
PodNet: Ensemble-based Classification of Podocytopathy on Kidney
Glomerular Images
George Oliveira Barros
1,6
, David Campos Wanderley
2
, Luciano Oliveira Rebouc¸as
3
,
Washington L. C. dos Santos
4
, Angelo A. Duarte
5
and Flavio de Barros Vidal
6
1
Instituto Federal Goiano, Posse-GO, Brazil
2
Instituto de Nefrologia, Faculdade de Sa
´
ude e Ecologia Humana, Belo Horizonte-MG, Brazil
3
Federal University of Bahia, Salvador-BA, Brazil
4
Instituto Gonc¸alo Moniz, Fiocruz, Salvador-BA, Brazil
5
State University of Feira de Santana, Feira de Santana-BA, Brazil
6
Department of Computer Science, University of Brasilia, Bras
´
ılia-DF, Brazil
Keywords:
Computational Pathology, Podocitopathy, Deep Learning, Glomeruli, Podocitopathy Data Set.
Abstract:
Podocyte lesions in renal glomeruli are identified by pathologists using visual analyses of kidney tissue sec-
tions (histological images). By applying automatic visual diagnosis systems, one may reduce the subjectivity
of analyses, accelerate the diagnosis process, and improve medical decision accuracy. Towards this direction,
we present here a new data set of renal glomeruli histological images for podocitopathy classification and a
deep neural network model. The data set consists of 835 digital images (374 with podocytopathy and 430
without podocytopathy), annotated by a group of pathologists. Our proposed method (called here PodNet) is
a classification method based on deep neural networks (pre-trained VGG19) used as features extractor from
images in different color spaces. We compared PodNet with other six state-of-the-art models in two data set
versions (RGB and gray level) and two different training contexts: pre-trained models (transfer learning from
Imagenet) and from-scratch, both with hyperparameters tuning. The proposed method achieved classification
results to 90.9% of f1-score, 88.9% precision, and 93.2% of recall in the final validation sets.
1 INTRODUCTION
Computational pathology is a research area that as-
sociates biological tissue analysis with digital image
processing and computer vision techniques (Srinidhi
et al., 2021). As a result of advances in image analy-
sis algorithms, the main approaches adopted are cur-
rently based on deep learning architectures (Deng
et al., 2020).
The difficulty of finding fully annotated medical
image data sets and various cases associated with the
complexity of the anatomy of the images’ biological
structures makes the development of histological im-
age analysis systems a challenging task.
Currently, there are many proposals in the litera-
ture for automatic histological image analysis systems
applied to different organs and diseases (Yari et al.,
2020; Candelero et al., 2020; Thomas et al., 2021).
However, there are little explored diseases, as podocy-
topathy.
Podocytes are cells of the kidneys’ visceral ep-
ithelium, and they are present in the internal struc-
ture of renal glomeruli (Chen et al., 2006). The pri-
mary function of podocytes is to restrict the passage
of proteins from the blood through the urine (Nagata,
2016). Podocyte lesions can compromise the ability
of a glomerulus to filter proteins, causing damage to
the glomerular structure, and are biomarkers of pro-
gressive glomerulosclerosis (Saga et al., 2021) (See
in Figure. 1 image with (a) and without (b) podocy-
topathy).
The diagnostic of the lesions in renal glomeruli
may substantially vary according to the experience of
the pathologist and a system that could automatically
classify such lesions could be of great help for pathol-
ogists. On one hand, these systems could reduce the
subjectivity of analyses, accelerate the diagnosis pro-
cess, and improve medical decision accuracy; on the
other hand, they could be used as a teaching tool for
training new pathologist (Jayapandian et al., 2021).
Barros, G., Wanderley, D., Rebouças, L., Santos, W., Duarte, A. and Vidal, F.
PodNet: Ensemble-based Classification of Podocytopathy on Kidney Glomerular Images.
DOI: 10.5220/0010828600003124
In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022) - Volume 5: VISAPP, pages
405-412
ISBN: 978-989-758-555-5; ISSN: 2184-4321
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
405

Figure 1: Disease example: (a) with podocytopathy (PAS
stain)(degeneration); (b) Without podocytopathy (PAS
stain). Some podocytes indicated by the arrows.
The evolution of artificial intelligence systems allow
them to achieve better results in many others areas,
as described in (Salas et al., 2019; Silva et al., 2020;
Abade et al., 2021; Abade. et al., 2019).
To visually recognize that a glomerulus has
podocytopathy, a pathologist needs to identify the
podocytes and then the lesions. However, podocytes
are easily confused with other intraglomerular cells
(endothelial and mesangial for example). Therefore,
a system that automatically classifies a glomerulus
image with podocytopathy can be a very useful aid
tool in the practice of nephropathologists (Maraszek
et al., 2020; Zimmermann et al., 2021; Zeng et al.,
2020; Govind et al., 2021b). We found few propos-
als for automatic podocyte analysis systems and none
of the studies found classify glomerulus images con-
cerning the presence of podocytopathy, but segment
podocytes to associate them with other diseases.
In this work, we propose a method, an artificial
intelligence system based on a convolutional neural
network (CNN) for the classification of histological
images of renal glomeruli with podocytopathy. We
also present a novel labeled public data set of renal
glomeruli images with podocytopathy.
2 RELATED WORKS
The literature review that we carried out for this work
focused on seeking two groups of researches: (i) clas-
sification of lesions in renal glomeruli and (ii) classi-
fication of glomerular podocytopathies.
Among the works on the classification of lesions
in glomeruli, the classification task is performed in
two ways: (i) in a single step, using images of iso-
lated glomeruli, or (ii) in two steps, performing the
segmentation of glomeruli in the Whole Slide Images
(WSI) before executing the classification. We present
both works below, focusing on the results of the clas-
sification task.
A recent work (Yang et al., 2021), classified the
glomeruli into five classes of lesion (including scle-
rosis) using the Densenet network (Huang et al.,
2018) and an LSTM (Long Short Term Memory) (Ul-
lah et al., 2018). The data set consisted of 1379
WSI (41886 glomeruli), stained in HE (15298), PAM
(5649), PAM(5641), and trichrome (5679). The best
result achieved for the task was 94.0% accuracy for
the different lesions in HE stained images. A previ-
ous work (Kannan et al., 2019), used 1706 images of
glomeruli, stained in trichrome to classify them into
four classes: (i) non-glomerulus, (ii) normal (iii) par-
tially sclerosed and (iv) globally sclerosed. The net-
work used was Inception v3 (Szegedy et al., 2015)
and the result obtained was a classifier capable of dis-
criminating non-glomerular images and images with
lesions with an accuracy of 92.67% ± 2.02%. In
the work of (Gallego et al., 2021), after segmenting
glomeruli with the U-Net network, they perform the
classification of glomeruli as normal or sclerotic. The
data set used had 51 tissue slides, stained in PAS (37)
and HE (14). The obtained results were of F1 of
94.0% (normal) and 76.0% (sclerosed).
Following the presented work in (Bukowy et al.,
2018), has proposed a method for identifying healthy
or injured glomeruli (without specifying the type
of lesion). The data set used had 87 WSI slides,
all stained with trichrome. The network used was
Alexnet and the average precision and recall were
96.94% and 96.79%, respectively. (Jiang et al., 2021)
classify glomeruli into 3 classes: normal, with sclero-
sis, or other lesions. The network was trained with
1123 snapshots, which are smaller portions of the
blade with one or more glomeruli. The results ob-
tained in the work were f1 scores of 91.4%, 089.6%,
68.1%, and 75.6%, for normal glomeruli, with sclero-
sis, global sclerosis, and other lesions, respectively.
In the work of (Uchino et al., 2020) 15888 im-
ages of renal glomeruli were used, distributed among
7 classes of histological lesion types: global sclero-
sis, segmental sclerosis, endocapillary proliferation,
mesangial matrix accumulation, mesangial cell pro-
liferation, crescent and structural membrane changes
basal. The network used was Inception v3 and the
best result obtained was an area under the curve (auc)
of 0.986 for classification of stained images in PAS
and 0.983 in PAMS. (Mathur et al., 2019) performs
two tasks: (i) classifies glomeruli as normal or ab-
normal and (ii) classifies regions of tissue without
glomerulus into three classes of fibrosis: mild, mod-
erate or severe. The data sets were composed of
patches of images extracted from tissue slides, total-
ing 935 images of glomeruli in data set 1 and 923 im-
ages of regions without glomerulus in data set 2. The
method used was a new proposed model, the Multi-
Gaze Attention Network (MGANet). The result ob-
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
406

tained was 87.25% and 81.47% accuracy for classifi-
cation of glomeruli and fibrosis, respectively.
As described in (Barros et al., 2017) a classifier
of renal glomeruli images regarding hypercellularity
was proposed. The result obtained was 88.3% accu-
racy. The data set had 811 images stained in PAS
and HE. Based on this work (Chagas et al., 2020)
performed the same task and data set, however, us-
ing convolutional neural networks as features extrac-
tor and classification with the SVM algorithm. In ad-
dition to the binary classification task, the authors per-
formed the classification of sub-lesions of hypercel-
lularity: mesangial, endocapillary, and both, reaching
an average accuracy of 82.0%. Both in the binary task
and the multiclass classification, the proposed method
surpassed the Xception, ResNet50, and Inception v3
networks, as well as the (Barros et al., 2017) method.
Among the works focusing on automatic podocyte
analysis, we found works on podocyte detection and
segmentation, however, none focused on podocy-
topathy, but to associate podocytes with other dis-
eases (Zeng et al., 2020; Govind et al., 2021b; Govind
et al., 2021a; Zimmermann et al., 2021; Maraszek
et al., 2020).
The work of the (Zeng et al., 2020) aimed to lo-
cate glomeruli, classify glomerular lesions, and iden-
tify and quantify different intrinsic glomerular cells.
For the task of classification of glomeruli, 1438 im-
ages of glomeruli were used. The method used in
the classification was the DenseNet-121 and LSTM-
SENet networks. In the internal cell segmentation
task (including podocytes), 460 images of glomeruli
were used, containing approximately 70 thousand an-
notated cells. The method used in the segmentation
of the internal cells was the 2D V-Net network. The
results obtained in the study were 95.0% for classi-
fication of the glomerular lesion, 88.2% for average
precision, and 87.9% for average recall for detection
of glomerular internal cells.
In (Govind et al., 2021a), the authors use PAS
stained WSI slides to detect and quantify podocytes
and correlate podocyte loss in diseased glomeruli.
The data set consisted of 122 slides stained in PAS,
with images originating from rat, mouse, and human
tissue. The results obtained were a sensitivity and
specificity of 0.80/0.80, 0.81/0.86, and 0.80/0.91 in
mice, rats, and humans, respectively. (Govind et al.,
2021b), perform the segmentation and quantification
of podocytes for recognition of Wilms’ tumor’ 1. The
data set used was composed of PAS stained images,
originating from mice. The proposed method uses
a GAN (Generative Adversarial Network) to convert
images of glomeruli to its immunofluorescence ver-
sion, where the segmentation of podocytes occurs by
traditional image processing methods. The result ob-
tained was 0.87 sensitivity and 0.93 specificity for de-
tecting podocytes.
In (Maraszek et al., 2020) the objective was also
to detect and quantify renal podocytes, however, to
associate them with the presence of diabetes melli-
tus. Like the work of (Govind et al., 2021b), the pro-
posed method used immunofluorescence images with
PAS versions of the same glomeruli. Immunofluo-
rescence imaging was processed with classical meth-
ods of digital image processing. The data set con-
sisted of 883 images of rat glomeruli. At the end
of the work, the authors calculated the damage to
the glomeruli through morphological analysis of the
podocytes and their intraglomerular distribution. The
results obtained had a sensitivity of 72.7%, specificity
of 99.9%, and accuracy of 95.9% in the location of
podocytes.
Finally, (Zimmermann et al., 2021) used a data
set with 1095 immunofluorescence images, contain-
ing a total of 27696 labeled podocytes. The aim of
the study was also to detect podocytes but to asso-
ciate them with the disease antineutrophil cytoplas-
mic antibody-associated glomerulonephritis (ANCA-
GN). The network used to segment glomeruli and
podocytes was the U-net. The Dice coefficient ob-
tained in the segmentation tasks of both glomeruli and
podocytes (0.92) was greater than 0.90.
3 MATERIAL AND METHODS
3.1 Data Set Preparation
The data set used in this work has 835 images of re-
nal glomeruli (340 with podocytopathy and 430 with-
out podocytopathy). All images were labeled by two
pathologists and were obtained from different institu-
tions and laboratories. The data set was available by
Instituto Gonc¸alo Moniz - FIOCRUZ and comprises
images stained in trichrome (173), Periodic Acid-
Schiff (PAS)(409), PAM (169), and Hematoxylin and
Eosin (H&E) (74). The images were captured using
whether cameras were attached to microscopy or by
digital scanners and came in different formats (JPG,
PNG, and TIF) and resolutions (from 238 × 201
to 1920 × 1440 pixels). The images labeled ”with
podocytopathy” have different podocyte lesion types:
hypertrophy, hyperplasia, and degeneration. Some
samples are depicted in Figure. 2. Additionally, both
in the group of images with podocytopathy and in the
group without podocytopathy there are other types of
associated lesions (hypercellularity, sclerosis, mem-
branous).
PodNet: Ensemble-based Classification of Podocytopathy on Kidney Glomerular Images
407
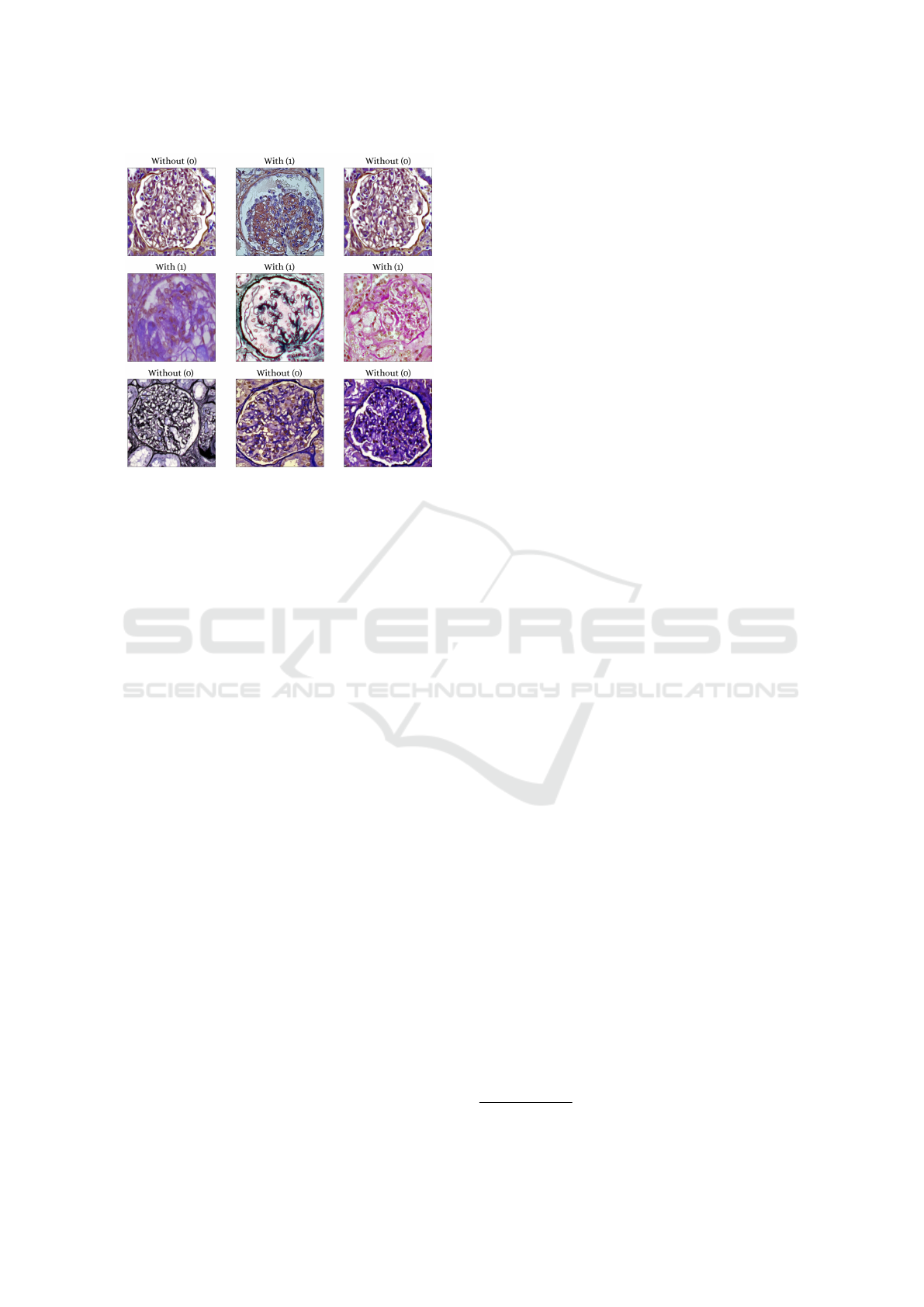
Figure 2: Examples of images that make up the data set of
glomeruli with podocytopathy and without podocytopathy.
3.2 Proposed Method (PodNet)
We built our solution over the hypothesis that when
converting the images, originally in the RGB color
space, to another color spaces that could isolate in-
formation from the stain used in the acquisition of
the images, associated with the extraction of features
through pre-trained convolutional neuron networks (a
practice adopted by (Chagas et al., 2020) and (Mathur
et al., 2019)), could extract complementary features,
which does not occur using an end-to-end network
with images in a single color space.
Traditional convolutional neural network architec-
tures receive images in only a certain color space.
However, given the possibility of isolating the infor-
mation of the stains with which the images were ob-
tained in channels of a color space, our hypothesis
that, associating features extracted from the images in
different color spaces could result in a classification
method more robust, is plausible.
The method is organized into three steps: Prepro-
cessing, feature extraction, and classification. The fig-
ure. 3 illustrates the architecture of the network and its
three steps.
In the first step, preprocessing, the images were
normalized (values between 0 and 1). Then, the con-
version from the RGB version to the HED and HDX
versions takes place. The HED (used in (Barros et al.,
2017)) convert into color space information relating
to Hematoxylin, Eosin, and DAB stains into its chan-
nels. The HDX color space converts into color spaces
channels of Hematoxylin and PAS information.
The second step is the extraction of features,
which is performed by processing the 3 images re-
sulting from the conversion to the VGG19 network
pre-trained with the Imagenet data set. The network
has been modified so that the output is the result of
max-pooling of the last convolutional layer. After
that, a flatten operation is performed, which generates
a vector of features for each image. Then, each of the
three feature vectors is scaled using the PCA algo-
rithm (Tipping and Bishop, 2006). The objective of
this operation, in addition to reducing the network’s
hyperparameters, is to speed up the training process
and eliminate unimportant features. Finally, the three
vectors are concatenated, resulting in a single vector.
In the classification step, there is a dense artifi-
cial neural network formed by an input layer of 350
neurons (features resulting from the PCA algorithm)
and 4 hidden layers (256, 128, 64 and 64 neurons, re-
spectively), with dropout regularization (0.1) between
each hidden layer. The hyperparameters of this net-
work were tuned with the Keras (Chollet et al., 2015)
grid search tool strategy. The network output is a neu-
ron with a sigmoid activation function.
3.3 Experiments Protocol
All the experiments reported in this work were made
using Python 3.6.8 programming language, Tensor-
flow 2.4
1
deep learning library for GPU, Docker envi-
ronment (Merkel, 2014), and Keras Tuner (O’Malley
et al., 2019) (for hyperparameters tuning), running on
NVidia Geforce 2080 TI (11 GB) GPU.
The classification results obtained with the pro-
posed method were compared with six other mod-
els based on deep learning architectures: Resnet101
v2, VGG19, Densenet201, Inception Resnet v2, In-
ception v3, and Xception. These architectures were
chosen, especially, because they represent different
models in terms of depth and learning strategy, which
allowed a broad analysis of the performance of con-
ventional networks for the execution of the proposed
task.
The training and validation of the networks was
carried out in two ways: (i) Generalization test and
(ii) Final validation. In the final validation the entire
data set was divided into 70% for training and 30%
for testing. In the generalization test, a 5-fold cross
validation was performed on the training set used in
the final validation (on the same set of 70% of the
data). In the training sets of the cross-validation and
the final validation set, the same data augmentation
was performed. The operations performed were: hori-
zontal and vertical flip, rotation (30, 90, 270 degrees),
1
More info.: https://www.tensorflow.org/.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
408

Figure 3: Proposal architecture. The proposed method (PodNet) extracts feature from the image in different color spaces with
the pre-trained VGG19 network.
brightness (range from 0.1 to 0.3) and random zoom
(from 0.2 to 0.5 times). These data augmentation op-
erations and value ranges were chosen because, in ad-
dition to enlarging the data set, they do not mischar-
acterize the original images, as occurs in some opera-
tions not suitable for medical images.
Before training the models, a hyperparameters
tuning was performed. The adjusted hyperparame-
ters were: Batch size (16, 32 and 62), number of neu-
rons from top dense layers (2048, 1024, 512 and 256),
learning rate (to 0.1 from 0.0000001), top dense lay-
ers activation functions (softmax, relu and tanh), op-
timizer (RMSprop, Adam, SGD and Adamax), mo-
mentum (0.3, 0.6 and 0.9) and loss function (binary
crossentropy and hinge).
Baseline networks were trained in two scenarios:
(1) Trained from scratch (with initialization of ran-
dom weights) and (2) trained with transfer learning
(with weights initialization of Imagenet data set (Rus-
sakovsky et al., 2015)). In the scenario trained from
scratch, the baseline networks were also trained with
the data set in the gray level version. This was done
to observe the effects of the absence of color informa-
tion over the performance of the networks.
In the transfer learning scenario, the weights of
networks trained on the Imagenet data set were loaded
into baseline networks (pre-trained networks). The
top layers of the baseline networks (with 1000 neu-
rons - originals classes number of the Imagenet data
set) were replaced by new layers: two dense layers
and one neuron (with sigmoid activation) for binary
classification (with and without injury). The last step
was to perform a tuning training of the added top lay-
ers by five epochs with all other layers frozen, and
finally, unfreezes and trains all layers of the networks.
We use the early stopping (monitor = validation
loss) as a strategy to stop training networks with pa-
tience 5 for all models (to avoid over fitting). The final
state of the baseline models was obtained by loading
the best weights obtained during net training.
4 RESULTS AND DISCUSSION
The metrics used to calculate the performance of the
networks were: precision, recall, and f1-score (James
et al., 2013). We also calculated the area under the
ROC curve of all evaluated models on final validation
set (see Baselines ROC curves in Figures: 5, 6 and
7. See Top 4 ROC curves models in Figure: 4). The
results show in Table 1 are the average of the met-
rics calculated for the five folds from cross-validation
(generalization set) and final validation results.
The complexity and diversity of data set images
and the fact that the discriminatory features are con-
tained in some nuclei, which are microscopic struc-
tures, contributed to the general results not reaching
absolute values. Additionally, even with data aug-
mentation operations, the data set is still small. The
method proposed presented satisfactory results when
compared to the other networks, with the highest F1
score in the final validation (90.0%) and the second
better on the generalization set (90.1%). The net-
work with the best values in the generalization set was
Resnet 101 v2 (90.2%) trained with transfer learning
with RGB data set version but presented a much lower
result in the f1 score final validation (84.4%), which
is a more relevant test set, as it has the largest number
of samples.
We also consider the results to be good, when
comparing the results of PodNet with other studies
on the classification of glomeruli concerning other
lesions, such as:(Mathur et al., 2019) (87.25% and
81.47% of f1 score for Fibrosis), (Gallego et al., 2021)
PodNet: Ensemble-based Classification of Podocytopathy on Kidney Glomerular Images
409
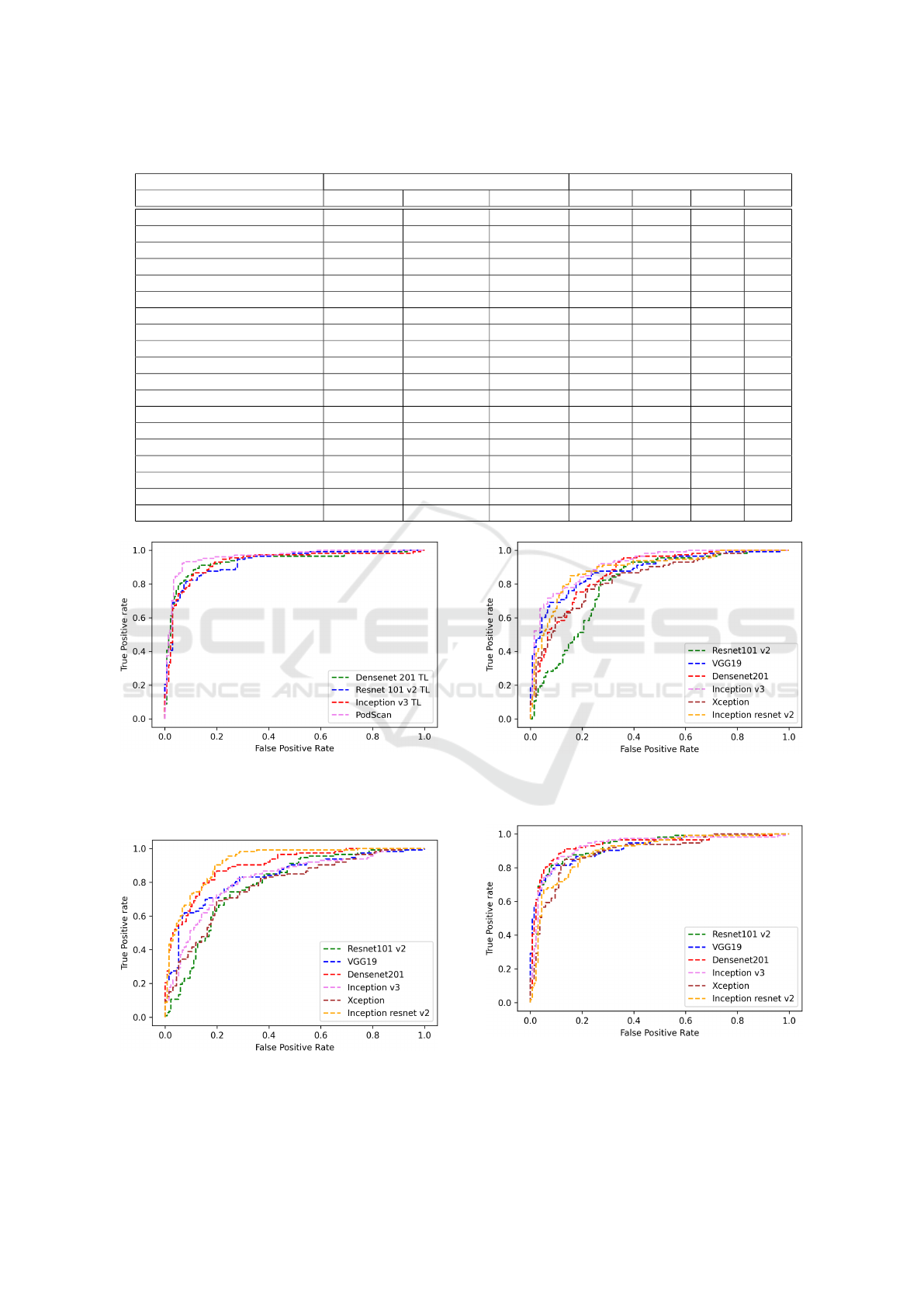
Table 1: Summary of results obtained in all models evaluated and ranked from F1-score in the final validation set.
Classification Generalization Set (average) Validation Set
Models Prec(%) Rec(%) F1(%) Prec(%) Rec(%) F1(%) AUC
Proposed method (PodNet) 90.6±3.07 89.6±1.36 90.1±1.70 88.9 93.2 90.9 0.959
Densenet201 TL (RGB) 90.0±3.66 90.0±5,31 88.0±3.89 85.0 91.0 87.8 0.935
Inception v3 TL (RGB) 87.0 ± 1.03 88.0 ± 8.93 87.4 ± 4.76 81.0 90.0 85.2 0.928
Resnet101 v2 TL (RGB) 94.0±2.65 86.0±7.44 90.2±3.54 83.00 86.0 84.4 0.927
VGG19 TL (RGB) 93.0±2.56 86.0±6.03 89.3±4.11 87.0 81.0 83.8 0.919
Xception TL (RGB) 89.0±2.66 90.0±8.26 88.4±4.69 82.0 84.0 82.9 0.893
Inception Resnet v2 FS (RGB) 86.0 ± 5.9 75.0 ± 12.02 80.4 ± 8.02 79.0 87.0 82.8 0.921
Inception Resnet v2 TL (RGB) 92.0±3.76 90.0±5.75 87.4±3.22 79.0 86.0 82.3 0.896
Densenet201 FS (RGB) 82.0 ± 7.9 83.0 ± 9.02 82.0 ± 8.52 75.0 88.0 80.9 0.895
Inception v3 FS (GL) 80.0 ± 6.94 67.0 ± 6.02 78.2 ± 8.02 77.0 84.0 80.3 0.915
Resnet101 v2 FS (GL) 80.0 ± 6.7 82.0 ± 1.79 76.7 ± 9.23 72.00 92.0 80.3 0.801
Inception Resnet v2 FS (GL) 79.0 ± 7.67 88.0 ± 9.60 83.2 ± 7.44 83.0 77.0 79.8 0.888
Densenet201 FS (GL) 72.0 ± 7.8 84.0 ± 1.36 88.0 ± 7.1 69.0 91.0 78.4 0.865
VGG19 FS (GL) 86.0±1.41 61.0±14.4 71.3±8,44 78.0 79.0 78.4 0.883
Xception FS (GL) 82.0 ± 5.02 69.0 ± 5.82 75.3 ± 5.28 70.0 80.0 74.6 0.835
Inception v3 FS (RGB) 83.0 ± 4.02 78.0 ± 10.0 80.4 ± 7.42 71.0 78.0 74.3 0.816
Resnet101 v2 FS (RGB) 72.0 ± 10.1 65.0 ± 14.0 69.6 ± 11.0 71.00 74.0 72.4 0.784
VGG19 FS (RGB) 89.0±4.80 80.0±9.80 84.3±5.80 87.0 61.0 71.8 0.833
Xception FS (RGB) 76.0 ± 5.92 69.0 ± 12.0 69.4 ± 8.80 71.0 69.0 69.9 0.781
Figure 4: ROC curves from top 4 models. The proposed
method is the best area under curve followed by three mod-
els trained with transfer learning in the RGB dataset ver-
sion.
Figure 5: ROC curves from baseline models trained with
from scratch in the RGB dataset version. The best model
this training context was Inception Resnet v2.
Figure 6: ROC curves from baseline models trained with
from scratch in the gray level dataset version. The best
model this training context was Inception v3.
Figure 7: ROC curves from baseline models trained with
transfer learning in the RGB dataset version. The best
model this training context was Densenet 201.
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
410

F1 score of 94.0% (normal) and 76.0% (sclerosed)
and (Kannan et al., 2019) 92.6% acc, also for scle-
rosis. About baselines results, when comparing the
networks trained from scratch with the data set in the
RGB and gray level version, we were unable to con-
clude whether the absence of color information of-
fered benefits or harm to the learning of networks,
given that there were architectures with better results
in the RGB version and others in the gray level ver-
sion.
5 CONCLUSIONS
In this work, we propose a method, called PodNet,
for the classification of histological images of renal
glomeruli with podocytopathy, and we present an un-
published public data set of histological images of re-
nal glomeruli with podocyte lesions. The proposed
method has better results when compared against well
know CNN networks. The experiments indicated that
deep neural networks are a promising approach for
supporting the development of a system to automat-
ically classification of podocytopathy in histological
images. The data set presented will continue to in-
crease with new images, and be made available to re-
searchers with academic interests. Additionally, the
studied lesions do not have a high incidence, we in-
tend to use new data augmentation strategies to solve
the low amount of available images. An ablation
study will be carried out to systematically analyze
the contributions of end-to-end training networks with
different color spaces, in the data set provided here
and other problems with multi-stain images. Finally,
we also highlight the possibility of segmentation and
classification of podocyte lesions, quantifying or cor-
relating them to diseases using models based on deep
learning.
ACKNOWLEDGEMENTS
Thanks to Instituto Federal Goiano - Campus Posse
and University of Brasilia to support this research
with computational and physical infrastructure. This
work be included on PathoSpotter project, that is
partially sponsored by the Fundac¸
˜
ao de Amparo
`
a
Pesquisa do Estado da Bahia (FAPESB), grants TO-
P0008/15 and TO-SUS0031/2018, and by the Inova
FIOCRUZ grant. Washington dos Santos and Luciano
Oliveira are research fellows of Conselho Nacional de
Desenvolvimento Cient
´
ıfico e Tecnol
´
ogico (CNPq),
grants 306779/2017 and 307550/2018-4, respectively.
REFERENCES
Abade, A., Ferreira, P. A., and de Barros Vidal, F. (2021).
Plant diseases recognition on images using convolu-
tional neural networks: A systematic review. Comput-
ers and Electronics in Agriculture, 185:106125.
Abade., A., S. de Almeida., A., and Vidal., F. (2019). Plant
diseases recognition from digital images using multi-
channel convolutional neural networks. In Proceed-
ings of the 14th International Joint Conference on
Computer Vision, Imaging and Computer Graphics
Theory and Applications - Volume 5: VISAPP,, pages
450–458. INSTICC, SciTePress.
Barros, G. O., Navarro, B., Duarte, A., and Dos-Santos, W.
L. C. (2017). PathoSpotter-K: A computational tool
for the automatic identification of glomerular lesions
in histological images of kidneys. Scientific Reports,
7(1):46769.
Bukowy, J. D., Dayton, A., Cloutier, D., Manis, A. D., Star-
uschenko, A., Lombard, J. H., Solberg Woods, L. C.,
Beard, D. A., and Cowley, A. W. (2018). Region-
Based Convolutional Neural Nets for Localization of
Glomeruli in Trichrome-Stained Whole Kidney Sec-
tions. Journal of the American Society of Nephrology,
29(8):2081–2088.
Candelero, D., Roberto, G., do Nascimento, M., Rozendo,
G., and Neves, L. (2020). Selection of cnn, haral-
ick and fractal features based on evolutionary algo-
rithms for classification of histological images. In
2020 IEEE International Conference on Bioinformat-
ics and Biomedicine (BIBM), pages 2709–2716, Los
Alamitos, CA, USA. IEEE Computer Society.
Chagas, P., Souza, L., Ara
´
ujo, I., Aldeman, N., Duarte, A.,
Angelo, M., Dos-Santos, W. L. C., and Oliveira, L.
(2020). Classification of glomerular hypercellularity
using convolutional features and support vector ma-
chine. Artificial Intelligence in Medicine, 103:101808.
Chen, H.-M., Liu, Z.-H., Zeng, C.-H., Li, S.-J., Wang, Q.-
W., and Li, L.-S. (2006). Podocyte lesions in patients
with obesity-related glomerulopathy. American jour-
nal of kidney diseases : the official journal of the Na-
tional Kidney Foundation, 48(5):772–779.
Chollet, F. et al. (2015). Keras.
Deng, S., Zhang, X., Yan, W., Chang, E. I.-C., Fan, Y., Lai,
M., and Xu, Y. (2020). Deep learning in digital pathol-
ogy image analysis: a survey. Frontiers of Medicine,
14(4):470–487.
Gallego, J., Swiderska-Chadaj, Z., Markiewicz, T., Ya-
mashita, M., Gabaldon, M. A., and Gertych, A.
(2021). A U-Net based framework to quantify
glomerulosclerosis in digitized PAS and H&E stained
human tissues. Computerized Medical Imaging and
Graphics, 89:101865.
Govind, D., Becker, J., Miecznikowski, J., Rosenberg, A.,
Dang, J., Tharaux, P. L., Yacoub, R., Thaiss, F., Hoyer,
P., Manthey, D., Lutnick, B., Worral, A., Mohammad,
I., Walavalkar, V., Tomaszewski, J., Jen, K.-Y., and
Sarder, P. (2021a). PodoSighter: A Cloud-Based Tool
for Label-Free Podocyte Detection in Kidney Whole
Slide Images. Journal of the American Society of
Nephrology.
PodNet: Ensemble-based Classification of Podocytopathy on Kidney Glomerular Images
411

Govind, D., Santo, B. A., Ginley, B., Yacoub, R., Rosen-
berg, A. Z., Jen, K.-Y., Walavalkar, V., Wilding, G. E.,
Worral, A. M., Mohammad, I., and Sarder, P. (2021b).
Automated detection and quantification of Wilms’ Tu-
mor 1-positive cells in murine diabetic kidney dis-
ease. In Tomaszewski, J. E. and Ward, A. D., editors,
Medical Imaging 2021: Digital Pathology, volume
11603, pages 76–82. International Society for Optics
and Photonics, SPIE.
Huang, G., Liu, Z., van der Maaten, L., and Weinberger,
K. Q. (2018). Densely connected convolutional net-
works.
James, G., Witten, D., Hastie, T., and Tibshirani, R. (2013).
An Introduction to Statistical Learning. Springer texts
in statistics.
Jayapandian, C. P., Chen, Y., Janowczyk, A. R., Palmer,
M. B., Cassol, C. A., Sekulic, M., Hodgin, J. B.,
Zee, J., Hewitt, S. M., and Toole, J. O. (2021). De-
velopment and evaluation of deep learning-based seg-
mentation of histologic structures in the kidney cortex
with multiple histologic stains. Kidney international,
99(1)(June 2020):86–101.
Jiang, L., Chen, W., Dong, B., Mei, K., Zhu, C.,
Liu, J., Cai, M., Yan, Y., Wang, G., and Zuo,
L. (2021). MACHINE LEARNING , COMPU-
TATIONAL PATHOLOGY , AND BIOPHYSICAL
IMAGING A Deep Learning-Based Approach for
Glomeruli Instance Segmentation from Multistained
Renal Biopsy Pathologic Images. The American Jour-
nal of Pathology, 191(8):1431–1441.
Kannan, S., Morgan, L. A., Liang, B., Cheung, M. G., Lin,
C. Q., Mun, D., Nader, R. G., Belghasem, M. E., Hen-
derson, J. M., Francis, J. M., Chitalia, V. C., and Ko-
lachalama, V. B. (2019). Segmentation of Glomeruli
Learning. Kidney International Reports, 4(7):955–
962.
Maraszek, K. E., Santo, B. A., Yacoub, R., Tomaszewski,
J. E., Mohammad, I., Worral, A. M., and Sarder,
P. (2020). The presence and location of podocytes
in glomeruli as affected by diabetes mellitus. In
Tomaszewski, J. E. and Ward, A. D., editors, Medi-
cal Imaging 2020: Digital Pathology, volume 11320,
pages 298–307. International Society for Optics and
Photonics, SPIE.
Mathur, P., Ayyar, M. P., Shah, R. R., and Sharma, S. G.
(2019). Exploring classification of histological dis-
ease biomarkers from renal biopsy images. In 2019
IEEE Winter Conference on Applications of Computer
Vision (WACV), pages 81–90.
Merkel, D. (2014). Docker: Lightweight linux containers
for consistent development and deployment. Linux J.,
2014(239).
Nagata, M. (2016). Podocyte injury and its consequences.
Kidney international, 89(6):1221–1230.
O’Malley, T., Bursztein, E., Long, J., Chollet, F., Jin,
H., Invernizzi, L., et al. (2019). Keras Tuner.
https://github.com/keras-team/keras-tuner.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh,
S., Ma, S., Huang, Z., Karpathy, A., Khosla, A.,
Bernstein, M., Berg, A. C., and Fei-Fei, L. (2015).
ImageNet Large Scale Visual Recognition Challenge.
International Journal of Computer Vision (IJCV),
115(3):211–252.
Saga, N., Sakamoto, K., Matsusaka, T., and Nagata, M.
(2021). Glomerular filtrate affects the dynamics of
podocyte detachment in a model of diffuse toxic
podocytopathy. Kidney International.
Salas, J., de Barros Vidal, F., and Martinez-Trinidad, F.
(2019). Deep learning: Current state. IEEE Latin
America Transactions, 17(12):1925–1945.
Silva, P., Luz, E., Silva, G., Moreira, G., Wanner, E., Vidal,
F., and Menotti, D. (2020). Towards better heartbeat
segmentation with deep learning classification. Scien-
tific Reports, 10(1):20701.
Srinidhi, C. L., Ciga, O., and Martel, A. L. (2021). Deep
neural network models for computational histopathol-
ogy: A survey. Medical Image Analysis, 67:101813.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2015). Rethinking the inception architecture for
computer vision.
Thomas, S. M., Lefevre, J. G., Baxter, G., and Hamil-
ton, N. A. (2021). Interpretable deep learning sys-
tems for multi-class segmentation and classification of
non-melanoma skin cancer. Medical Image Analysis,
68:101915.
Tipping, M. E. and Bishop, C. M. (2006). Mixtures of
Probabilistic Principal Component Analysers. Neural
Computation 11(2).
Uchino, E., Suzuki, K., Sato, N., Kojima, R., and Tamada,
Y. (2020). Classification of glomerular pathological
findings using deep learning and nephrologist – AI
collective intelligence approach. International Jour-
nal of Medical Informatics, pages 1–14.
Ullah, A., Ahmad, J., Muhammad, K., Sajjad, M., and Baik,
S. W. (2018). Action recognition in video sequences
using deep bi-directional lstm with cnn features. IEEE
Access, 6:1155–1166.
Yang, C.-K., Lee, C.-Y., Wang, H.-S., Huang, S.-C., Liang,
P.-I., Chen, J.-S., Kuo, C.-F., Tu, K.-H., Yeh, C.-Y.,
and Chen, T.-D. (2021). Glomerular disease classifi-
cation and lesion identification by machine learning.
Biomedical Journal.
Yari, Y., Nguyen, T. V., and Nguyen, H. T. (2020). Deep
learning applied for histological diagnosis of breast
cancer. IEEE Access, 8:162432–162448.
Zeng, C., Nan, Y., Xu, F., Lei, Q., Li, F., Chen, T., Liang,
S., Hou, X., Lv, B., Liang, D., Luo, W., Lv, C., Li, X.,
Xie, G., and Liu, Z. (2020). Identification of glomeru-
lar lesions and intrinsic glomerular cell types in kid-
ney diseases via deep learning. The Journal of Pathol-
ogy, 252(1):e5491.
Zimmermann, M., Klaus, M., Wong, M. N., Thebille, A.-
K., Gernhold, L., Kuppe, C., Halder, M., Kranz, J.,
Wanner, N., Braun, F., Wulf, S., Wiech, T., Panzer,
U., Krebs, C. F., Hoxha, E., Kramann, R., Huber,
T. B., Bonn, S., and Puelles, V. G. (2021). Deep learn-
ing–based molecular morphometrics for kidney biop-
sies. JCI Insight, 6(7).
VISAPP 2022 - 17th International Conference on Computer Vision Theory and Applications
412
