
Harena Semantics: A Framework to Support Semantic Annotation in
Citizen Science Systems
Fagner Leal Pantoja
1 a
, Marco Antonio de Carvalho Filho
2 b
and Andr
´
e Santanch
`
e
1 c
1
Institute of Computing, University of Campinas, Campinas, Brazil
2
Faculty of Veterinary Medical Education, Utrecht University, Utrecht, The Netherlands
Keywords:
Semantic Web, Natural Language Processing, Machine Learning, Citizen Science, Clinical Reasoning.
Abstract:
We propose a new approach to support human agents to annotate semantic concepts in free-text sentences in
the biomedical domain. Using our markdown-derived language called Versum, authors can easily annotate
relevant terms while producing content for Citizen Science systems. Besides, an embedded Automatic Anno-
tation Mechanism suggests semantic concepts for the author. It implements a Named Entity Recognition task
using a hybrid approach: (1) a Transformer-based Deep Neural Network and (2) an Ontology-based method.
We conducted a case study running over content produced in the Harena e-learning system, which intends
to teach Clinical Reasoning to students using Clinical Cases. Results of this pilot evaluation suggest the po-
tential of Harena Semantics to engage volunteers in the production of semantic, agent-centered resources on
crowdsourcing systems.
1 INTRODUCTION
In clinical learning environments, professors use
Clinical Cases as pedagogical resources to teach stu-
dents to solve problems and, consequently, to develop
their clinical reasoning capacities. Usually, Clini-
cal Cases have fictional narratives inspired by real
situations interconnected in a network of unantici-
pated events commonly occurring in a clinical envi-
ronment. This complex information comprises a valu-
able health knowledge source.
Despite the potential that Clinical Cases have to
become an unprecedented Knowledge Base, there are
open challenges concerning: (1) how to handle and
process free-text information contained in the case
narrative; and (2) how to integrate and interrelate
complex information fragmented across a plethora of
documents on the Web.
Envisaging these challenges, we propose Harena
Semantics to construct and curate Clinical Cases de-
livering two main contributions:
• Versum: a markdown-based script language that
enables authors to annotate semantic concepts in-
side natural language texts. Via Versum, the se-
a
https://orcid.org/0000-0003-1784-5512
b
https://orcid.org/0000-0001-7008-4092
c
https://orcid.org/0000-0002-1756-4852
mantic annotation may be done manually by a hu-
man agent (e.g., professors, learners), automati-
cally by some computer-assisted method, or by a
mixture of these methods.
• An Annotation Mechanism which automatically
recognizes relevant clinical concepts within a
given sentence following a hybrid approach com-
posed of two independent algorithms: (1) a
Transformer-based Named Entity Recognition
(NER) task implemented as a Deep Neural Net-
work (Vaswani et al., 2017) and (2) an Ontology-
based NER to link terms from free-text sentences
to ontology-related concepts, formally defined as
knowledge graph, which comprises a network of
interconnected semantic resources.
We conducted a case study of our framework run-
ning over the Harena
1
system (de Menezes Mota
et al., 2019), an e-learning environment, based on
cases resolution, which is used as a supporting peda-
gogical tool in Emergency Medicine courses. Harena
represents Clinical Cases in a Virtual Patient for-
mat (Cook and Triola, 2009).
The Harena environment comprises a Knowledge
Base of clinical cases, besides two complementary
Web-based modes: (1) Interface Author, to enable one
1
https://jacinto.harena.org/
336
Pantoja, F., Filho, M. and Santanchè, A.
Harena Semantics: A Framework to Support Semantic Annotation in Citizen Science Systems.
DOI: 10.5220/0010785300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 336-343
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

to create and curate artificial Clinical Cases inspired
by real-life clinical encounters; and (2) Interface
Player, to execute the simulations of the Clinical
Cases narratives.
Through our approach, the Harena system adheres
to the Semantic Web, enabling authors to produce
semantic annotations over free-text content of their
Clinical Cases. The authors of Clinical Cases (i.e.,
the Harena system users) are also Citizen Scientists
who embed medical knowledge in Clinical Case nar-
ratives.
Citizen Science projects promote the collection
and analysis of scientific data by members of the gen-
eral public and professional scientists. By adhering
to the Semantic Web, Citizen Science systems enable
content producers to add a higher level of abstraction
to the crowdsourced information. Harena Semantics
enables volunteer users to produce agent-centered re-
sources (e.g., Clinical Cases and beyond) and there-
fore cooperate on the gradual building of an intercon-
nected network of Knowledge Bases. Agent-centered
resource engagement is one of the three types of en-
gagement in citizen science projects (Jackson et al.,
2020) which have the potential to increase engage-
ment in the early stages of training in a volunteer
learning scenario. Preliminary results of this study
reinforce the claim about the need for mechanisms to
engage users in the production of agent-centered re-
sources.
The remaining of this paper is organized as fol-
lows: Section 2 gives some background foundations
and related work; Section 3 describes the Harena Se-
mantics framework and some results of the NER task
evaluation; Section 4 presents a case study of our ap-
proach running over the Harena system; Section 5
presents our concluding remarks.
2 FOUNDATIONS AND RELATED
WORK
The scope of this review section is twofold. First,
we give a background about Clinical Cases, Vir-
tual Patients, and Semantic Web applied to the Clin-
ical Reasoning research field. Lastly, we briefly
present some concepts of Natural Language Process-
ing (NLP), Named Entity Recognition (NER), and the
just arrived Word Embeddings.
2.1 Virtual Patient
In the health context, there is a wide spectrum of
strategies to simulate patients for students’ train-
ing (Cook and Triola, 2009). The adopted strategy
depends on the available resources, the goal expected
from the training, the level of structure in the data and
the desired expressiveness of the clinical narrative of
simulation.
Virtual Patients (VP) are designed to present sce-
narios and narratives of a Clinical Case, guided by
computers. They represent the Clinical Case in a
graph of states affording structured guidance (Cook
and Triola, 2009).
By taking advantage of Semantic Web abstraction,
OLabX (extended OpenLabyrinth) uses mEducator
schema to discover, retrieve, share, and reuse medi-
cal educational resources (Dafli et al., 2015). Hege et
al. present a tool to foster the acquisition of clinical
reasoning skills through Virtual Patients and Concept
Maps (Hege et al., 2017).
Our approach differs from related work, as it de-
parts from a markdown-derived language, apt for hu-
man writing, reading, and annotation, combining it
with automatically generated superimposed annota-
tions.
2.2 Named Entity Recognition
Named Entity Recognition (NER) is a Natural Lan-
guage Processing (NLP) task to identify and clas-
sify entity types, such as People, Organization
and Location, . In the biomedical domain, research
works focus on Gene, Protein, Disease, Chemical,
Anatomy, etc.
There are many approaches to implement NER
tasks. Recent works using statistical approaches have
leveraged the NER state-of-the-art by using Deep
Neural Networks to learn Word Embeddings (Col-
lobert and Weston, 2008; Devlin et al., 2018). These
neural language models encode syntactic and seman-
tic information in vectors known as embeds in such
a way that those embeds with similar meanings have
similar representations. They feed the algorithm that
decides if it should tag a term within the given sen-
tence as a named entity.
Google released BERT (Bidirectional Encoder
Representations from Transformers) (Devlin et al.,
2018), an implementation of the Transformer archi-
tecture introduced on the paper “Attention is all you
need” (Vaswani et al., 2017). BERT is pre-trained
on the Masked Language Model (MLM), an unsu-
pervised NLP task whose objective is to predict the
hidden word in a given input sentence. MLM is
an expensive task since it requires millions of sam-
ple sentences. The pre-training phase produces the
Word Embeddings as a by-product of the task ob-
jective. One can easily reuse BERT embeddings by
Fine-Tuning (a Transfer Learning technique) them in
Harena Semantics: A Framework to Support Semantic Annotation in Citizen Science Systems
337

a downstream task (e.g., NER, Question-Answering,
Natural Language Inference). Generally, Fine-Tuning
is an inexpensive method since it requires a small la-
beled dataset.
BERT produces context-aware Word Embeddings
through the Attention Mechanism, which detects the
most representative parts in the whole sentence. The
Attention Mechanism is a procedure to capture the
sentence context based on the statistical relationship
between the current word and every other word in
the input sentence, providing a bidirectional context
– i.e., interpreting the sentence considering the pre-
vious context (left-right direction) and posterior con-
text (right-left direction). This bidirectional context
leveraged the state-of-the-art of NER methods since
the previous works were capable of dealing with the
context in just one direction (Devlin et al., 2018).
The NLP community sees the rising of transformer-
based Word Embeddings as a revolution in this re-
search field.
There are works (Alsentzer et al., 2019; Lee et al.,
2020; Akhtyamova et al., 2020) specializing BERT-
embeddings to the Biomedical and clinical domains.
(Lee et al., 2020) introduce BioBERT, a BERT-based
model specialized in biomedical language. BioBERT
is pre-trained on large-scale biomedical corpora com-
posed of PubMed abstracts and PMC full-text articles.
BioBERT outperformed the state-of-the-art models in
a bunch of experiments over NER tasks. BioBERT
trained several models to recognize different named
entities (e.g., one model to handle diseases, another
one to deal with proteins, and so on). We based our
NER implementation on the BioBERT model, extend-
ing it to recognize multiple entities (anatomy, chemi-
cals, and disease) in a single model.
Recent works attempted to join Word Embeddings
and Semantic Web ideas. We intend to contribute
to this endeavor by investigating how to superim-
pose NER annotations produced by these distinct, al-
though related, research areas. According to our liter-
ature search, there is not a standardized, established
definition of ontology-based NER methods. Some
works define them as Concept Normalization (Mif-
tahutdinov et al., 2021; Do
˘
gan et al., 2014), Entity
Linking (Basaldella et al., 2020), Entity Typing (Choi
et al., 2018), and so on.
(Kim et al., 2019) present BERN, a neu-
ral biomedical multi-type NER tool based on
the BioBERT model. BERN is equipped with
probability-based decision rules to treat overlapping
entities (polysemy – for instance, one can tag andro-
gen as gene or chemical) and synonyms (i.e., terms
described by multiple names). BERN normalizes the
recognized entities assigning an ID (linking to con-
trolled vocabularies) to each recognized entity.
Other initiatives also address NER extending
BERT, refining the embeddings, or fine-tunning with
specialized datasets (Basaldella et al., 2020; Lyu and
Zhong, 2021; Miftahutdinov et al., 2021).
3 HARENA SEMANTICS
This section presents Harena Semantics, a framework
consisting of two complementary components: (1) a
markdown-based language called Versum to enable
Citizen Scientists to create and curate Clinical Cases
adherents to Semantic Web; and (2) a hybrid approach
to perform a Named Entity Recognition (NER) task to
annotate Clinical Cases with semantic concept labels.
3.1 Versum
Versum enables one to add semantic structure into the
free-text content of Clinical Cases, aiming to allow
easy integration of semantic annotations into clinical
narratives. By making explicit the semantic of Clini-
cal Cases, Versum creates pedagogical resources ad-
herent to the Semantic Web while providing a step
forward to a machine-interpretable representation of
the natural human language.
In a previous research paper, Menezes et al. devel-
oped the first version of Versum following the Narra-
tive Design approach, which provides elements to en-
able scenario building and flow control of narratives.
In this paper, we release the Annotation Mecha-
nism as an improvement feature of Versum. The pro-
cess of semantic annotation using the Versum syntax
is straightforward through a predefined set of reserved
markups to add high-level structured information to
the Clinical Case.
Using Versum markups, one can annotate a
text fragment as a semantic concept enclosing it
between the curly braces { } followed by the
concept label between parenthesis, e.g., {heart
attack}(disease). Moreover, one can link a
free-text entity to Knowledge Bases – e.g., ontolo-
gies, controlled vocabularies, taxonomies, thesaurus
– through the namespaces markup. As an example of
Versum usage, Figure 1 shows:
1. A Clinical Case narrative in free-text format. This
Clinical Case was authored in the Harena system.
2. The Semantic Clinical Case produced from the
original free-text Clinical Case. By annotating
with Versum, the narrative becomes more struc-
tured and semantically enriched. These annota-
tions could be manually made by a human agent
HEALTHINF 2022 - 15th International Conference on Health Informatics
338
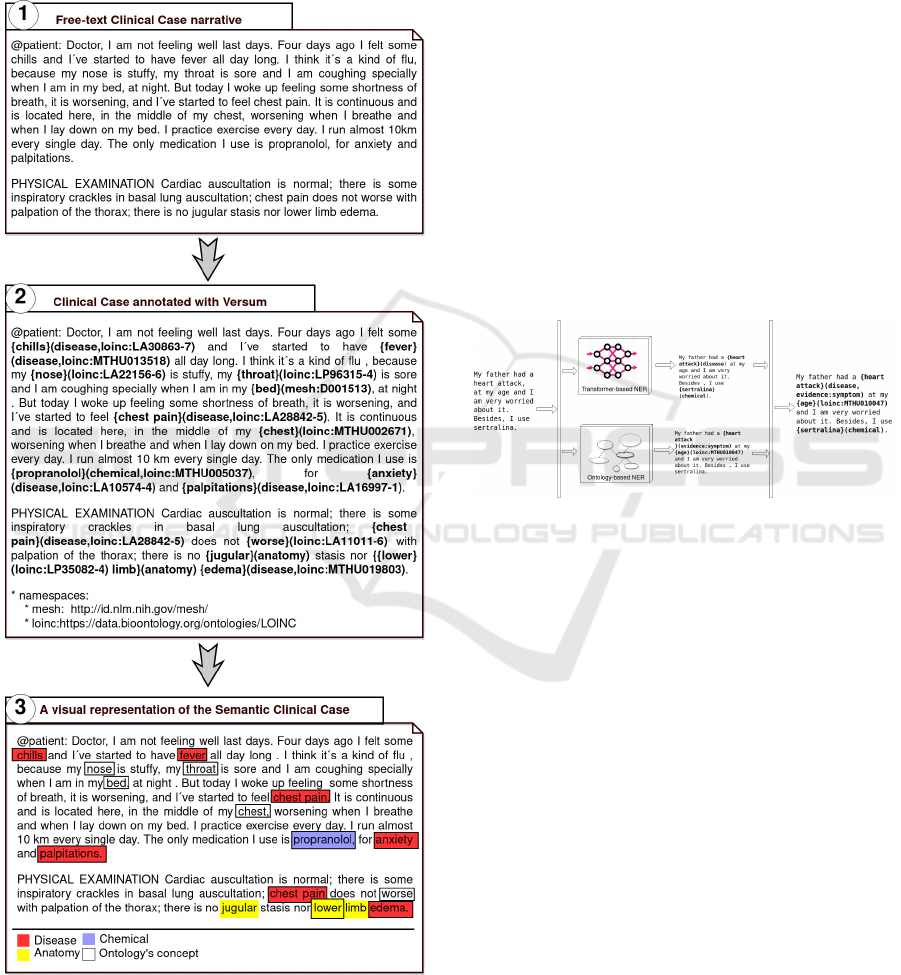
or by an automatic process (like the Automatic
Annotation Mechanism depicted in the next Sec-
tion 3.2).
3. A visual representation of the Semantic Clinical
Case highlighting the semantic concepts anno-
tated.
Figure 1: (1) A Clinical Case narrative in free-text format;
(2) The Semantic Clinical Case produced from 1; (3) A vi-
sual representation of the Semantic Clinical Case.
3.2 Automatic Annotation Mechanism
Harena Semantics provides a mechanism to automat-
ically annotate the concepts within the Clinical Case
through a hybrid approach:
1. Transformer-based Named Entity Recognition
(NER) task to assign labels to clinical terms
within a given sentence. This method is based
on the Transformer architecture, a Deep Neural
Network capable of capturing linguistic features
based on statistical inferences.
2. Ontology-based Named Entity Recognition task
to link from free-text terms to concepts formally
defined on biomedical ontologies.
As output, the Automatic Annotation mechanism
produces the class labels to terms of the sentence
given as input, as depicted by Figure 2. The labels
may be (1) ontology concepts, (2) named entities, or
(3) a combination of them.
Figure 2: The hybrid approach to NER.
3.2.1 Transformer-based Named Entity
Recognition
We developed a Transformer-based NER that attends
to the contextual information – i.e., syntactic and se-
mantic aspects related to the context of a sentence –
to decide if it should label or not a given term as a
named entity.
This method classifies the sentence terms accord-
ing to the clinical domain-specific labels Anatomy,
Chemical, and Disease. In order to train the
model, we generated a small labeled corpus called
ACD (Anatomy, Chemical, Disease) from the con-
catenation of the two pre-existing labeled data sets:
BC5CDR (Li et al., 2016) and AnatEM (Pyysalo and
Ananiadou, 2014). The Disease class in BC5CDR
also includes disease mentions (Do
˘
gan and Lu, 2012),
which comprises Signs and Symptoms. According
to (Crichton et al., 2017), BC5CDR and AnatEM do
not exhibit a significant overlap between the training
sentences of one dataset and the test sentences of the
other one (it would expose the training algorithm to
sentences of the validation set), which indicates the
Harena Semantics: A Framework to Support Semantic Annotation in Citizen Science Systems
339

feasibility of concatenating them. Table 1 provides
some metrics about the ACD corpus.
Table 1: ACD corpus statistics.
Corpus # Sentences Entities # Annotations
Training Test Training Test
ACD 21,223 10,867 Anatomy 9,085 4,616
Chemical 10,550 5,378
Disease 8,428 4,424
We reused the BioBERT-embeddings which are
pre-trained on an unsupervised Masked language
Modeling task specialized on biomedical domain (see
Section 2.2 for more details). Then, we trained our su-
pervised NER algorithm by adjusting the BioBERT-
embeddings through a Transfer Learning technique
called Fine-tuning. Therefore, this NER algorithm is
a semi-supervised method once it reuses the embed-
dings produced by an unsupervised source task (i.e.,
Masked Language Modeling) in a supervised target
task (i.e., NER).
This proposed Deep Neural Network – called En-
voy – is a stack of 12 BioBERT layers plus an extra
Fine-Tuning layer liable for specializing the network
(by adjusting the parameters) to recognize the label of
each term inside the given input sentence.
The Fine-Tuning process involves adding an ex-
tra linear layer to the top of the pre-trained neural
model and adjusting its weights for each sentence on
the training dataset. Each neuron on the first layer
processes a token of the input given sentence and for-
wards it to the next neuron layer. This process is re-
peated for each sample sentence on the training data
set.
We released our NER model (specialized to rec-
ognize anatomy, chemicals, and diseases) at Hug-
gingface Model Hub: https://huggingface.co/fagner/
envoy. The open-source code to extend the BioBERT
language model by fine-tuning it in a multi-class NER
model is available at our fork of BioBERT: https:
//github.com/faguim/biobert-pytorch.
A REST-based implementation of the Seman-
tics framework can be deployed as a docker con-
tainer: https://github.com/datasci4health-incubator/
harena-semantics.
Model Evaluation. To validate our approach, we
conducted an intrinsic evaluation (Velupillai et al.,
2018) measuring the performance of the model at per-
forming the NER task objective. The experimental
setup is as follows:
• Pre-trained Model: We experimented with both
biobert
base cased
2
(containing 768 hidden
2
https://huggingface.co/dmis-lab/
states, 12 neuron layers and totaling 100 million
parameters) and biobert large cased
3
(24 lay-
ers, 1024 hidden states).
• Learning rate 5e
−6
using AdamW optimizer, cho-
sen by considering a threshold between perfor-
mance and stability, since high learning rate in-
creases performance while incurs instability on
the training (Chiu et al., 2016).
• Batch size: 32 sentences/batch (i.e., 4 sentences ∗
128 tokens = 512 tokens/batch).
Among the two versions adopted in the evaluation,
the envoy large version accomplished 85,8% of suc-
cess on the f1 score, while envoy base 85,5% .
Although envoy large present better results
(lower error rate and higher f1-score), we released
biobert base as the official version of Harena Se-
mantics due to its smaller model size (biobert base
is 432 MB, while biobert large is 1,5 GB) which
facilitates the deployment of Harena Semantics in per-
sonal pcs. Another relevant feature of smaller mod-
els is their stability on the training process (Mosbach
et al., 2020).
3.2.2 Ontology-based Named Entity Recognition
This rule-based NER method uses ontologies as
source information to label the sentence terms with
the concept labels formally defined on biomedical on-
tologies. The algorithm looks for matches (exacts
or partials) between the free-text sentence terms and
ontology concepts. It provides two modes to match
against ontologies:
• External ontologies: This mode uses the bion-
tology annotator to match terms against ontolo-
gies stored on the open repository Bioportal (Noy
et al., 2009).
• Local ontologies: This mode uses a RDF database
to store RDF triples. We developed an API called
OntoMatch to query the RDF triples through
the RDFLib python library. Ontomatch enables
matches based on a range of metrics such as Lev-
enshtein, Jaccard, Cosine etc.
4 CASE STUDY ON THE HARENA
SYSTEM
To get a pilot evaluation of Harena Semantics, we
conducted a case study running it over the Harena sys-
biobert-base-cased-v1.1
3
https://huggingface.co/dmis-lab/
biobert-large-cased-v1.1/tree/main
HEALTHINF 2022 - 15th International Conference on Health Informatics
340

tem (de Menezes Mota et al., 2019). The Medicine
course from University of Campinas uses Harena as
a supporting pedagogical tool to situate the individ-
uals in an e-learning environment, which simulates
the context of an Emergency Care Unit (de Araujo
Guerra Grangeia et al., 2016).
This section presents a process to construct se-
mantically rich Virtual Patients (VPs). We intend to
reinforce the feasibility of a global knowledge net-
work connecting the information scattered in different
Virtual Patient systems.
This research paper intends to explore ways of in-
creasing the underlying structure of Virtual Patients
towards the glimpse of the Semantic Web. More
structured information can be interpreted by ma-
chines, expanding the possibilities of application: (i)
it becomes easier to find, reuse, and group cases and
parts of cases – e.g., it becomes possible to query:
cases in which the patient experienced shortness of
breath; cases where the ECG was fundamental to di-
agnose a heart disease; (ii) data from cases can be
used beyond the scope of training as a Citizen Sci-
ence data source.
Our approach focuses on building a Semantic Vir-
tual Patient from free-text Clinical Case narratives.
The deployment of a Semantic Virtual Patient poten-
tially facilitates intelligent searches, complex queries,
and easy exchange between institutions. As detailed
in previous sections, Harena Semantics identifies clin-
ical concepts and links them to ontology concepts
through the Automatic Annotation Mechanism and
Versum tags. Therefore, it creates a RDF graph repre-
senting key knowledge about the virtual patient and
integrating it to an interconnected network of con-
cepts envisioned by the Semantic Web research area.
To evaluate the feasibility of creating and curat-
ing Semantic Virtual Patient using our framework, we
departed from Virtual Patients manually annotated by
doctors. These annotations are part of the case ra-
tionale, they relate relevant symptoms to the problem
(disease) narrated on the Clinical Case.
At the authoring process, the author tags relevant
symptoms and indicate whether they are directly re-
lated to the clinical case (e.g., arterial hypertension
and acute onset of chest pain), or key to the diagno-
sis (e.g., pain radiating to neck and back), or just dis-
tractors to the learner which mislead her to a wrong
direction (e.g., symmetric radial pulses is a specific
sign but present in only one third of the patients). The
diagnosis, which will be presented in the final of the
case presentation as a feedback is also annotated.
Harena can superimpose several layers of anno-
tation in the same text content and combine them
throughout superimposed contexts.
It is possible to assign a context to any segment
of text surrounding it by double curly braces {{ }}.
Each context can receive an identifier prefixed by at
sign @. Segments with the same identifier must re-
fer to the same textual content, even though they can
afford distinct superimposed annotations. For exam-
ple, the three following contexts refer to the same text
fragment through the identifier @symp01:
{{@symp01 / e v i d e n c e : f i n d i n g r e l e v a n c e
mesh:D000784
A man , 52 y e a r s old , r e p o r t s he i s
f e e l i n g { v e r y s t r o n g c h e s t p a i n } /
e v i d e n c e : c o r r o b o r a t e f i n d i n g / .
}}
{{@symp01
A man , 52 y e a r s old , r e p o r t s he i s
{ f e e l i n g } ( loinc:MTHU021518 ) { v e r y
s t r o n g } ( l o i n c : LA284 4 1 −6)
{ c h e s t p a i n } ( lo i nc:LA 2 8 8 4 2 −5).
}}
{{@symp01
A man , 52 y e a r s o l d , r e p o r t s he i s
f e e l i n g v e r y s t r o n g { c h e s t } ( anatomy )
{ p a i n } ( d i s e a s e )
}}
The first copy of the segment was annotated by
physicians, as previously described. Besides the con-
text id, it is possible to specify the target of the anno-
tations. In this case, evidence:finding relevance
mesh:D000784 indicates that the following annota-
tions point to the relevance of the symptom to the
Aortic Dissection disease (mesh:D000784). The sec-
ond copy was annotated by the ontology-based anno-
tation mechanism and the third by the Transformer-
based mechanism.
These superimposed annotations are transformed
into an RDF Graph (Schreiber and Raimond, 2014)
as shown in Figure 3. A uniquely identified RDF re-
source (node in the RDF graph) is associated with
each word. When the annotation refers to a word –
e.g., the chest is annotated with Anatomy (which in
turn refers to mesh:D000715) – the related RDF node
is connected by a skos:related association. SKOS
- Simple Knowledge Organization System is a data
model for knowledge organization (Miles and Bech-
hofer, 2009).
When annotations refer to a sentence with more
than one word – e.g., the sentence chest pain anno-
tated by the concept Chest pain in the LOINC Doc-
ument Ontology (loinc:LA28842-5) – a node aggre-
gating the sentence’s words is created and related to
the concept. An aggregation will reuse already ag-
gregated nodes whenever is possible, as in the case
of the node that aggregates “very strong” and “chest
Harena Semantics: A Framework to Support Semantic Annotation in Citizen Science Systems
341

Figure 3: The RDF graph representing a case of Aortic Dis-
section, built with the support of Harena Semantics.
Figure 4: Key elements of a Semantic Virtual Patient RDF
Graph extracted from a Clinical Case.
pain” aggregated nodes.
Following this process, annotated content will
converge to a semantic RDF profile of a virtual pa-
tient, as shown in Figure 4. The diagram shows a sim-
plified version of the graph presenting an overview
of what we call: Semantic Virtual Patient. The case
in the example is related to an aortic dissection – a
dangerous injury to the innermost layer of the aorta,
which puts the life of the patient at risk.
In the long term, the produced semantic clinical
cases could be used to grasp knowledge from the un-
structured text within the clinical narrative. The nar-
rative scripts in a machine-interpretable format enable
sharing, versioning, and crowdsourcing. Such capa-
bilities are needed for a system with clinical case data.
5 CONCLUSION
This paper presented Harena Semantics, a framework
to enable Citizen Scientists to create semantic annota-
tions directly into the text narrative of Clinical Cases.
By adopting our approach, the data crowdsourced in
Citizen Science systems may incorporate the informa-
tion gathered in the Knowledge Network envisioned
by the Semantic Web research area.
The introduced Automatic Annotation Mech-
anism benefits both from Rule-based (the
ontology-based NER) and Statistical Learning
(the Transformer-based NER) approaches. Our NER
task presents results comparable to the state-of-the-
art works in such research area. Our approach to
superimpose annotations enables to combine human
and automatic annotations to produce a knowledge
network representing our Semantic Virtual Patient.
The technology stack presented in this paper could
serve several purposes. As future works, we intend to
implement a search engine to retrieve Clinical Cases
aided by the support of semantic information. Be-
sides, the Semantic Virtual Patient can also be used
to train inference systems that automatically gener-
ate feedback to the users of Learning Environments.
These educational resources must be adherent to ped-
agogy practices, therefore it is necessary to develop
approaches to involve experts, professors, and scien-
tists in the creation of these resources. The Harena
Semantics is an initiative engaged in such effort.
ACKNOWLEDGEMENTS
This study was financed in part by CNPq grant num-
ber 428459/2018-8. We thank dr. Tiago Grangeia for
the production and annotation of the Clinical Cases
addressed in this work.
REFERENCES
Akhtyamova, L., Mart
´
ınez, P., Verspoor, K., and Cardiff,
J. (2020). testing contextualized word embeddings to
improve ner in spanish clinical case narratives. IEEE
Access, 8:164717–164726.
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H.,
Jin, D., Naumann, T., and McDermott, M. (2019).
Publicly available clinical bert embeddings. arXiv
preprint arXiv:1904.03323.
Basaldella, M., Liu, F., Shareghi, E., and Collier, N. (2020).
Cometa: A corpus for medical entity linking in the
social media. arXiv preprint arXiv:2010.03295.
Chiu, B., Crichton, G., Korhonen, A., and Pyysalo, S.
(2016). How to train good word embeddings for
biomedical nlp. In Proceedings of the 15th workshop
on biomedical natural language processing, pages
166–174.
Choi, E., Levy, O., Choi, Y., and Zettlemoyer, L.
(2018). Ultra-fine entity typing. arXiv preprint
arXiv:1807.04905.
Collobert, R. and Weston, J. (2008). A unified architec-
ture for natural language processing: Deep neural net-
works with multitask learning. In Proceedings of the
25th international conference on Machine learning,
pages 160–167.
HEALTHINF 2022 - 15th International Conference on Health Informatics
342

Cook, D. A. and Triola, M. M. (2009). Virtual patients:
a critical literature review and proposed next steps.
Medical Education, 43(4):303–311.
Crichton, G., Pyysalo, S., Chiu, B., and Korhonen, A.
(2017). A neural network multi-task learning ap-
proach to biomedical named entity recognition. BMC
bioinformatics, 18(1):1–14.
Dafli, E., Antoniou, P., Ioannidis, L., Dombros, N., Topps,
D., and Bamidis, P. D. (2015). Virtual patients on the
semantic web: a proof-of-application study. Journal
of medical Internet research, 17(1):e3933.
de Araujo Guerra Grangeia, T., de Jorge, B., Franci,
D., Martins Santos, T., Vellutini Setubal, M. S.,
Schweller, M., and de Carvalho-Filho, M. A. (2016).
Cognitive load and self-determination theories applied
to e-learning: impact on students’ participation and
academic performance. PloS one, 11(3):e0152462.
de Menezes Mota, M. F., Pantoja, F. L., Mota, M. S.,
Grangeia, T. d. A. G., de Carvalho Filho, M. A., and
Santanch
`
e, A. (2019). Analytical design of clinical
cases for educational games. In Joint International
Conference on Entertainment Computing and Serious
Games, pages 353–365. Springer.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Do
˘
gan, R. I. and Lu, Z. (2012). An improved corpus of
disease mentions in PubMed citations. In Proc. 2012
Workshop on Biomedical Natural Language Process-
ing, pages 91–99.
Do
˘
gan, R. I., Leaman, R., and Lu, Z. (2014). Ncbi disease
corapus: a resource for disease name recognition and
concept normalization. Journal of biomedical infor-
matics, 47:1–10.
Hege, I., Kononowicz, A. A., and Adler, M. (2017). A clin-
ical reasoning tool for virtual patients: design-based
research study. JMIR medical education, 3(2):e8100.
Jackson, C. B., Østerlund, C., Crowston, K., Harandi, M.,
and Trouille, L. (2020). Shifting forms of engage-
ment: Volunteer learning in online citizen science.
Proceedings of the ACM on Human-Computer Inter-
action, 4(CSCW1):1–19.
Kim, D., Lee, J., So, C. H., Jeon, H., Jeong, M., Choi, Y.,
Yoon, W., Sung, M., and Kang, J. (2019). A neural
named entity recognition and multi-type normaliza-
tion tool for biomedical text mining. IEEE Access,
7:73729–73740.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H.,
Leaman, R., Davis, A. P., Mattingly, C. J., Wiegers,
T. C., and Lu, Z. (2016). Biocreative v cdr task cor-
pus: a resource for chemical disease relation extrac-
tion. Database, 2016.
Lyu, Y. and Zhong, J. (2021). Dsmer: A deep semantic
matching based framework for named entity recog-
nition. In European Conference on Information Re-
trieval, pages 419–432. Springer.
Miftahutdinov, Z., Kadurin, A., Kudrin, R., and Tutubalina,
E. (2021). Drug and disease interpretation learn-
ing with biomedical entity representation transformer.
arXiv preprint arXiv:2101.09311.
Miles, A. and Bechhofer, S. (2009). SKOS Simple Knowl-
edge Organization System Reference. Technical re-
port.
Mosbach, M., Andriushchenko, M., and Klakow, D. (2020).
On the stability of fine-tuning bert: Misconceptions,
explanations, and strong baselines. arXiv preprint
arXiv:2006.04884.
Noy, N. F., Shah, N. H., Whetzel, P. L., Dai, B., Dorf, M.,
Griffith, N., Jonquet, C., Rubin, D. L., Storey, M.-A.,
Chute, C. G., et al. (2009). Bioportal: ontologies and
integrated data resources at the click of a mouse. Nu-
cleic acids research, 37(suppl 2):W170–W173.
Pyysalo, S. and Ananiadou, S. (2014). Anatomical entity
mention recognition at literature scale. Bioinformat-
ics, 30(6):868–875.
Schreiber, G. and Raimond, Y. (2014). RDF 1.1 Primer.
Technical report.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, L., and Polosukhin, I.
(2017). Attention is all you need. arXiv preprint
arXiv:1706.03762.
Velupillai, S., Suominen, H., Liakata, M., Roberts, A.,
Shah, A. D., Morley, K., Osborn, D., Hayes, J., Stew-
art, R., Downs, J., et al. (2018). Using clinical natu-
ral language processing for health outcomes research:
overview and actionable suggestions for future ad-
vances. Journal of biomedical informatics, 88:11–19.
Harena Semantics: A Framework to Support Semantic Annotation in Citizen Science Systems
343
