
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided
Attention Networks
Gurbandurdy Dovletov
1 a
, Duc Duy Pham
1
, Josef Pauli
1 b
, Marcel Gratz
2,3 c
and Harald H. Quick
2,3
1
Intelligent Systems Group, Faculty of Engineering, University of Duisburg-Essen, Duisburg, Germany
2
High-Field and Hybrid MR Imaging, University Hospital Essen, University of Duisburg-Essen, Essen, Germany
3
Erwin L. Hahn Institute for MR Imaging, University of Duisburg-Essen, Essen, Germany
Keywords:
Image-to-Image Translation, Pseudo-CT, Attention Mechanism, U-Net, Generative Adversarial Network.
Abstract:
In this paper, we propose 2D MRI-based pseudo-CT (pCT) generation approaches that are inspired by U-Net
and generative adversarial networks (GANs) and that additionally utilize coarse bone segmentation guided
attention (SGA) mechanisms for better image synthesis. We first introduce and formulate SGA and its ex-
tended version (E-SGA), then we embed them into our baseline U-Net and conditional Wasserstein GAN
(cWGAN) architectures. Since manual bone annotations are expensive, we derive coarse bone segmentations
from CT/pCT images via thresholding and utilize them during the training phase to guide image-to-image
translation attention networks. For inference, no additional segmentations are required. The performance of
the proposed methods regarding the image generation quality is evaluated on the publicly available RIRE data
set. Since MR and CT image pairs in this data set are not correctly aligned with each other, we also briefly
describe the applied image registration procedure. The results of our experiments are compared to baseline
U-Net and conditional Wasserstein GAN implementations and demonstrate improvements for bone regions.
1 INTRODUCTION
Positron emission tomography (PET) is an imaging
technique that reveals physiological and biochemi-
cal processes of tissue and organs by measuring their
metabolic activity. The technique is based on the de-
tection of radioactivity emitted in opposite directions
after a small amount of a radioactive tracer is injected
into a peripheral vein (Paans, 2006). When traveling
through some tissue or hardware parts such as the pa-
tient table these photons lose their energy and as a
consequence, only a part of them reaches detectors.
In order to compensate for such physical behavior,
an attenuation correction (AC) procedure is required
(Ollinger and Fessler, 1997). In stand-alone PET sys-
tems, AC is performed based on a transmission scan
acquired using external rod sources.
A lack of anatomical information in a pure PET
initiated the development of combined PET and com-
puted tomography (CT) systems in a single gantry,
a
https://orcid.org/0000-0002-2401-8745
b
https://orcid.org/0000-0003-0363-6410
c
https://orcid.org/0000-0001-9723-5233
also known as PET/CT. The additional CT examina-
tion in such systems allows deriving attenuation cor-
rection maps (µ-maps) directly from Hounsfield units
(HU) by scaling the acquired CT image to the energy
level of a PET image (Beyer et al., 2000). Obtained
in such a way µ-maps are later used for attention cor-
rection of PET scans. The main application areas
of PET/CT are cardiology, neurology, and oncology.
According to (Brady et al., 2008), in oncology, it is
by far the most intensively used diagnostic and stag-
ing device.
Being able to provide an excellent soft tissue con-
trast, PET/MR combination was introduced, which
moreover goes without the need of additional radia-
tion as it is the case for PET/CT. Such systems do not
provide a straightforward way to obtain µ-maps for
AC by energy scaling like it is possible in stand-alone
PET or PET/CT combination (Keereman et al., 2013).
Unlike PET and CT, MRI uses physical mechanisms
that do not allow for a direct derivation or correlation
of image intensities with electron density.
Various approaches have been proposed to resolve
this issue. In atlas-based methods, µ-maps are calcu-
lated based on prior registration of atlas images to a
Dovletov, G., Pham, D., Pauli, J., Gratz, M. and Quick, H.
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided Attention Networks.
DOI: 10.5220/0010849200003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 2: BIOIMAGING, pages 131-140
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
131

target image with subsequent utilization of achieved
transformations. (Burgos et al., 2014) propose to gen-
erate synthetic CTs by locally matching target MR
images to a database of MR-CT pairs and deriving
the corresponding µ-maps from them. An alterna-
tive approach is a segmentation-based AC, where dis-
crete linear attenuation coefficients (LACs) are as-
signed to each achieved segmentation class (Berker
et al., 2012). In order to take advantage of both
segmentation- and atlas-based approaches, (Paulus
et al., 2015) propose to first derive µ-maps via seg-
menting a Dixon image into air, fat, lung, and soft tis-
sue classes. After that, atlas-based continuous µ-maps
for each of major body bones (femur, hip, spine, and
skull) are derived and fused with the existing µ-maps
from the previous step.
One other possible way to derive µ-maps for PET
AC is to first generate a pseudo-CT (pCT) image from
an MRI and then scale it to PET’s energy level in the
same way as it is done for PET/CT machines. This
MRI-based pCT generation task, however, introduces
some challenges, since the MR modality is sensitive
to proton density and thus not capable of distinguish-
ing between air and lungs. Moreover, compact bone
is also indistinguishable as its signal vanishes, due to
the comparably short relaxation time, before it can be
acquired.
Previously proposed methods have already
demonstrated the feasibility of generating pCT from
MR images using deep learning methods. (Liu et al.,
2017) propose a pipeline for PET attenuation correc-
tion, where an encoder-decoder like architecture is
first employed to segment an MR image into three
classes, namely, air, bone, and soft tissue. The ob-
tained segmentation is later converted to a pseudo-CT
image via mapping each class ID to its corresponding
statistical HU value. (Nie et al., 2016) propose to
use a 3D fully convolution network (FCN) for a
direct non-linear mapping from MRI to CT domain.
They utilize a patch-based training procedure for
better preservation of the neighborhood information
in predicted pseudo-CT images. Moreover, an
additional image gradient difference loss term was
used to retain the sharpness of synthesized CTs. In
order to generate even more realistic images, (Nie
et al., 2017) extend their existing architecture with an
additional discriminator and employ the adversarial
training approach from (Goodfellow et al., 2014).
(Han, 2017) adapts and modifies the well-known
U-Net (Ronneberger et al., 2015) architecture by
changing the number of convolutional layers in such
a way, that allows to initialize the feature extraction
part of the proposed architecture from a pretrained
VGG-16 model. (Wolterink et al., 2017) propose to
utilize the state-of-the-art Cycle-GAN (Zhu et al.,
2017) image-to-image translation model, which is
the extension of GAN (Goodfellow et al., 2014) and
involves the simultaneous training of two generators
and two discriminator models.
There have been several attempts to improve
the bone quality of generated (synthesized) pseudo-
CT images with the help of additional information.
(Leynes et al., 2018) propose to utilize zero-echo-time
(ZTE) images additionally to fat and water maps de-
rived from a 2-echo Dixon MRI (in-phase and out-
of-phase) sequence to capture bone information, and
thus, generate more accurate pseudo-CT images. The
main limitations of their work, however, are the long
ZTE image acquisition time (Mecheter et al., 2020)
and UTE/ZTE’s limited availability, since they are
not standard clinical sequences. Dixon-type pulse se-
quences, on contrary, are increasingly popular and
are offered, nowadays, by nearly every manufacturer.
To this end, (Torrado-Carvajal et al., 2019) employ a
U-Net architecture with four Dixon images, where fat
and water maps are explicitly used as additional in-
puts to 2-echo Dixon MRI. Alternatively, (Qi et al.,
2020) utilize a stack of images from four different se-
quences (T1, T2, T1C, and T1DixonC-Water) as an
input to their U-Net/GAN-based networks.
This paper makes several contributions. We in-
troduce and formulate segmentation guided attention
(SGA) mechanisms that can be adapted and used for
different image-to-image translation tasks. We utilize
the proposed SGA and its extended version (E-SGA)
for the task of MRI-based pseudo-CT generation. The
proposed attention mechanisms are based on coarse
bone masks, which are derived from CT/pCT im-
ages in a fully automatic manner. These masks are
only used during the training phase and thus are not
required during the inference. We demonstrate that
these attention networks are capable of producing su-
perior quality pCTs, especially for bone regions.
2 METHODOLOGY
In this section, we first introduce and formulate the
segmentation guided attention mechanism and its ex-
tended version. After that, we introduce U-Net-
based and conditional Wasserstein GAN-based atten-
tion networks for the MRI-based pseudo-CT genera-
tion task.
2.1 Segmentation Guided Attention
The main goal of the image-to-image translation task
is to transform input images from a source domain
BIOIMAGING 2022 - 9th International Conference on Bioimaging
132

X to a target domain Y via a mapping function
G : X → Y . Neural networks can learn such non-
linear G mapping functions and, thus, can be used to
generate (predict) ˆy from the input image x, as fol-
lows:
ˆy = G(x) (1)
Pixel-wise loss functions, such as L1 or L2 norm, are
playing an essential role while training such image-
to-image translation networks. L1 loss, for example,
can be mathematically formalized as follows:
L
1
(y, ˆy) =
1
M · N
M
∑
i=1
N
∑
j=1
ky
i j
− ˆy
i j
k
1
(2)
where ˆy denotes the output image of size M × N pre-
dicted by a neural network and y is the corresponding
ground truth (GT) image. This loss function is cal-
culated as the mean absolute error (MAE) between
GT and prediction image over all (i, j) positions. As
a consequence, while training, a neural network pays
attention to the entire input image.
The proposed segmentation guided attention
(SGA) loss, in contrast, focuses specifically on pro-
vided regions of interest (ROI) and helps image-to-
image translation networks to achieve better perfor-
mance for them. We formulate the SGA loss function
as follows:
L
SGA
(y, ˆy, seg
y
) =
γ(seg
y
) ·
1
M · N
M
∑
i=1
N
∑
j=1
seg
y
i j
· ky
i j
− ˆy
i j
k
1
(3)
where seg
y
is a binary mask derived from the target
domain image y. Multiplication of seg
y
i j
with its cor-
responding pixel-wise difference at position (i, j) pe-
nalizes an error only for the provided area of interest.
We propose to use an additional weighting function
γ(.) which is dependent on seg
y
and can be calculated
as follows:
γ(seg
y
) =
M · N
(
∑
M
i=1
∑
N
j=1
seg
y
i j
) + ε
(4)
where ε is needed to deal with the cases of division
by 0, when seg
y
i j
is 0 at each of its positions. This
ensures that L
SGA
stays on approximately the same
range independent of ROI mask properties.
It can be noticed that the SGA loss is a generaliza-
tion of the L
1
loss from Equation 2. When the ROI
mask contains 1’s at each of its positions L
SGA
is ap-
proximately equal to L
1
, since γ(seg
y
) is equal to 1.
The proposed segmentation guided attention loss
can be used as a separate, alternative pixel-wise func-
tion or in combination with existing ones. Moreover,
an alternative SGA loss can also be formulated based
on the L2 norm.
2.2 Extended-SGA
The previously proposed L
SGA
loss definition can be
extended to L
E-SGA
, which considers two segmenta-
tion masks simultaneously. In addition to seg
y
, which
is based on a ground truth image, a segmentation seg
ˆy
,
based on a predicted image ˆy, is utilized. Thus, the
extended-SGA (E-SGA) loss is defined as follows:
L
E-SGA
(y, ˆy, seg
y
, seg
ˆy
) = γ
E
(seg
y
, seg
ˆy
)
·
1
M · N
M
∑
i=1
N
∑
j=1
kseg
y
i j
· y
i j
− seg
ˆy
i j
· ˆy
i j
k
1
(5)
where γ
E
(.) is a weighting function:
γ
E
(seg
y
, seg
ˆy
) =
2 · M · N
∑
M
i=1
∑
N
j=1
(seg
y
i j
+ seg
ˆy
i j
)
+ ε
(6)
which is dependent on seg
y
and seg
ˆy
. In contrast to
SGA, minimization of the extended loss during the
training not only enforces networks to generate better
quality seg
y
regions but also constrains them to pay
attention to seg
ˆy
regions.
It can be noticed that L
E-SGA
is equivalent to L
SGA
from Equation 3 when seg
y
and seg
ˆy
are identical.
2.3 U-Nets with Additional SGA/E-SGA
The previously introduced loss functions can be uti-
lized for MRI-based pseudo-CT generation task. Our
proposed network topologies are based on the U-Net
architecture, which was originally proposed to ad-
dress semantic segmentation tasks in the medical im-
age computing domain (Ronneberger et al., 2015). It
is a fully convolutional network (FCN) which con-
sists of a contracting (encoding) path followed by an
expanding (decoding) path with additional skip con-
nections between layers of the same size, as can be
seen on the left part of Figure 1 and Figure 2. While
the encoding path of the network behaves similar to
a traditional convolutional neural network (CNN) and
learns to extract hierarchical features from an input,
the decoding path is responsible for reconstructing an
output image gradually by adding more details at each
following resolution level.
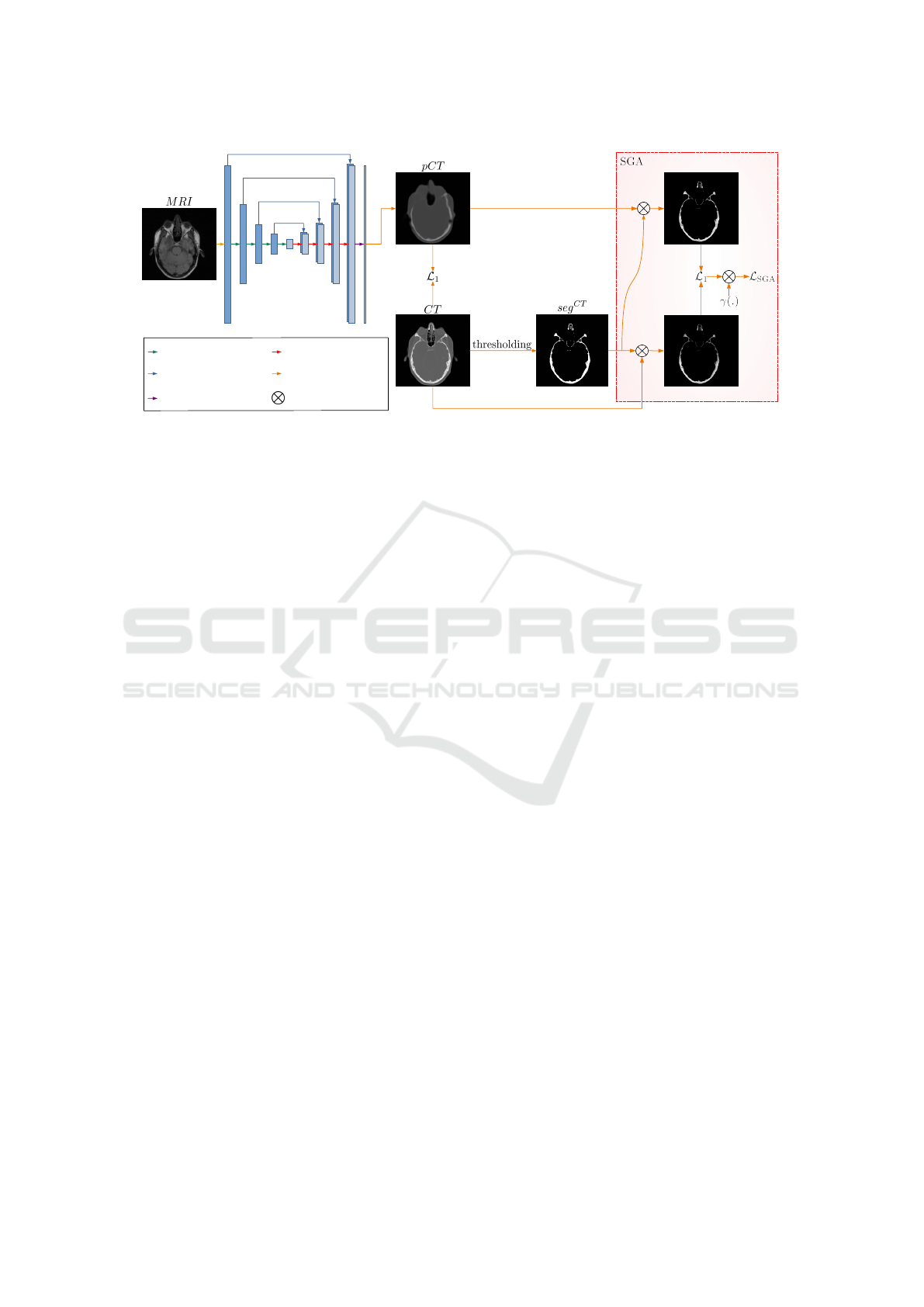
The proposed segmentation guided U-Net
SGA
ap-
proach is schematically shown in Figure 1. By prop-
agating an input MR image through the network we
generate a pCT image. The global L
1
loss func-
tion (see Equation 2) compares the synthesized pCT
to its corresponding ground truth CT image. In U-
Net
SGA
we enforce the network to pay particular at-
tention to the bone area via utilizing the previously
introduced L
SGA
loss term (see Equation 3). There-
fore, we first multiply both generated pCT and its
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided Attention Networks
133
2xConvolution,
followed by Max-Pooling
Skip Connection
(Concatenation)
Data Flow Arrow
(no operation)
Transposed Convolution,
followed by 2xConvolution
Element-wise multiplication
32
64
128
256
512
1x1 Convolution
Figure 1: The proposed segmentation guided U-Net
SGA
approach for MRI-based pseudo-CT generation. While L
1
loss
considers entire pCT and CT images, L
SGA
loss concentrates exclusively on the bone area from CT images.
corresponding CT images with the binary bone mask
(seg
CT
, in Figure 1), and then calculate the difference
between the resulting images. We propose to derive
the required bone masks from CT via thresholding,
which is described in more detail in Subsection 3.2.
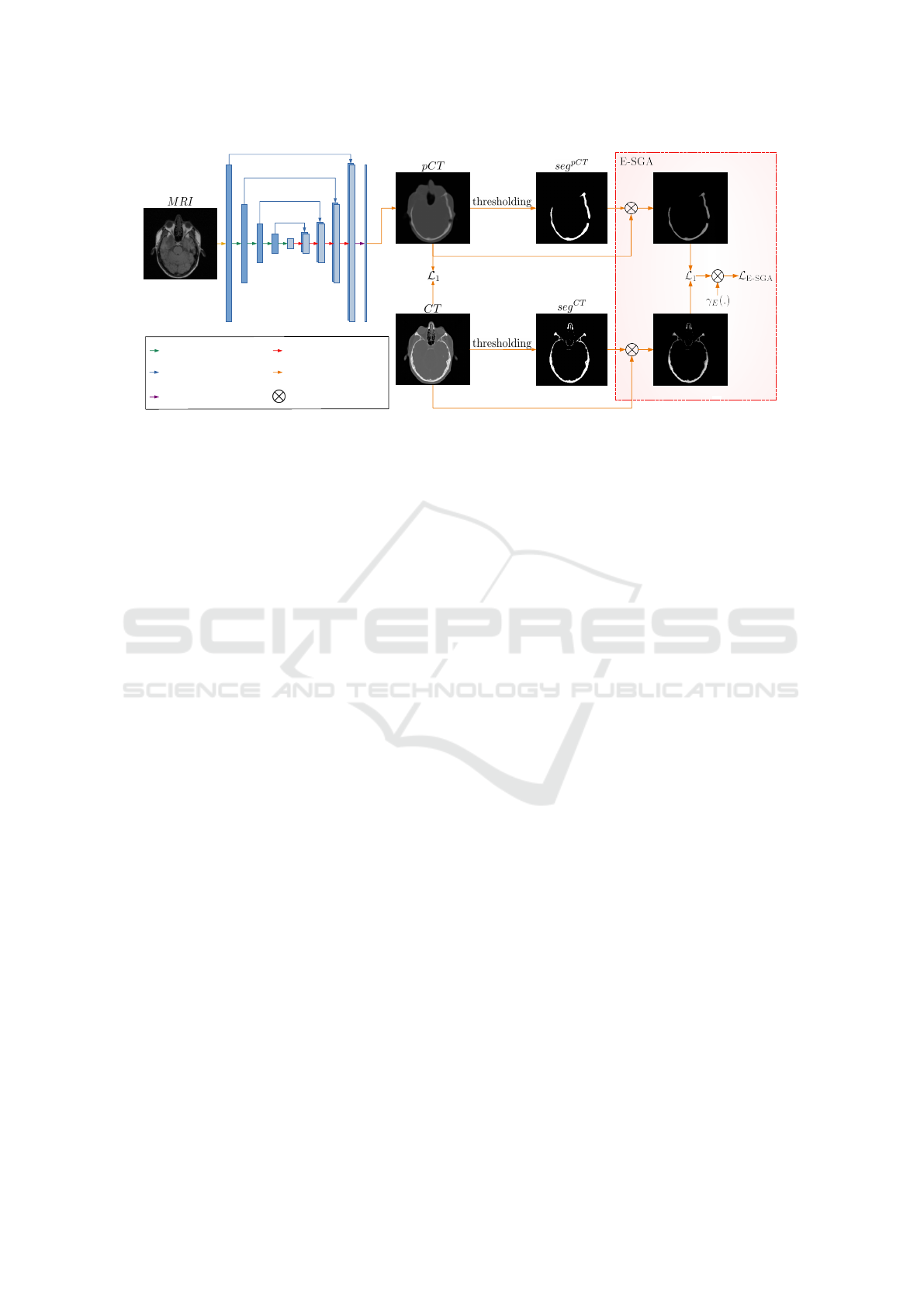
We extend the proposed SGA-based U-Net approach
and introduce U-Net
E-SGA
, which is schematically
shown in Figure 2. This approach uses the additional
L
E-SGA
loss term (see Equation 6). In comparison
to U-Net
SGA
, this method requires additional seg
pCT
bone masks from predicted pCTs. We propose to de-
rive them in a similar manner as seg
CT
images from
CTs. Thus, the total objective for U-Net
SGA/E-SGA
consists of two loss functions and is defined as fol-
lows:
L
Unet
SGA/E-SGA
= L
1
+ αL
SGA/E-SGA
(7)
where α is a hyperparameter and can be used to con-
trol the relative importance of the two objectives.
2.4 Conditional Wasserstein GANs with
Additional SGA/E-SGA
MRI-based pseudo-CT images can also be synthe-
sized by using generative models, such as genera-
tive adversarial networks (GANs) (Goodfellow et al.,
2014). The simplistic GAN framework consists of
generator G and discriminator D networks. While the
generator attempts to create data similar to those in
the training set, the discriminator inspects it and tries
to correctly identify whether it is real (from the train-
ing set) or fake (generated by G). Both networks are
trained together by taking turns and using the adver-
sarial loss function, which is defined as follows:
L
adv
= E
y
[log(D(y))] + E
x
[log(1 − D(G(x)))] (8)
with x and y representing images from source and tar-
get domain, correspondingly. In other words, they are
playing a minimax game against each other: while G
attempts to maximize L
adv
and thus fool D, the latter
one tries to not be fooled by minimizing the adversar-
ial loss.
Our proposed network topologies are based on the
conditional Wasserstein GAN (cWGAN) training ap-
proach, which is an extension of Wasserstein GAN
(Arjovsky et al., 2017). In contrast to traditional
GANs, Wasserstein GAN can improve the stability
of the learning process and can help to get rid of the
mode collapse problem. This is achieved by using
a different adversarial loss function, which approxi-
mates the Earth Mover’s Distance, and changing the
role of the discriminator from a binary classifier into
a critic C, which predicts scores of how real or fake
provided images are looking. For faster convergence
and better preservation of structural information, MR
image-based conditioning on the corresponding critic
is utilized. Conditional Wasserstein GAN objective
can be formalized as follows:
L
cW
= E
x,y
[C(x, y)]− E
x
[C(x, G(x))] (9)
with x and y representing MR and CT images, cor-
respondingly. Thus, while G tries to minimize L
cW
against adversarial critic C, the latter one attempts to
maximize the same objective. We use U-Net as the
generator in our baseline cWGAN approach. For the
critic network, CNN architecture is utilized, which is
described in more detail in Subsection 3.2). Anal-
ogously to the U-Net-based extensions, we also in-
troduce two extensions of the cWGAN. The pro-
posed cWGAN
SGA
and cWGAN
E-SGA
approaches
additionally include previously introduced segmenta-
tion guided attention loss terms while training their
corresponding U-Net generator networks. For the
BIOIMAGING 2022 - 9th International Conference on Bioimaging
134
2xConvolution,
followed by Max-Pooling
Skip Connection
(Concatenation)
Data Flow Arrow
(no operation)
Transposed Convolution,
followed by 2xConvolution
Element-wise multiplication1x1 Convolution
32
64
128
256
512
Figure 2: The proposed extended segmentation guided U-Net
E-SGA
approach for MRI-based pseudo-CT generation. While
L
1
loss considers the entire pCT and CT images, L
E-SGA
loss concentrates exclusively on bone areas from both CT and pCT.
sake of shortness, we only depict cWGAN
E-SGA
ap-
proach in Figure 3. Thus, our final objectives for gen-
erators G and critics C of cWGAN
SGA/E-SGA
can be
expressed as:
L
G
= (L
1
+ αL
SGA/E-SGA
) + λL
cW
L
C
= λL
cW
(10)
where λ denotes the weighting factor of the condi-
tional Wasserstein GAN objective.
3 EXPERIMENTS
In this section, data set information including utilized
registration procedure and following data preparation
are introduced. After that, implementation details of
proposed architectures and evaluation metrics are out-
lined.
3.1 Data
We evaluated the proposed architecture on the pub-
licly available Retrospective Image Registration Ex-
periment (RIRE) Project data set (West et al., 1997),
that provides different modality images of the head
area for 16 patients. For our experiments, we chose
to use T1-weighted MR images of size 256 × 256
in combination with corresponding CTs of size
512 × 512. Both images fit into 12 bits data represen-
tation, meaning that CT’s intensities range between
−1024 HU and 3071 HU while MR’s are ranging
from 0 and 4095. Some of the CT volumes include
the patient’s table.
The RIRE project was originally designed to com-
pare CT-MR and PET-MR registration techniques.
Since the ground truth data is not provided, we used
Mattes’s mutual-information-based multi-resolution
algorithm (Mattes et al., 2003) implemented in Sim-
pleITK (Lowekamp et al., 2013; Yaniv et al., 2018)
framework to register CT and MR volume pairs. Dur-
ing the registration procedure, CT was chosen as a
fixed volume, while the corresponding MR volume
was considered as a moving one. Optimization of mu-
tual information between MR and CT volumes was
done via gradient descent with a learning rate set to
0.01 value. While solving the optimization task, we
utilized linear interpolation to deform the MR image.
Since ground truth transformations are not available,
we qualitatively inspected the registered MR-CT vol-
ume pairs and validated the utilized registration pro-
cedure.
All registered volumes were first brought to
isotropic 1×1 × 1 mm
3
voxel spacing. After that, de-
pending on the obtained spatial resolution, cropping
from the center point of the image or padding around
its borders was utilized in order to achieve the same
field of view. Finally, we resized the achieved MR
and CT slices to the size of 256 × 256 pixels.
After the final visual inspection of the data set,
we identified MR/CT slices that did not have valid
CT/MR counterparts. These image pairs were located
either on the top or at the bottom of the registered
volumes when looking along the longitudinal axis.
The main reason for their occurrence at these spe-
cific locations is not the low-quality registration, but
rather the different fields of view of initial (not reg-
istered) MR and CT scans. Thus, we excluded them
and ended up with 553 valid MR-CT image pairs in
total.
3.2 Implementation Details
The experiments were conducted in a 4-fold cross-
validation manner with one set used for each testing
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided Attention Networks
135

2xConvolution,
followed by Max-Pooling
Skip Connection
(Concatenation)
Data Flow Arrow
(no operation)
Transposed Convolution,
followed by 2xConvolution
Element-wise multiplication
1x1 Convolution
32
64
128
256
512
Strided Convolution
16
64
128
256
512
1
32
Figure 3: The proposed conditional Wasserstein GAN with extended segmentation guided attention (cWGAN
E-SGA
) for MRI-
based pseudo-CT generation.
and three sets used for training and validation. During
our experiments, we increased the number of training
samples via data augmentation by means of random
rotations (in the range of ±7.5 degrees), scaling (with
factors in the range [1; 1.15]), and horizontal flipping
(with the probability of 50%).
3.2.1 U-Nets
Baseline U-Net. In our baseline U-Net implemen-
tation, we started with 32 convolutional kernels in the
first resolution level and we used two subsequent con-
volutional layers with zero padding at each resolution
level. We chose to use kernels of size 5 × 5 at each
convolutional layer and windows of size 2×2 for each
following max pooling step. The number of convo-
lutional filters was doubled at each following image
resolution. In the expanding path, we used transposed
convolutions with kernels of size 2 × 2 with a stride
size of 2 pixels in both directions. At each upsam-
ple step the number of output features was reduced
twice compared to the corresponding number of input
channels. As a final layer, we used 1 × 1 convolution
in order to get a single channel output image. We did
not normalize MR and CT images as we did not ob-
serve any performance gain compared to the network
trained with normalized inputs.
U-Nets with SGA/E-SGA. We used the same
U-Net structure, as described previously, for both pro-
posed segmentation guided approaches. For a given
MRI-based pseudo-CT generation task we are partic-
ularly interested in an improved bone quality synthe-
sis. Therefore, regions of interest required for SGA
calculations should also represent bone areas.
Since manual annotation of bones from CTs is
expensive and time consuming, we generated coarse
bone masks by applying global threshold-based seg-
(a) MR. (b) CT. (c) Bone mask.
Figure 4: Exemplary MR, CT and coarse bone mask images
for Patient 002 (slice #7).
mentation approach to ground truth CT images:
seg
y
i j
=
(
1 if y
i j
> T
0 otherwise
(11)
where T is a threshold value.
We set the threshold value to 350 HU, which is
approximately in the same range as stated in the liter-
ature (Buzug, 2009; Chougule et al., 2018). Figure 4
shows exemplary MR, CT and the obtained from CT
coarse bone segmentation mask. In a similar man-
ner and with the same threshold value we derived
seg
ˆy
, segmentation masks from synthesized ˆy images,
which are needed for E-SGA loss term calculations.
We set α to 1 when calculating the total loss func-
tions for both U-Net
SGA
and U-Net
E-SGA
approaches.
The intuition behind this choice is that we want to
achieve a better synthesis quality for bone areas, how-
ever, we want to keep a good performance for other
parts of the image as well.
Training U-Nets. We trained our U-Net models for
100 epochs with a learning rate of 0.01 using the
Adam optimizer with β
1
= 0.9 and β
2
= 0.999 as sug-
gested by (Kingma and Ba, 2014). Mini-batches of
size 16 were used for all our experiments.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
136

3.2.2 Conditional WGANs
Baseline cWGAN. We used the previously intro-
duced baseline U-Net as a generator in our baseline
conditional Wasserstein GAN architecture. A hyper-
bolic tangent layer was added on top of it as the fi-
nal activation layer. This ensured that the output in-
tensities landed in the range between −1 and 1. In
our critic architecture, we started with 16 convolu-
tional kernels in the first resolution level. We chose
to use kernels of size 4 × 4 with a stride of 2 pix-
els and 1 pixel padding in both directions, which al-
lowed us to avoid max pooling layers, as was sug-
gested by (Radford et al., 2015). We applied the
LeakyReLU non-linear activation function after each
convolutional layer. The number of filters was dou-
bled at each following image resolution. In order to
make the training procedure more stable, we applied
a batch normalization at each resolution level, except
for the first one. Strided convolutions were repeated
until we obtained a single scalar value as an output
per input image.
Input images were normalized to the range of −1
and 1 before being fed into our networks. This was
achieved by applying the following min-max normal-
ization equations for MR and CT images:
MR
normalized
= 2 ·
MR
4095
− 1 (12)
CT
normalized
= 2 ·
CT + 1024
4095
− 1 (13)
During the inference, synthesized pseudo-CT images
were mapped back to the original intensity range
based on the same normalization scheme, as in Equa-
tion 13.
CWGANs with SGA/E-SGA. The same baseline
conditional WGAN architecture, as described pre-
viously, was utilized in our segmentation guided
cWGAN
SGA
and cWGAN
E-SGA
approaches. Since
their corresponding generator networks synthesize
normalized pCT images, the previously utilized
350 HU threshold value was also mapped to the range
between −1 and 1 using the Equation 13, which led
to the value of −0.329.
With similar intuition behind as for segmentation
guided U-Nets, we set α to 1 when calculating the
total loss objectives of the generator networks.
Training cWGANs. We used a two times update
rule for training our cWGANs as it was proposed by
(Heusel et al., 2017). Our models were trained for
2000 epochs with learning rates of 0.0002 and 0.0004
for generators and discriminators, correspondingly.
(a) MR. (b) CT. (c) Head mask.
Figure 5: Exemplary MR, CT and head mask from MR im-
ages for Patient 002 (slice #2).
During the training, we utilized an additional gradi-
ent penalty as suggested by (Wu et al., 2018). Net-
works were optimized using the Adam optimizer and
mini-batches of size 16.
3.3 Evaluation Metrics
For evaluation, the obtained 2D pseudo-CT images
were first stacked to build 3D volumes, and then only
compared to the desired ground truth CT volumes.
We chose mean squared error (MSE) and mean ab-
solute error (MAE) as pixel-wise quality metrics.
Moreover, we calculated MSE and MAE val-
ues only for specific regions of interest, such as for
bone and head areas (MSE
bone/head
and MAE
bone/head
).
Head masks were derived from MR images via Otsu’s
thresholding followed by subsequent morphological
operations. First, a morphological opening with a
disk-shaped structuring element of a 5 px radius was
used to remove small artifacts from initial segmen-
tations. Next, a closing operation (radius of 25 px)
was utilized to fill the holes in nasal areas. Finally, a
morphological dilation (radius of 5 px) was applied to
slightly increase the total shape of the segments. An
exemplary head mask is depicted in Figure 5.
To better evaluate the quality of generated pseudo-
CT images we additionally calculated peak signal-to-
noise ratio (PSNR) as follows:
PSNR(y, ˆy) = 10 · log
10
MAX
2
I
MSE(y, ˆy)
(14)
where MAX
I
is the maximum possible intensity value.
We also calculated the structural similarity index
measure (SSIM) (Wang et al., 2004) between gener-
ated pCTs and corresponding CTs as follows:
SSIM(y, ˆy) =
(2µ
y
µ
ˆy
+C
1
)(2σ
y ˆy
+C
2
)
(µ
2
y
+ µ
2
ˆy
+C
1
)(σ
2
y
+ σ
2
ˆy
+C
2
)
(15)
where µ
ˆy
and µ
y
denote mean values of pCT and CT
images, respectively; σ
y
and σ
ˆy
are corresponding
variances; σ
y ˆy
is the covariance between pCT and CT
image; and C
1
= (k
1
L)
2
and C
2
= (k
2
L)
2
are two vari-
ables to stabilize the division with weak denomina-
tors. Here, k
1
= 0.01, k
2
= 0.03, and L = MAX
I
rep-
resents the dynamic range of CT/pCT intensities.
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided Attention Networks
137

4 RESULTS
Table 1 shows averaged evaluation metrics for
both baseline and segmentation guided architectures.
Baseline U-Net implementation achieved MAE of
101 ± 35 HU and MSE of 69139 ± 27664 HU
2
when considering entire pseudo-CT images. Re-
garding the quality of image synthesis, it reached a
PSNR of 24.3 ± 1.9 dB and SSIM of 79.6 ± 6.8 %.
The proposed U-Net
SGA
performed significantly bet-
ter than baseline U-Net when considering only the
bone regions (column ”Bone Area” in Table 1). With
327 ± 46 HU and 180162 ± 56182 HU
2
, over 45%
reduction of MAE
bone
and around 66% reduction of
MSE
bone
have been achieved. This improvement is
the expected behavior since the SGA mechanism has
been formulated in a such way, that it pays more atten-
tion to bone regions from ground truth images. How-
ever, U-Net
SGA
’s performance is worse than base-
line when considering the overall MAE and MSE
metrics (columns ”Entire Image” and ”Head Area”
in Table 1). This could be due to the suboptimal
choice of α when calculating the total loss function
of U-Net
SGA
. Although U-Net
E-SGA
was not able
to improve bone metrics as drastically as U-Net
SGA
,
values still appeared to be better than for the base-
line architecture. Regarding the averaged metrics for
the bone area, U-Net
E-SGA
yield 454 ± 88 HU and
345229 ± 112172 HU
2
for MAE and MSE, which is
around 141 HU and 187466 HU
2
gain over the base-
line U-Net implementation. In contrast to the net-
work with SGA, U-Net
E-SGA
was able to pay atten-
tion to bones, however, was still able to retain the im-
age quality for other parts of the image too. MSE
and MAE values for entire images and head areas are
in the same range as for the baseline U-Net. Exem-
plary pseudo-CT generation results for Patient 007
(slice #1) from baseline U-Net, U-Net
SGA
and from
U-Net
E-SGA
are shown in Figure 7 (b-d). Thus,
with regard to the proposed two segmentation guided
mechanisms, U-Net
E-SGA
seems to yield better perfor-
mance than baseline U-Net and U-Net
SGA
. The same
conclusion can be drawn from the difference images,
which are shown in Figure 8 (first row).
The baseline conditional Wasserstein GAN per-
formed slightly worse than the baseline U-Net when
comparing the entire pCTs, however, the generated
images are looking qualitatively better. As expected
Table 1: Averaged MAE, MSE, PSNR and SSIM metrics for baselines and segmentation guided networks. While MAE and
MSE values (for entire images, head areas and bone areas) are given in HU and HU
2
, PSNR and SSIM values are reported in
dB and %, respectively.
Entire Image Head Area Bone Area
Name MAE MSE MAE MSE MAE MSE PSNR SSIM
U-Net 101±35 69139±27664 180±30 131393±38343 595±120 532695±198330 24.3±1.9 79.6±6.8
U-Net
SGA
128±34 83695±28792 257±42 192630±45684 327±46 180162±56182 23.2±1.5 77.5±6.2
U-Net
E-SGA
108±35 67528±27680 191±32 138309±37912 454±88 345229±112172 24.3±1.8 79.3±6.6
cWGAN 113±37 80507±31839 202±34 154101±42147 493±90 408417±131774 23.7±1.9 77.2±7.3
cWGAN
SGA
117±38 81997±31843 206±34 154975±40802 485±95 394859±129604 23.6±1.9 74.8±7.2
cWGAN
E-SGA
110±43 76425±35158 195±39 144978±44869 473±101 377988±133286 24.0±2.2 77.2±8.1
(a) MR (b) U-Net (c) U-Net
SGA
(d) U-Net
E-SGA
(e) CT
(f) cWGAN (g) cWGAN
SGA
(h) cWGAN
E-SGA
Figure 6: Exemplary MR, CT and MRI-based pseudo-CTs from U-Nets (b-d) and cWGANs (f-h) for Patient 007 (slice #1).
BIOIMAGING 2022 - 9th International Conference on Bioimaging
138

Figure 7: Differences images between CTs and correspond-
ing pCTs for Patient 007 (slice #1). From upper left to
bottom right: U-Net, U-Net
SGA
, U-Net
E-SGA
, cWGAN,
cWGAN
SGA
and cWGAN
E-SGA
. Red color represents un-
derestimated regions, while blue one highlights overesti-
mated areas.
and as it can be seen by comparing (b) and (f) im-
ages from Figure 7, cWGAN based approach was able
to preserve more anatomical bone structures and to
generate more realistic pCT images, due to the assis-
tance of an additional critic network. While consid-
ering only ROIs, it improved MAE
bone
and MSE
bone
metrics. Thus, cWGAN yields 493 ± 90 HU and
408417 ± 131774 HU
2
, which is around 102 HU
and 124278 HU
2
gain over the baseline U-Net. With
485 ± 95 HU and 394859 ± 129604 HU
2
for
bone regions, conditional WGAN with SGA per-
formed slightly better than the baseline cWGAN,
while its extended versions improved the metrics even
further. Considering MAE
bone
and MSE
bone
met-
rics, cWGAN
E-SGA
yield approximately 20 HU and
30429 HU
2
improvement over the baseline cWGAN.
Averaged SSIM values for cWGANs are in general
slightly lower than for U-Nets, due to attempts of gen-
erative models to synthesize the patient’s table. This
can be confirmed by inspecting the difference images
in Figure 8 (second row), where the table partially ap-
pears, although it does not exist in the corresponding
ground truth image from Figure 7. The same reason
lies behind the slightly higher errors for whole im-
ages (compared to the baseline U-Net and U-net
SGA
).
MAE
head
and MSE
head
values correlate with their cor-
responding metrics when considering the entire pCT
images. Thus, comparing the obtained results to the
desired GTs, significant qualitative and quantitative
improvements for bone regions in favor of the E-SGA
approach can be observed. It is worth mentioning
that cWGAN
SGA
delivered slightly better overall re-
sults when compared to the corresponding baseline,
whereas U-Net
SGA
was better only for bone regions.
We argue that this was mainly achieved due to the
contribution of the additional critic network which
did not allow the corresponding generator to pay too
much attention to the regions of interest.
5 CONCLUSION
In this work, we present 2D MRI-based pseudo-CT
generation approaches with the additional segmenta-
tion guided attention mechanisms. We defined our to-
tal loss functions as a combination of global and local
loss terms, whereas the second one enforces networks
to pay particular attention to bone areas while gener-
ating pCT images. From the evaluation results, we
observe that segmentation guided approaches yield
improvements compared to baseline U-Net and con-
ditional Wasserstein GAN architectures. As a re-
sult, more precise µ-maps for attenuation correction
of PET image in PET/MR systems could be derived.
Despite the apparent simplicity, segmentation guided
attention allows networks to focus more on specific
regions of interest, and as a consequence, achieve
better performances for them. We believe that the
proposed segmentation guidance can also be helpful
when training cycle GAN-based architectures. We are
currently in process of investigating this hypothesis.
REFERENCES
Arjovsky, M., Chintala, S., and Bottou, L. (2017). Wasser-
stein generative adversarial networks. In Interna-
tional conference on machine learning, pages 214–
223. PMLR.
Berker, Y., Franke, J., Salomon, A., Palmowski, M.,
Donker, H. C., Temur, Y., Mottaghy, F. M., Kuhl,
C., Izquierdo-Garcia, D., Fayad, Z. A., et al.
(2012). MRI-based attenuation correction for hy-
brid PET/MRI systems: a 4-class tissue segmen-
tation technique using a combined ultrashort-echo-
time/dixon mri sequence. Journal of nuclear
medicine, 53(5):796–804.
Beyer, T., Townsend, D. W., Brun, T., Kinahan, P. E., Char-
ron, M., Roddy, R., Jerin, J., Young, J., Byars, L.,
Nutt, R., et al. (2000). A combined PET/CT scanner
for clinical oncology. Journal of nuclear medicine,
41(8):1369–1379.
Brady, Z., Taylor, M., Haynes, M., Whitaker, M., Mullen,
A., Clews, L., Partridge, M., Hicks, R., and Trapp, J.
(2008). The clinical application of PET/CT: a contem-
porary review. Australasian Physics & Engineering
Sciences in Medicine, 31(2):90–109.
Burgos, N., Cardoso, M. J., Thielemans, K., Modat, M.,
Pedemonte, S., Dickson, J., Barnes, A., Ahmed, R.,
Mahoney, C. J., Schott, J. M., et al. (2014). Atten-
uation correction synthesis for hybrid PET-MR scan-
ners: application to brain studies. IEEE transactions
on medical imaging, 33(12):2332–2341.
Buzug, T. M. (2009). Computed tomography: from photon
statistics to modern cone-beam CT.
Chougule, V., Mulay, A., and Ahuja, B. (2018). Clini-
cal case study: spine modeling for minimum invasive
Improved MRI-based Pseudo-CT Synthesis via Segmentation Guided Attention Networks
139

spine surgeries (MISS) using rapid prototyping. Bone
(CT), 226:3071.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. (2014). Generative adversarial nets. Advances
in neural information processing systems, 27.
Han, X. (2017). MR-based synthetic CT generation using a
deep convolutional neural network method. Medical
physics, 44(4):1408–1419.
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., and
Hochreiter, S. (2017). Gans trained by a two time-
scale update rule converge to a local nash equilibrium.
Advances in neural information processing systems,
30.
Keereman, V., Mollet, P., Berker, Y., Schulz, V., and Van-
denberghe, S. (2013). Challenges and current methods
for attenuation correction in PET/MR. Magnetic Res-
onance Materials in Physics, Biology and Medicine,
26(1):81–98.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Leynes, A. P., Yang, J., Wiesinger, F., Kaushik, S. S.,
Shanbhag, D. D., Seo, Y., Hope, T. A., and Larson,
P. E. (2018). Zero-echo-time and dixon deep pseudo-
ct (ZeDD CT): direct generation of pseudo-CT images
for pelvic PET/MRI attenuation correction using deep
convolutional neural networks with multiparametric
MRI. Journal of Nuclear Medicine, 59(5):852–858.
Liu, F., Jang, H., Kijowski, R., Bradshaw, T., and McMil-
lan, A. B. (2017). Deep learning MR imaging–based
attenuation correction for PET/MR imaging. Radiol-
ogy, 286(2):676–684.
Lowekamp, B. C., Chen, D. T., Ib
´
a
˜
nez, L., and Blezek, D.
(2013). The design of SimpleITK. Frontiers in neu-
roinformatics, 7:45.
Mattes, D., Haynor, D. R., Vesselle, H., Lewellen, T. K.,
and Eubank, W. (2003). PET-CT image registration in
the chest using free-form deformations. IEEE trans-
actions on medical imaging, 22(1):120–128.
Mecheter, I., Alic, L., Abbod, M., Amira, A., and Ji, J.
(2020). MR image-based attenuation correction of
brain PET imaging: Review of literature on machine
learning approaches for segmentation. Journal of Dig-
ital Imaging, 33(5):1224.
Nie, D., Cao, X., Gao, Y., Wang, L., and Shen, D. (2016).
Estimating CT image from MRI data using 3D fully
convolutional networks. In Deep Learning and Data
Labeling for Medical Applications, pages 170–178.
Springer.
Nie, D., Trullo, R., Lian, J., Petitjean, C., Ruan, S., Wang,
Q., and Shen, D. (2017). Medical image synthesis
with context-aware generative adversarial networks.
In International conference on medical image comput-
ing and computer-assisted intervention, pages 417–
425. Springer.
Ollinger, J. M. and Fessler, J. A. (1997). Positron-
emission tomography. Ieee signal processing maga-
zine, 14(1):43–55.
Paans, A. M. (2006). Positron emission tomography.
Paulus, D. H., Quick, H. H., Geppert, C., Fenchel, M.,
Zhan, Y., Hermosillo, G., Faul, D., Boada, F.,
Friedman, K. P., and Koesters, T. (2015). Whole-
body PET/MR imaging: quantitative evaluation of a
novel model-based MR attenuation correction method
including bone. Journal of Nuclear Medicine,
56(7):1061–1066.
Qi, M., Li, Y., Wu, A., Jia, Q., Li, B., Sun, W., Dai, Z., Lu,
X., Zhou, L., Deng, X., et al. (2020). Multi-sequence
MR image-based synthetic CT generation using a gen-
erative adversarial network for head and neck mri-
only radiotherapy. Medical physics, 47(4):1880–1894.
Radford, A., Metz, L., and Chintala, S. (2015). Unsu-
pervised representation learning with deep convolu-
tional generative adversarial networks. arXiv preprint
arXiv:1511.06434.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. In International Conference on Medical
image computing and computer-assisted intervention,
pages 234–241. Springer.
Torrado-Carvajal, A., Vera-Olmos, J., Izquierdo-Garcia, D.,
Catalano, O. A., Morales, M. A., Margolin, J., Sori-
celli, A., Salvatore, M., Malpica, N., and Catana,
C. (2019). Dixon-VIBE deep learning (DIVIDE)
pseudo-CT synthesis for pelvis PET/MR attenuation
correction. Journal of nuclear medicine, 60(3):429–
435.
Wang, Z., Bovik, A. C., Sheikh, H. R., and Simoncelli, E. P.
(2004). Image quality assessment: from error visi-
bility to structural similarity. IEEE transactions on
image processing, 13(4):600–612.
West, J., Fitzpatrick, J. M., Wang, M. Y., Dawant, B. M.,
Maurer Jr, C. R., Kessler, R. M., Maciunas, R. J., Bar-
illot, C., Lemoine, D., Collignon, A., et al. (1997).
Comparison and evaluation of retrospective inter-
modality brain image registration techniques. Journal
of computer assisted tomography, 21(4):554–568.
Wolterink, J. M., Dinkla, A. M., Savenije, M. H., Seevinck,
P. R., van den Berg, C. A., and I
ˇ
sgum, I. (2017). Deep
mr to ct synthesis using unpaired data. In Interna-
tional Workshop on Simulation and Synthesis in Med-
ical Imaging, pages 14–23. Springer.
Wu, J., Huang, Z., Thoma, J., Acharya, D., and Van Gool, L.
(2018). Wasserstein divergence for gans. In Proceed-
ings of the European Conference on Computer Vision
(ECCV), pages 653–668.
Yaniv, Z., Lowekamp, B. C., Johnson, H. J., and Beare, R.
(2018). SimpleITK image-analysis notebooks: a col-
laborative environment for education and reproducible
research. Journal of digital imaging, 31(3):290–303.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Proceedings of
the IEEE international conference on computer vi-
sion, pages 2223–2232.
BIOIMAGING 2022 - 9th International Conference on Bioimaging
140
