GA-mod, adopting the same
locus-based representation, initialization, crossover
and mutation operators which optimizes the modular-
ity value. In particular, population initialization con-
nects a node with one of its nn nearest neighbors, in-
stead of a random neighbor.
We also compare OmeGAnet to two benchmarks:
Louvain (Blondel et al., 2008) and Infomap (Rosvall
and Bergstrom, 2008).
The Louvain method is based on a greedy mod-
ularity optimization approach. First, the algorithm
identifies small communities by locally optimizing
modularity. Then, it builds a new network whose
nodes are the communities previously found, and
these steps are repeated until a hierarchy of high-
modularity communities is obtained.
Infomap exploits the principles of information the-
ory by defining the community detection problem as
the problem of finding a description of minimum in-
formation of a random walk on the graph. The method
maximizes the Minimum Description Length as ob-
jective function by quickly providing an approxima-
tion of the optimal solution.
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
32
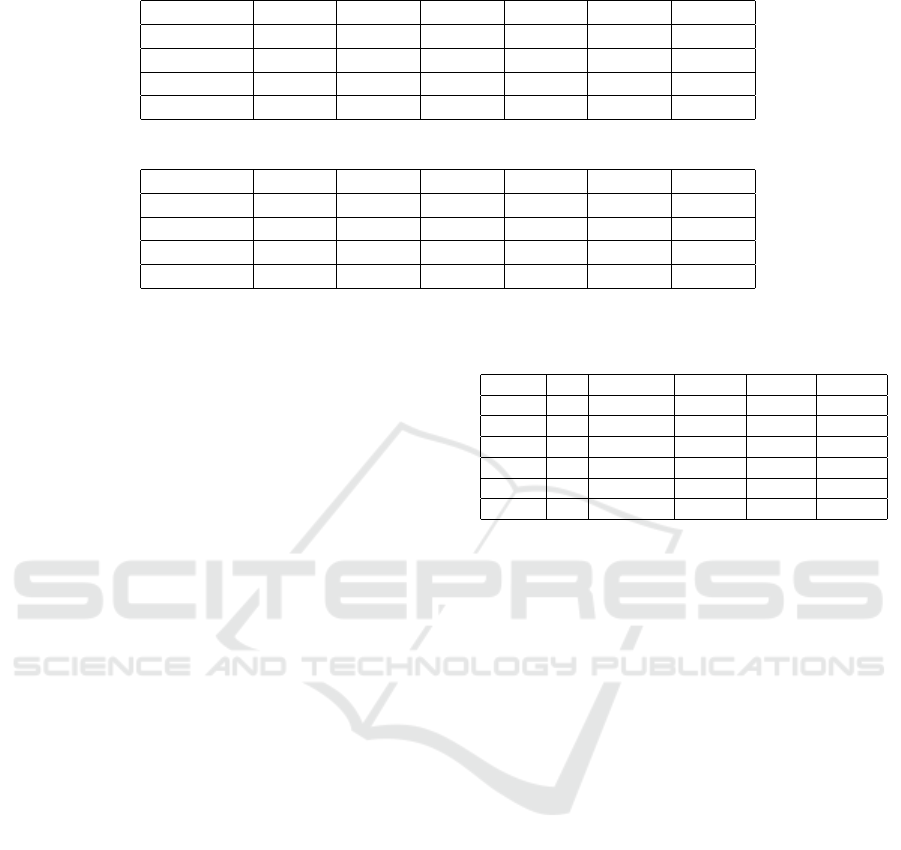
Table 2: NMI results for the LFR-128 networks.
µ = 0.1 µ = 0.2 µ = 0.3 µ = 0.4 µ = 0.5 µ = 0.6
OmeGAnet 1 1 0.8049 0.3308 0.1314 0.0698
GA-mod 1 1 0.6967 0.2988 0.117 0.06
Louvain 0.8793 0.8716 0.5555 0.3228 0.0865 0.055
In f omap 1 1 0.6728 0.2875 0.1162 0.0624
Table 3: Modularity results for the LFR-128 networks.
µ = 0.1 µ = 0.2 µ = 0.3 µ = 0.4 µ = 0.5 µ = 0.6
OmeGAnet 0.7038 0.5446 0.3983 0.3308 0.2999 0.2854
GA-mod 0.7038 0.5446 0.3908 0.3104 0.3223 0.3035
Louvain 0.6268 0.4661 0.3733 0.314 0.3356 0.3086
In f omap 0.7038 0.5446 0.4101 0.2875 0.3386 0.3289
4.4 Results
The first experiment has been carried out on a pool of
synthetic network with 128 nodes.
Figure 1 shows an instance of a network gener-
ated with a mixing parameter µ = 0.2. Figure 1 (a)
shows the community structure of the ground-truth
composed by four communities colored in red, green,
magenta and blue. Figure 1 (b), shows the reduced
graph with nn=5, the value for this parameter able to
produce the better results.
It is worth pointing out that, by considering the
subset of neighbors having the highest similarity (i.e.
the lowest distance), the underlying communities are
more visible and the structure is clearer. Here, the
few interlinks between communities make, for exam-
ple, more visible the communities in green and in red.
Table 2 shows the NMI results for the LFR-128 net-
works.
For the genetic algorithms OmeGAnet and GA-
mod we have set maximum number of generations
100, population size 700, nn=5, crossover fraction 0.9
and mutation rate 0.1. Each value has been averaged
over 10 runs of the method. For communities with
clear structure (µ = 0.1 and µ = 0.2), the two genetic
algorithms and Infomap match the ground-truth cor-
rectly identifying the underlying communities.
Louvain, on the contrary, achieves only 0.8793
and 0.8716 for µ = 0.1 and µ = 0.2, respectively.
As µ increases and the structure of the communi-
ties changes becoming less clear, OmeGAnet always
outperforms all the other contestant methods. From
µ = 0.3 to µ = 0.6 the NMI significantly decreases
achieving very low values for µ = 0.6.
In Table 3, the modularity values are reported. For
µ = 0.1 and µ = 0.2 the modularity value achieved
with NMI=1 is 0.7038, as can be observed for
OmeGAnet, GA-mod, and Infomap. For µ = 0.3,
µ = 0.5 and µ = 0.6 the highest modularity is ob-
Table 4: Number of communities for the LFR-128 net-
works.
GT OmeGAnet GA-mod Louvain Infomap
µ = 0.1 4 4 4 5 4
µ = 0.2 4 4 4 3 4
µ = 0.3 4 4 5 5 7
µ = 0.4 4 4 7 7 9
µ = 0.5 4 4 7 7 6
µ = 0.6 4 4 7 7 9
tained by In f omap while for the other approaches op-
timizing modularity the highest value is obtained by
OmeGAnet for mu = 0.4.
We point out that our focus is to obtain the high-
est NMI value for the community partition found and
this not always corresponds to the highest modularity
value as can be observed.
In Table 4, we finally show the number of com-
munities obtained for a single run of a network. In
the second column, the number of classes for the
ground- truth (GT) is reported. For each µ the ground-
truth is composed by 4 communities. OmeGAnet is
always able to split the network in 4 groups while
the other schemes fragment the communities result-
ing in a higher number of communities from µ = 0.3
to µ = 0.6. For µ = 0.4, for example, GA-mod and
Louvain find 7 communities, while Infomap even 9.
In the second experiment, we tested OmeGAnet
on the real-world networks described above for which
the true number of communities is known.
Table 5 shows the NMI results obtained by setting
for the genetic algorithms maximum number of gen-
erations 100, population size 500, nn = 4, crossover
fraction 0.9 and mutation rate 0.1.
In Tables 6 and 7 the modularity and the number
of communities are reported. Also for this experi-
ment, the NMI and the modularity values are aver-
aged over 10 runs while the number of communities
refers to a single run.
In terms of NMI, OmeGAnet outperforms the
An Effective Resistance based Genetic Algorithm for Community Detection
33

Figure 2: The Zachary Karate Club (a) and its reduced graph with nn = 4 (b).
Figure 3: The Bottlenose Dolphins (a) and its reduced graph with nn = 4 (b).
other contestant methods also in the real-world net-
working scenarios considered. For Karate, for ex-
ample, the two communities are correctly identified
while GA-mod achieves only 0.5485, Louvain 0.5195
and Infomap 0.6995 of NMI.
Figure 2 shows the Karate original graph (a) and
its reduced version (b) where we can observe that the
communities are much more clear than in the original
network. Also for the Dolphins dataset, OmeGAnet
significantly outperforms the other methods.
Figure 3 shows how the reduction of the initial
graph better separates the communities. The modular-
ity results, show again that a high modularity does not
correspond always to a high NMI. In fact, for Karate,
for example, the modularity value giving the ground-
truth is the one found by OmeGAnet, 0.3715, which
is the lowest.
Looking at the number of communities, it is worth
pointing out that OmeGAnet matches the number of
underlying communities in most of the cases. The
other algorithms, on the contrary, produce partitions
with a higher number of communities. For Dolphins,
for example, where the network is divided into two
groups, GA-mod, Louvain and Infomap find 4, 10 and
6 communities, respectively.
Table 5: NMI results for the real-world networks with max-
Gen=100, popSize=500 and nn=4.
OmeGAnet GA-mod Louvain Infomap
Karate 1 0.5485 0.5195 0.6995
Books 0.6313 0.5338 0.4142 0.5369
Football 0.9326 0.9151 0.9269 0.9242
Dolphins 0.8888 0.5749 0.5169 0.5197
Table 6: Modularity results for the real-world networks.
OmeGAnet GA-mod Louvain Infomap
Karate 0.3715 0.4033 0.402 0.402
Books 0.4546 0.4793 0.4833 0.5268
Football 0.5976 0.6008 0.601 0.6005
Dolphins 0.3787 0.5124 0.4952 0.5146
Table 7: Number of communities for the real-world net-
works.
GT OmeGAnet GA-mod Louvain Infomap
Karate 2 2 4 3 3
Books 3 2 3 8 5
Football 12 12 11 12 12
Dolphins 2 2 4 10 6
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
34

5 CONCLUSIONS
We proposed OmeGAnet, a new method based on ge-
netic algorithms for dividing the nodes of an undi-
rected and connected graph in communities.
We considered the graph as an electric circuit and
computed for each couple of connected nodes the ef-
fective resistance. We then exploited this distance for
weighting the graph and searching communities with
high weighted modularity.
By performing several experiments on both syn-
thetic and real-world networks, the results show
that the proposed methodology is promising since it
clearly outperforms both a standard GA-based algo-
rithm running on the original adjacency matrix of
the graph, and the state-of-the-art approaches Louvain
and Infomap.
It is worth pointing out that the choice of the
parameter nn plays an important role on the perfor-
mance of OmeGAnet. In the current implementation
we experimentally set it and found that low values of
nn allow to obtain good results.
However, more study is necessary to find a general
criterion which allows a good setting of this parame-
ter. In fact, the network sparsification is crucial for
improving the quality of the community division ob-
tained by the approach.
(Yan et al., 2018) proposed a measure that esti-
mates the variation of spectral properties of the graph
when edges are removed. They showed that the struc-
ture of real weighted networks is very robust under
weight thresholding when edges are removed if their
weight is below a threshold value computed with such
a measure.
This research line could be a starting point deserv-
ing deeper investigation which could be beneficial for
determining the minimum number of nearest neigh-
bors to consider when building the sparse similarity
weighted matrix
e
Ω.
REFERENCES
Avrachenkov, K., Chebotarev, P., and Rubanov, D. (2017).
Kernels on graphs as proximity measures. In Inter-
national workshop on algorithms and models for the
web-graph, pages 27–41. Springer.
Avrachenkov, K., Chebotarev, P., and Rubanov, D. (2019).
Similarities on graphs: Kernels versus proximity mea-
sures. European Journal of Combinatorics, 80:47–56.
Aynulin, R. (2019). Impact of network topology on ef-
ficiency of proximity measures for community de-
tection. In International Conference on Complex
Networks and Their Applications, pages 188–197.
Springer.
Barrat, A., Barthelemy, M., Pastor-Satorras, R., and Vespig-
nani, A. (2004). The architecture of complex weighted
networks. In Proc. National Academy of Science,
pages 101,3747.
Blondel, V. D., Guillaume, J.-L., Lambiotte, R., and Lefeb-
vre, E. (2008). Fast unfolding of communities in large
networks. Journal of statistical mechanics: theory
and experiment, 2008(10):P10008.
Chandra, A. K., Raghavan, P., Ruzzo, W. L., Smolensky,
R., and Tiwari, P. (1996). The electrical resistance of
a graph captures its commute and cover times. Com-
putational Complexity, 6(4):312–340.
Deza, M. M. and Deza, E. (2009). Encyclopedia of dis-
tances. In Encyclopedia of distances, pages 1–583.
Springer.
Dijkstra, E. W. et al. (1959). A note on two problems
in connexion with graphs. Numerische mathematik,
1(1):269–271.
Doyle, P. and Snell, J. (1989). Random walks and electric
networks. Mathematical Association of America.
Fortunato, S. and Hric, D. (2016). Community detection in
networks: A user guide. Physics reports, 659:1–44.
Ghosh, A., Boyd, S., and Saberi, A. (2008). Minimizing
effective resistance of a graph. SIAM Rev., 50(1):37–
66.
Girvan, M. and Newman, M. E. (2002). Community struc-
ture in social and biological networks. Proceedings of
the national academy of sciences, 99(12):7821–7826.
Goldberg, D. E. (1989). Genetic algorithms in search. Op-
timization, and MachineLearning.
Klein, D. J. and Randi
´
c, M. (1993). Resistance distance.
Journal of mathematical chemistry, 12(1):81–95.
Lancichinetti, A., Fortunato, S., and Radicchi, F. (2008).
Benchmark graphs for testing community detection
algorithms. Physical review E, 78(4):046110.
Park, Y. and Song, M. (1998). A genetic algorithm for clus-
tering problems. In Proceedings of the third annual
conference on genetic programming, volume 1998,
pages 568–575.
Pizzuti, C. (2018). Evolutionary computation for commu-
nity detection in networks: a review. IEEE Transac-
tions on Evolutionary Computation, 22(3):464–483.
Radicchi, F., Ramasco, J. J., and Fortunato, S. (2011). Infor-
mation filtering in complex weighted networks. Phys-
ical Review E, E83:046101.
Rosvall, M. and Bergstrom, C. T. (2008). Maps of random
walks on complex networks reveal community struc-
ture. Proceedings of the National Academy of Sci-
ences, 105(4):1118–1123.
Saerens, M., Fouss, F., Yen, L., and Dupont, P. (2004). The
principal components analysis of a graph, and its re-
lationships to spectral clustering. In European confer-
ence on machine learning, pages 371–383. Springer.
Sommer, F., Fouss, F., and Saerens, M. (2016). Compari-
son of graph node distances on clustering tasks. In In-
ternational Conference on Artificial Neural Networks,
pages 192–201. Springer.
Spielman, D. A. and Srivastava, N. (1996). Graph spar-
sification by effective resistances. Siam Journal on
Computing, (40):1913.
An Effective Resistance based Genetic Algorithm for Community Detection
35

Tumminello, M., Aste, T., Matteo, T. D., , and Mantegna,
R. N. (2005). A tool for filtering information in com-
plex systems. In Proc. National Academy of Science,
pages 102,10421.
Van Mieghem, P. (2010). Graph spectra for complex net-
works. Cambridge University Press.
Van Mieghem, P., Devriendt, K., and Cetinay, H. (2017).
Pseudoinverse of the laplacian and best spreader node
in a network. Physical Review E, 96(3):032311.
Yan, X., Jeub, L. G. S., Flammini, A., Radicchi, F., and
Fortunato, S. (2018). Weight thresholding on complex
networks. Physical Review E, E98:042304.
Yen, L., Fouss, F., Decaestecker, C., Francq, P., and
Saerens, M. (2007). Graph nodes clustering based
on the commute-time kernel. In Pacific-Asia Con-
ference on Knowledge Discovery and Data Mining,
pages 1037–1045. Springer.
Yen, L., Fouss, F., Decaestecker, C., Francq, P., and
Saerens, M. (2009). Graph nodes clustering with the
sigmoid commute-time kernel: A comparative study.
Data & Knowledge Engineering, 68(3):338–
361.
ECTA 2021 - 13th International Conference on Evolutionary Computation Theory and Applications
36
