
Biomedical Text Mining: Applicability of Machine Learning-based
Natural Language Processing in Medical Database
Nafiseh Mollaei
a
, Catia Cepeda
b
, Joao Rodrigues
c
and Hugo Gamboa
d
Department of Physics, Faculdade de Ciencias e Tecnologia da Universidade Nova de Lisboa, Monte de Caparica,
2892-516, Caparica, Portugal
Keywords:
Natural Language Processing, Machine Learning, Medical Text Mining, Biomedical Science, Clinical Notes.
Abstract:
Machine learning has demonstrated superior performance in solving many problems in various fields of
medicine compared to non-machine learning approaches. The aim of this review is to understand how Machine
Learning-based Natural Language Processing (ML-NLP) has been applied to the clinical notes databases. Op-
timization algorithms are listed as examples to demonstrate the simplicity and effectiveness of their applica-
tions for clinical notes database. We reviewed the literature in clinical applications of ML-NLP, particularly
techniques of deep learning such as mainly in pathology reports of diabetes, schizophrenia, cancer and cardi-
ology, where NLP either on a classical algorithm or with deep learning has been actively adopted. We covered
60 different studies in this domain, focusing on a wide range of medical perspective based algorithms. Ma-
chine learning-based approaches combine the benefits of health systems with the expertise and experience of
human well-being. From this review, it is clear that these techniques can improve the quantification of diagno-
sis and prognosis of cases and may create tools to assist patients during diagnosis and treatment. We complete
this work by providing guidelines on the applicability of ML-NLP by describing the most relevant libraries to
extract medical expressions from clinical reports text that can support clinical decision-making.
1 INTRODUCTION
Machine learning (ML) is a branch of Artificial In-
telligence (AI) that is derived from the study of pat-
tern recognition and computational learning theory. It
produces models that can learn from vast amounts of
data and make predictions based on that data (Provost
and Kohavi, 1998). Since ML models can learn from
data without the use of rules (Rodrigues et al., 2020),
non-ML refers to statistical modeling using mathe-
matical equations to formalize relationships between
variables in data (Gago et al., 2017). Additionally,
Deep Learning (DL) is a type of ML technology that
uses artificial neural networks to learn representations
(Goodfellow et al., 2016). Considering the signifi-
cant advantages of ML techniques in medical areas
(Ojo and Olanrewaju, 2019; Mollaei et al., 2021),
ML has been increasingly and widely used in a va-
riety of areas in medicine, with increased perspec-
tives of application in the coming decades (Sun et al.,
a
https://orcid.org/0000-0002-8332-489X
b
https://orcid.org/0000-0002-2998-976X
c
https://orcid.org/0000-0001-7320-511X
d
https://orcid.org/0000-0002-4022-7424
2017; Wang et al., 2017a; Wang et al., 2017b; Fer-
reira et al., 2020). Advanced analytical techniques
to extract informative features from clinical notes
and model underlying relationships that cannot be
modelled with traditional statistical tools could trans-
form biomedical research as they include techniques
such as speech recognition (Mitrofan and Ion, 2017)
and automated cancer detection (Halilaj et al., 2018).
Traditional ML methods were based on one dataset
and one task. Interestingly, the new generation of
ML consists of a) data: self-supervised, graphs, and
multi-model; b)systems: transfer learning, few shot
learning, federated learning, and mixture of experts;
c)techniques: transformers, deep reinforcement learn-
ing, and Generative Adversarial Networks (GANs)
(Thornton et al., 2021).
One of the other interesting fields of AI which was
introduced in 1950 is Natural Language Processing
(NLP) (Nadkarni et al., 2011). The way we communi-
cate through language has radically changed over the
past few years. With technological development, new
instruments have been used by humans to communi-
cate. Communication between humans and comput-
ers has never been as broad or as global as it is now.
Mollaei, N., Cepeda, C., Rodrigues, J. and Gamboa, H.
Biomedical Text Mining: Applicability of Machine Learning-based Natural Language Processing in Medical Database.
DOI: 10.5220/0010819500003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 4: BIOSIGNALS, pages 159-166
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
159

How does a computer comprehend the structure, orga-
nization, and even meaning of what is written? In that
regard, NLP is the computational science developed
to be this bridge between natural language and com-
puter language. NLP can be applied to all domains,
from our day-to-day tasks in auto-completion or spam
filtering, to medi-cal text analysis. In general, NLP
techniques include text generation, text classification,
text pro-cessing, and text comprehension. Currently,
ML strategies, both supervised (using sample input-
output pairs to learn a function that maps an input to
an output) and unsupervised (algorithms to find pat-
terns in data sets with unclassified or unlabeled data
points), dominate the main strategies for NLP appli-
cations. Not only NLP can use in medical database, it
can then be used in a broad set of applications, such
as machine translation, grammar check, spam filters,
sentiment analysis, etc. For example; based on unbal-
anced data on Adverse Drug Reactions (ADRs) in real
Electronic Health Records (EHRs) from the Spanish
population, the hybrid approach presented a combina-
tion of rule-based and machine learning-based tech-
niques. In a highly skewed categorization environ-
ment, recall and precision complement the elements
for knowledge-based and inferred models, accord-
ing to both intra-sentence and inter-sentence ADRs
(Casillas et al., 2016). DL has been applied to clini-
cal notes in Spanish and Swedish for medical named
entity recognition (Weegar et al., 2019). Other stud-
ies can evaluate medical texts like (Pardo et al., 2004)
producing rhetorical structures named DiZer of scien-
tific texts based upon the Rhetorical Structure Theory
(Rino et al., 2004). This work gives insight into the
concept of how text mining helps medical decision-
making. Hence, we listed a variety of biomedi-
cal text applications; 1)Large-scale classification of
biomedical documents; 2)Classification of biomedi-
cal databases using ML; 3)Collecting relevant docu-
ment databases and their methods; 4)How the delivery
of the retrieved information made concise and user-
understandable by critical libraries? While applica-
tions of ML-NLP procedures include medical notes, a
critical evaluation of studies that use ML-NLP meth-
ods remains difficult. As the biomedical field be-
comes data-intense and the use of ML, particularly
DL, continues to increase, good practices for con-
ducting and reporting research at the intersection of
biomedical text and ML are needed to ensure that con-
clusions are valid and reproducible. Researchers can
use the findings to develop an intuition for assigning
ML-NLP issues more effectively than they can using
traditional statistics. We also aim to investigate more
visible ML efforts and propose standards to increase
the quality and impact of future research in this excit-
ing area. To achieve this goal, we first review appli-
cations of ML encountered in the literature. Then, we
outline best practices for reporting the results of ML-
NLP analyses that focus on various diseases, such
as pathology reports, diabetes, schizophrenia, various
cancers, and cardiology. In the discussion that fol-
lowed, we debate some topics to overcome the chal-
lenges faced by biomedical text analysis. And, Sec-
tion 6 highlights opportunities where emerging tech-
niques are likely to have a significant impact in up-
coming years.
2 SEARCH OF METHODS
We conducted a search for original research articles
published up to September 7, 2021. For this rea-
son, using the arXiv
1
, medRxiv
2
, IEEE Xplore
3
,
Scopus
4
, ACM Digital Library
5
, PubMed
6
and
Google scholar database
7
(2004-) was considered.
This search used the phrase ”natural language pro-
cessing in the medical text domain” to get a sense
of how academics are using NLP to evaluate medical
texts. NLP, ML, Text Mining, Biomedical Science,
Medical Texts or Clinical Notes, and ”not related”
were the five categories in which the works were clas-
sified. These keywords were taken into account based
on the published papers’ titles and abstracts. The
automatic search (Figure 1) of 103 papers was fi-
nally selected. ML articles were investigated in two
categories: mathematical articles and papers dealing
with the diagnosis and prognosis of diseases affect-
ing health and well-being (Srinivasagopalan et al.,
2019). Overall, authors manually culled 60 papers
from those 62 to find 60 that discussed the use of
NLP in medical textual data. To find relevant stud-
ies, we employed search phrases from three separate
categories: 1)Identify clinical factors and patterns to
screen for such learning tasks. 2)ML terms, such as
DL (Akl et al., 2019), and neural network (Zhang and
Zhou, 2006) by taking into account their libraries.
3) medical terminology, such as pathology reports
(Brimo et al., 2010; Chang et al., 2019), diabetes (Xu
et al., 2014), schizophrenia (Srinivasagopalan et al.,
2019), various cancers (Fakoor et al., 2013; Sun et al.,
2017), and cardiology (Weissman et al., 2018). At
least one term from each of these three categories ap-
1
https://arxiv.org/
2
https://www.medrxiv.org/
3
https://ieeexplore.ieee.org/Xplore/home.jsp
4
https://www.scopus.com/
5
https://dl.acm.org/
6
https://pubmed.ncbi.nlm.nih.gov/
7
https://scholar.google.com
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
160
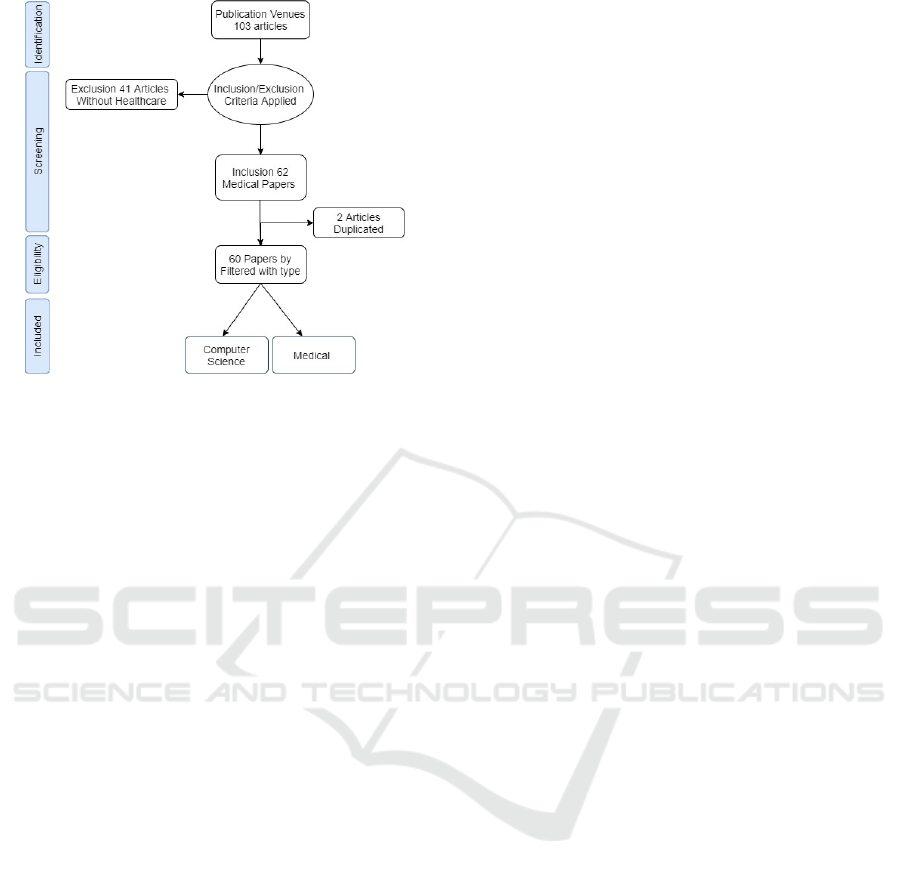
Figure 1: Preferred reporting items for reviews flowchart
detailing how selected articles based on identification,
screening, eligibility and included.
peared in the title or abstract of 60 considered arti-
cles. We also considered the following: dissertations,
conference proceedings, non-English articles, studies
whose primary outcome was not one of supervised or
unsupervised learning, and studies that utilized ML
followed by traditional inferential statistics. Among
these 60 articles, ten of them are considered the main
ones. Table 1 shows a brief summary of the tools of
these studies with their GitHub. It is worthwhile to
mention that table 2 has these tools, whether solo or
in combination.
3 RESULT
We found only 10 empirical papers that matched our
inclusion criterion. The use of inconsistent report-
ing methods on classifier performance made it im-
practical to compare model performance across stud-
ies. We did not provide commentary on models’ rel-
ative performance or a corresponding meta-analytic
examination of findings. Table 2 shows the most
important studies which are related to ML-NLP pa-
pers in clinical notes in terms of four questions for-
mulated in this table of ML-NLP: 1)What are the
medical studies addressed in this study?; 2)What are
the most frequently adopted combination methods?;
3)What are the database used?; 4)What are the solo
ML-NLP/ML/NLP methods?
Wang et al. introduced a new multi-task
learning framework using char-level neural models
for BioNER (Biomedical named entity recognition)
(Wang et al., 2019). A Bi-directional Long Short-
Term Memory-Conditional random field (BiLSTM-
CRF) model (Habibi et al., 2017; Lample et al., 2016)
is represented with an additional context-dependent
BiLSTM (Liu et al., 2018) layer for modeling charac-
ter sequences. A prominent advantage of this multi-
task model is that it was inputted from different 15
datasets that can effectively share both character- and
word-level representations by reusing parameters in
the corresponding BiLSTM units. DL has provided
a new advanced approach such as deep unified net-
works (DUNs), a new mesh-like network structure
of DL designed to avoid overfitting in comparison
with other neural networks (Rav
`
ı et al., 2016). Go-
las et al. improved the 30-day readmission risk for
this (magnetic resonance imaging) EMR-based pre-
diction model for heart failure patients on discharge
from hospital admission. By reducing features via
KL divergence, Logistic Regression (LR), Gradient
Boosting (GB), and Maxout networks cannot outper-
form AUC performance (Golas et al., 2018). Weng et
al. (Weng et al., 2017) applied the Integrating Data
for Analysis, Anonymization, and Sharing (iDASH)
data repository and Massachusetts General Hospital
datasets to the clinical NLP system, Clinical Text
and Knowledge Extraction (cTAKES) analysis with
a Unified Medical Language System (UMLS). These
databases can be found in the fields of neurology, car-
diology, and endocrinology. Furthermore, the ”nltk”
program was used to normalize the lexicon (word to-
kenization and stemming process) (Agarwal, 2015).
Hybrid bag-of-words (Marafino et al., 2014) with
UMLS concepts are restricted to five semantic groups,
with ”tf-idf” (Salton and Buckley, 1988) weighting
and linear Support Vector Machine (SVM) algorithm
(Fan et al., 2008) yielded the best performing clas-
sifier for medical sub-domain classification in the
”iDASH” database. In parallel, ”scikit-learn” pack-
age was selected for the supervised learning algo-
rithms implementation and model evaluation. ”gen-
sim” was used for document embedding. ”Tensor-
Flow” and ”Keras” were adopted for building deep
neural networks and neural word embedding. Con-
volutional Neural Network (CNN) (Xu et al., 2016)
and CRNN (Rakhlin, 2016) pre-trained fastText word
embedding with Adam optimization, (Kingma and
Ba, 2014) performed better than other top-performing
shallow supervised learning algorithms, such as lin-
ear SVM and regularized multinomial logistic regres-
sion, at document classification. A gated scaled dot
product is presented based on another neural network
for biomedical question answering tasks by the use of
transfer learning methods (Du et al., 2020). BioBERT
was the hidden representation of each token for ques-
tions and passages. The performance of the model
is pre-trained on the large-scale SQuAD dataset, and
Biomedical Text Mining: Applicability of Machine Learning-based Natural Language Processing in Medical Database
161

Table 1: Summary of the tools of ten main empirical papers.
Tools Description
bioBert W
A language model representing with Google-AI with deep bidirectional representations from unlabeled
medical text by jointly conditioning on both left and right context in all layers
skip-gram W
A method for learning high-quality distributed vector representations that capture a large number of syntactic
and semantic word associations.
fastText W
An algorithm for generating word vectors is provided by Facebook. This model can generate vectors for words
that have not been trained in the steps using character N-grams, which has been able to have better results than
previous models.
TF-IDF W A matrix with weights over word occurrences
Bag of Words W
A method to generate representations of sentences using representations of its component. This information is
stored in a matrix of data with rows and columns for each unique word. In this case, the features extracted are
purely statistical, but can provide a relevant measure of differences between documents.
BioNER W A NLP methods that recognizes specific names in medical texts
UMLS W Understanding the meaning of the language of biomedicine and health
Apache cTAKES W Extracting clinical information from electronic health record unstructured text
fine-tuning is implemented on the small target data set
of BioASQ. Conversely, a novel unsupervised deep
feature learning method (Miotto et al., 2016) used to
retrieve a general-purpose patient representation from
EHR data that facilitates clinical predictive modeling.
A lexical scanner with a syntactic method in seventy-
eight diseases such as diabetes, schizophrenia, and
various cancers is used by implementing principal
component analysis (PCA), three-layer stacked de-
noising auto-encoders (”Deep-patient”), K-Means,
Gaussian Mixture Model (GMM) Independent Com-
ponent Analysis (ICA) and supervised learning with
Random Forest. Some of the most recent studies
on how DL was used in text by keyword extrac-
tion algorithms is based on Bidirectional Encoder,
CNN, and Long Short-Term Memory (LTSM) (Kim
et al., 2020). In other words, the Bayes classifier and
the two feature-based key phrase extractors named
Wingnus (Nguyen and Luong, 2010). By using NLP
(Zhang et al., 2019) and text mining (Sung et al.,
2020) with association rules, automatically recog-
nize stressor entities and classify psychiatric stres-
sors from clinical text using NLP–based methods.
Other examples of extracting association rules from
medical records are: (Lakshmi and Vadivu, 2017)
by discovering the correlation between diseases, dis-
eases and symptoms, diseases and medicines, Nat-
ural Language Enhanced Association Rules Mining
(NEARM) (Ren et al., 2018) captured the relations
between lamentation text and natural language pat-
terns in the combining of the knowledge base. Re-
garding i2b2 clinical NLP challenge, (Chen et al.,
2015b) identified heart disease risk factors in clinical
texts over time with a hybrid pipeline system based on
machine learning-based and rule-based approaches.
Chen et al. (Chen et al., 2015a) classified the in-
jury text narrative with the evaluation of the feasibility
of matrix auto-encoders (Kingma and Welling, 2014)
factorization (NNMF-SVM) achieved the best perfor-
mance for both external cause and major injury factor
classification; Decision Tree (DT), Neural Network
(NN), and K-Nearest Neighbors (KNN).
4 DISCUSSION
This review summarized studies that used ML and
NLP to improve the understanding and processing of
biomedical text. The findings from 60 studies pub-
lished since 2004 were considered in the current re-
view. Based on this body of work, we concluded that
ML-NLP has shown promise in greatly boosting clin-
ical note text mining. Despite a recent growth in the
use of ML-NLP, we find that their application in this
field of research is still limited. We hope to identify
gaps in the literature and offer strategies to expand
on current findings so that researchers can fully grasp
ML-NLP’s promise. In recent advances in the size
of data sets, DL is crucial to developing novel algo-
rithms. In (Wang et al., 2020; Jang and Cho, 2019)
took a broad view of NLP techniques, from produc-
ing dependency parses to text-based event prediction.
clinical context is applied by BERT, but also the appli-
cability of GRU (e.g. (Belo et al., 2017) in signal pro-
cessing) can be introduced in biomedical text mining.
There are DL methods used in (Wang et al., 2020)
that can also be used in biomedical text mining, such
as incremental learning, (Sarwar et al., 2019), varia-
tional and GAN, (Mirza et al., 2014), but they were
not considered in the evaluated studies.
Additionally, aside from Adam optimization,
(Kingma and Ba, 2014) deep neural networks (Akl
et al., 2019) can be used to optimize hyperparame-
ters in clinical literature. On the one hand, most of
the studies in Table 2 utilized DL to improve their
algorithms. But, on the other hand, in (Chen et al.,
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
162
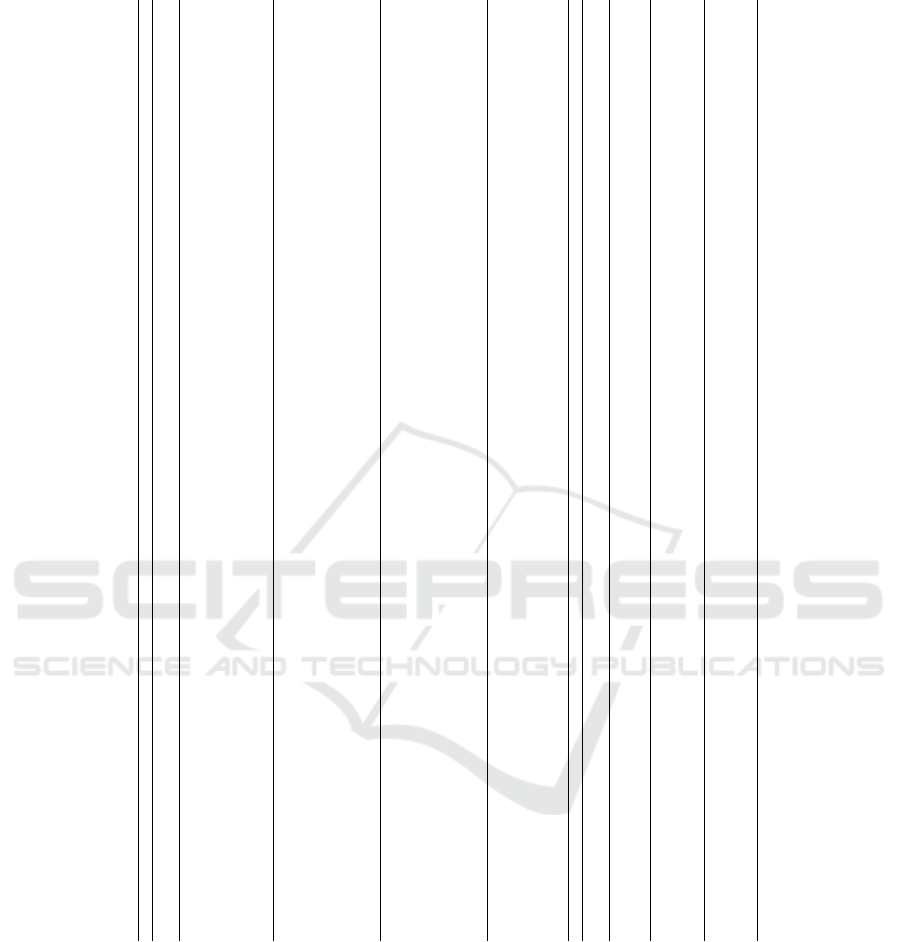
Table 2: The ML-NLP studies which used clinical notes in Biomedical text mining.
Authors, year Paper Type NLP Approach Text Description Disease Stastiscal Analysis
(Wang et al., 2019) Mathematics
BioNER and word embedding
with skip-gram model
Huge biomedical text in different areas - DL) LSTM
(Golas et al., 2018) Medical bag-of-word, Part-of-speech tagging
Physician notes and discharge
summaries cause of admission,
the patients’ hospital course and
discharge conclusions, and
instructions, Heart failure,
DUN, Logistic Regression,
Gradient Boosting, Maxout network
Heart failure
ML) Logistic
Regression,
Gradient Boosting, Maxout
network/ DL) DUN
(Weng et al., 2017) Medical
cTAKES, UMLS, ”bag-of-words”,
TF-IDF
Specialist-written notes and created;
an automated mapping script;
which allows the
mapping between note authors and
they’re medical specialization using
the PartnersEnterprise data,
warehouse (EDW),
physician database
Neurology, Cardiology,
and Endocrinology
ML) Support Vector Machine
DL) CRNN, CNN, and
LSTM With
Adam optimization
algorithm
(Du et al., 2020)
Biomedical
Question
Answering
-
Factoid questions on a set of
Wikipedia articles,
where the answer to every question is
a text segment
from the corresponding passage,
With 100,000+ question-answer pairs on
500+ articles from PubMed abstracts
and possible answers
-
Transfer Learning and
DL) BioBERT
and LSTM
(Miotto et al., 2016) Medical
A lexical scanner with
syntactic method
Diagnoses (ICD-9codes),
medications,
procedures, and lab tests,
free-text clinical notes
78 diseases
Unsupervised deep feature
learning ML)K-Means, Gaussian
mixture model,
PCA, independent component analysis
”ICA” and supervised learning with
Random Forest
(Kim et al., 2020) Medical - Pathology reports Pathology DL) BERT, CNN, LTSM
(Zhang et al., 2019) Medical Bag-of-word, POS tagging, n-grams
Suicidal behaiviors and sychiatric
Stressors text in EHR
Psychiatric Stress
Unsupervisd word
representation features
(Lakshmi and Vadivu, 2017) Medical
Stemming, POS tagging, Parsing, Stop
words removal, synonym finding, semantic
analysis, Negative scope identification
Disease, medicines, symptoms
text in EHR
Many diseases
ML) Apriori, Apriori Tid,
Close, RElim,
Eclat, FP-Growth
(Chen et al., 2015a) Medical TF-IDF and Binery weghing
Injury narrative
text
-
ML) SVM, KNN, NN,
AdaBoost, DT,
Naive Bayes algorithm,
NNMF
(Chen et al., 2015b) Medical
TF–IDF, n-grams, bag-of-words,
part-of-speech (POS) tags
Heart disease and
diabetes-associated
risk factors risk
factors in clinical texts
Heart disease, diabetes
ML) SSVMs, SVMs,
and CRFs,
2019) concluded that DL is useful for some tasks,
such as classifying medical images, but it is not ap-
propriate for other clinical data challenges. Accord-
ing to our experience across numerous clinical chal-
lenges, conventional, off-the-shelf ML approaches
can be trained faster and have overall higher perfor-
mance. ML and NLP hold great promise for improv-
ing clinical decision-making and accelerating rehabil-
itation programs (Sung et al., 2020), primarily for di-
agnosis and treatment of disease prognosis. To enable
proper use of advanced analytical approaches, open-
source databases, applications, and discussions must
be actively encouraged within the medical commu-
nity. Application in chronic disease (Sheikhalishahi
et al., 2019) was described in review publications as
re-enacting medical decision-making, which can aid
comprehension of how and why a decision is made.
In biology and medicine, (Ching et al., 2018) exam-
ined various biomedical problems in terms of patient
classification, fundamental biological processes, and
treatment of patients. Furthermore, Wu et al. (Wu
et al., 2020) investigated the use of DL as a baseline
Biomedical Text Mining: Applicability of Machine Learning-based Natural Language Processing in Medical Database
163

for NLP research, as well as the use of DL-based NLP
in the medical field is being conducted.
5 LIMITATION
Authors proposed medical publications introducing
ML-NLP and its applications for other research pur-
poses. The purpose of this article was to supply med-
ical references with an introduction to NLP and an in-
vestigation of current applications of NLP that may be
of interest to investigators. A limitation ubiquitous in
any study of a survey is publication bias. This paper
just took into account published manuscripts, which
may support a biased representation of the scope and
success of the study. Some investigators may submit
source code for software to online platforms such as
GitHub.
6 CONCLUSION & FUTURE
WORK
The main purpose of this work was to present the ad-
vances and novel developments in the area of ML-
NLP, the state of the art being developed, but also
the fundamentals and main strategies that have al-
ways been used until now. For this demonstration,
an exhaustive list of tools (Table 1), coupled with
their applicability, is presented. Hence, the extention
of tease tools that are used for ML-NLP applications
(Table 2) in the biomedical domain. With the help of
these tools, this work shows, explains, and exempli-
fies all the steps to solve most types of problems in
the ML-NLP field. Additionally, the investigation of
ML-NLP on topics like pathology reports of diabetes,
schizophrenia, cancer, and cardiology can be useful
to the healthcare area. This work was also a chal-
lenge to search for solutions that could contribute to
the development of new strategies for the analysis of
a symbolic representation of time series.
ACKNOWLEDGEMENTS
This work was partly supported by science and
technology foundation (FCT), under projects OP-
ERATOR (ref. 04/SI/2019) and PREVOCU-
PAI (DSAIPA/AI/0105/2019), and Ph.D. grants
PD/BDE/142816/2018 and PD/BDE/142973/2018.
The authors have no conflicts of interest to report.
REFERENCES
Agarwal, V. (2015). Research on data preprocessing and
categorization technique for smartphone review anal-
ysis. International Journal of Computer Applications,
975:8887.
Akl, A., El-Henawy, I., Salah, A., and Li, K. (2019). Op-
timizing deep neural networks hyperparameter posi-
tions and values. Journal of Intelligent & Fuzzy Sys-
tems, 37(5):6665–6681.
Belo, D., Rodrigues, J., Vaz, J. R., Pezarat-Correia, P., and
Gamboa, H. (2017). Biosignals learning and synthesis
using deep neural networks. Biomedical engineering
online, 16(1):1–17.
Brimo, F., Schultz, L., and Epstein, J. I. (2010). The
value of mandatory second opinion pathology review
of prostate needle biopsy interpretation before radical
prostatectomy. The Journal of urology, 184(1):126–
130.
Casillas, A., P
´
erez, A., Oronoz, M., Gojenola, K., and San-
tiso, S. (2016). Learning to extract adverse drug reac-
tion events from electronic health records in spanish.
Expert Systems with Applications, 61:235–245.
Chang, H. Y., Jung, C. K., Woo, J. I., Lee, S., Cho, J., Kim,
S. W., and Kwak, T.-Y. (2019). Artificial intelligence
in pathology. Journal of pathology and translational
medicine, 53(1):1.
Chen, D., Liu, S., Kingsbury, P., Sohn, S., Storlie, C. B.,
Habermann, E. B., Naessens, J. M., Larson, D. W.,
and Liu, H. (2019). Deep learning and alternative
learning strategies for retrospective real-world clinical
data. NPJ digital medicine, 2(1):1–5.
Chen, L., Vallmuur, K., and Nayak, R. (2015a). In-
jury narrative text classification using factorization
model. BMC medical informatics and decision mak-
ing, 15(1):1–12.
Chen, Q., Li, H., Tang, B., Wang, X., Liu, X., Liu, Z., Liu,
S., Wang, W., Deng, Q., Zhu, S., et al. (2015b). An
automatic system to identify heart disease risk factors
in clinical texts over time. Journal of biomedical in-
formatics, 58:S158–S163.
Ching, T., Himmelstein, D. S., Beaulieu-Jones, B. K.,
Kalinin, A. A., Do, B. T., Way, G. P., Ferrero, E.,
Agapow, P.-M., Zietz, M., Hoffman, M. M., et al.
(2018). Opportunities and obstacles for deep learn-
ing in biology and medicine. Journal of The Royal
Society Interface, 15(141):20170387.
Du, Y., Pei, B., Zhao, X., and Ji, J. (2020). Deep scaled dot-
product attention based domain adaptation model for
biomedical question answering. Methods, 173:69–74.
Fakoor, R., Ladhak, F., Nazi, A., and Huber, M. (2013).
Using deep learning to enhance cancer diagnosis and
classification. In Proceedings of the international con-
ference on machine learning, volume 28. ACM, New
York, USA.
Fan, R.-E., Chang, K.-W., Hsieh, C.-J., Wang, X.-R., and
Lin, C.-J. (2008). Liblinear: A library for large lin-
ear classification. the Journal of machine Learning
research, 9:1871–1874.
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
164

Ferreira, F., Gago, M., Mollaei, N., Bicho, E., Sousa, N.,
Gama, J., and Ferreira, C. (2020). Objective graphi-
cal clustering of spatiotemporal gait pattern in patients
with parkinsonism. In AIP Conference Proceedings,
volume 2293, page 420101. AIP Publishing LLC.
Gago, M., Ferreira, F. R., Mollaei, N., Rodrigues, M.,
Sousa, N., Bicho, E., and Rodrigues, P. (2017). Gait
analysis as a complementary tool in the levodopa dose
decision in vascular parkinson’s disease.
Golas, S. B., Shibahara, T., Agboola, S., Otaki, H., Sato,
J., Nakae, T., Hisamitsu, T., Kojima, G., Felsted, J.,
Kakarmath, S., et al. (2018). A machine learning
model to predict the risk of 30-day readmissions in
patients with heart failure: a retrospective analysis of
electronic medical records data. BMC medical infor-
matics and decision making, 18(1):1–17.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
learning. MIT press.
Habibi, M., Weber, L., Neves, M., Wiegandt, D. L., and
Leser, U. (2017). Deep learning with word embed-
dings improves biomedical named entity recognition.
Bioinformatics, 33(14):i37–i48.
Halilaj, E., Rajagopal, A., Fiterau, M., Hicks, J. L., Hastie,
T. J., and Delp, S. L. (2018). Machine learning in
human movement biomechanics: Best practices, com-
mon pitfalls, and new opportunities. Journal of biome-
chanics, 81:1–11.
Jang, H.-J. and Cho, K.-O. (2019). Applications of deep
learning for the analysis of medical data. Archives of
pharmacal research, 42(6):492–504.
Kim, Y., Lee, J. H., Choi, S., Lee, J. M., Kim, J.-H., Seok,
J., and Joo, H. J. (2020). Validation of deep learn-
ing natural language processing algorithm for key-
word extraction from pathology reports in electronic
health records. Scientific Reports, 10(1):1–9.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Kingma, D. P. and Welling, M. (2014). Stochastic gradi-
ent vb and the variational auto-encoder. In Second In-
ternational Conference on Learning Representations,
ICLR, volume 19.
Lakshmi, K. S. and Vadivu, G. (2017). Extracting associ-
ation rules from medical health records using multi-
criteria decision analysis. Procedia Computer Sci-
ence, 115:290–295.
Lample, G., Ballesteros, M., Subramanian, S., Kawakami,
K., and Dyer, C. (2016). Neural architectures
for named entity recognition. arXiv preprint
arXiv:1603.01360.
Liu, L., Shang, J., Ren, X., Xu, F., Gui, H., Peng, J.,
and Han, J. (2018). Empower sequence labeling with
task-aware neural language model. In Proceedings of
the AAAI Conference on Artificial Intelligence, vol-
ume 32.
Marafino, B. J., Davies, J. M., Bardach, N. S., Dean, M. L.,
and Dudley, R. A. (2014). N-gram support vector ma-
chines for scalable procedure and diagnosis classifica-
tion, with applications to clinical free text data from
the intensive care unit. Journal of the American Med-
ical Informatics Association, 21(5):871–875.
Miotto, R., Li, L., Kidd, B. A., and Dudley, J. T. (2016).
Deep patient: an unsupervised representation to pre-
dict the future of patients from the electronic health
records. Scientific reports, 6(1):1–10.
Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville,
A., Bengio, Y., et al. (2014). Generative adversarial
nets. Proceedings of Advances in Neural Information
Processing Systems.
Mitrofan, M. and Ion, R. (2017). Adapting the ttl roma-
nian pos tagger to the biomedical domain. In Biomed-
icalNLP@ RANLP, pages 8–14.
Mollaei, N., Londral, A. R., Cepeda, C., Azevedo, S., San-
tos, J. P., Coelho, P., Fragata, J., and Gamboa, H.
(2021). Length of stay prediction in acute intensive
care unit in cardiothoracic surgery patients. In 2021
Seventh International conference on Bio Signals, Im-
ages, and Instrumentation (ICBSII), pages 1–5.
Nadkarni, P. M., Ohno-Machado, L., and Chapman, W. W.
(2011). Natural language processing: an introduction.
Journal of the American Medical Informatics Associ-
ation, 18(5):544–551.
Nguyen, T. D. and Luong, M.-T. (2010). Wingnus:
Keyphrase extraction utilizing document logical struc-
ture. In Proceedings of the 5th international workshop
on semantic evaluation, pages 166–169.
Ojo, A. K. and Olanrewaju, A. B. (2019). Knowledge dis-
covery in medical database using machine learning
techniques. International Journal of Computer Ap-
plications, 975:8887.
Pardo, T. A. S., Nunes, M. d. G. V., and Rino, L. H. M.
(2004). Dizer: An automatic discourse analyzer for
brazilian portuguese. In Brazilian Symposium on Ar-
tificial Intelligence, pages 224–234. Springer.
Provost, F. and Kohavi, R. (1998). Glossary of terms. Jour-
nal of Machine Learning, 30(2-3):271–274.
Rakhlin, A. (2016). Convolutional neural networks for sen-
tence classification. GitHub.
Rav
`
ı, D., Wong, C., Deligianni, F., Berthelot, M., Andreu-
Perez, J., Lo, B., and Yang, G.-Z. (2016). Deep learn-
ing for health informatics. IEEE journal of biomedical
and health informatics, 21(1):4–21.
Ren, S., Li, Z., Wang, H., Li, Y., Shen, K., and Cheng, S.
(2018). Nearm: Natural language enhanced associa-
tion rules mining. In 2018 IEEE International Con-
ference on Data Mining Workshops (ICDMW), pages
438–445. IEEE.
Rino, L. H. M., Pardo, T. A. S., Silla, C. N., Kaestner, C.
A. A., and Pombo, M. (2004). A comparison of auto-
matic summarizers of texts in brazilian portuguese. In
Brazilian Symposium on Artificial Intelligence, pages
235–244. Springer.
Rodrigues, J., Gamboa, H., Mollaei, N., Os
´
orio, D.,
Assunc¸
˜
ao, A., Fuj
˜
ao, C., and Carnide, F. (2020). A ge-
netic algorithm to design job rotation schedules with
low risk exposure. In Doctoral Conference on Com-
puting, Electrical and Industrial Systems, pages 395–
402. Springer.
Biomedical Text Mining: Applicability of Machine Learning-based Natural Language Processing in Medical Database
165

Salton, G. and Buckley, C. (1988). Term-weighting ap-
proaches in automatic text retrieval. Information pro-
cessing & management, 24(5):513–523.
Sarwar, S. S., Ankit, A., and Roy, K. (2019). Incremental
learning in deep convolutional neural networks using
partial network sharing. IEEE Access, 8:4615–4628.
Sheikhalishahi, S., Miotto, R., Dudley, J. T., Lavelli, A.,
Rinaldi, F., and Osmani, V. (2019). Natural lan-
guage processing of clinical notes on chronic dis-
eases: systematic review. JMIR medical informatics,
7(2):e12239.
Srinivasagopalan, S., Barry, J., Gurupur, V., and
Thankachan, S. (2019). A deep learning approach
for diagnosing schizophrenic patients. Journal of
Experimental & Theoretical Artificial Intelligence,
31(6):803–816.
Sun, W., Tseng, T.-L. B., Zhang, J., and Qian, W. (2017).
Enhancing deep convolutional neural network scheme
for breast cancer diagnosis with unlabeled data. Com-
puterized Medical Imaging and Graphics, 57:4–9.
Sung, S.-F., Lin, C.-Y., and Hu, Y.-H. (2020). Emr-
based phenotyping of ischemic stroke using super-
vised machine learning and text mining techniques.
IEEE journal of biomedical and health informatics,
24(10):2922–2931.
Thornton, J. M., Laskowski, R. A., and Borkakoti, N.
(2021). Alphafold heralds a data-driven revolu-
tion in biology and medicine. Nature Medicine,
27(10):1666–1669.
Wang, C., Elazab, A., Wu, J., and Hu, Q. (2017a). Lung
nodule classification using deep feature fusion in chest
radiography. Computerized Medical Imaging and
Graphics, 57:10–18.
Wang, X., Yang, W., Weinreb, J., Han, J., Li, Q., Kong,
X., Yan, Y., Ke, Z., Luo, B., Liu, T., et al. (2017b).
Searching for prostate cancer by fully automated mag-
netic resonance imaging classification: deep learning
versus non-deep learning. Scientific reports, 7(1):1–8.
Wang, X., Zhang, Y., Ren, X., Zhang, Y., Zitnik, M.,
Shang, J., Langlotz, C., and Han, J. (2019). Cross-type
biomedical named entity recognition with deep multi-
task learning. Bioinformatics, 35(10):1745–1752.
Wang, X., Zhao, Y., and Pourpanah, F. (2020). Recent ad-
vances in deep learning.
Weegar, R., P
´
erez, A., Casillas, A., and Oronoz, M. (2019).
Recent advances in swedish and spanish medical en-
tity recognition in clinical texts using deep neural ap-
proaches. BMC medical informatics and decision
making, 19(7):1–14.
Weissman, G. E., Hubbard, R. A., Ungar, L. H., Harhay,
M. O., Greene, C. S., Himes, B. E., and Halpern,
S. D. (2018). Inclusion of unstructured clinical text
improves early prediction of death or prolonged icu
stay. Critical care medicine, 46(7):1125.
Weng, W.-H., Wagholikar, K. B., McCray, A. T., Szolovits,
P., and Chueh, H. C. (2017). Medical subdomain clas-
sification of clinical notes using a machine learning-
based natural language processing approach. BMC
medical informatics and decision making, 17(1):1–13.
Wu, S., Roberts, K., Datta, S., Du, J., Ji, Z., Si, Y., Soni,
S., Wang, Q., Wei, Q., Xiang, Y., et al. (2020). Deep
learning in clinical natural language processing: a me-
thodical review. Journal of the American Medical In-
formatics Association, 27(3):457–470.
Xu, J., Chen, D., Qiu, X., and Huang, X. (2016).
Cached long short-term memory neural networks
for document-level sentiment classification. arXiv
preprint arXiv:1610.04989.
Xu, R., Li, L., and Wang, Q. (2014). driskkb: a large-scale
disease-disease risk relationship knowledge base con-
structed from biomedical text. BMC bioinformatics,
15(1):1–13.
Zhang, M.-L. and Zhou, Z.-H. (2006). Multilabel neu-
ral networks with applications to functional genomics
and text categorization. IEEE transactions on Knowl-
edge and Data Engineering, 18(10):1338–1351.
Zhang, Y., Zhang, O. R., Li, R., Flores, A., Selek, S., Zhang,
X. Y., and Xu, H. (2019). Psychiatric stressor recogni-
tion from clinical notes to reveal association with sui-
cide. Health informatics journal, 25(4):1846–1862.
BIOSIGNALS 2022 - 15th International Conference on Bio-inspired Systems and Signal Processing
166
