
Classifying Breast Cytological Images using Deep Learning
Architectures
Hasnae Zerouaoui
1a
and Ali Idri
1,2 b
1
Modeling, Simulation and Data Analysis, Mohammed VI Polytechnic University,
Benguerir, Morocco
2
Software Project Management Research Team, ENSIAS, Mohammed V University in Rabat, Morocco
Keywords: Computer-aided Diagnosis, Breast Cancer, Classification, Deep Convolutional Neural Networks, Image
Processing, Histological Images.
Abstract: Breast cancer (BC) is a leading cause of death among women worldwide. It remains a critical challenge,
causing over 10 million deaths globally in 2020. Medical images analysis is the most promising research area
since it provides facilities for diagnosing several diseases such as breast cancer. The present paper carries out
an empirical evaluation of recent deep Convolutional Neural Network (CNN) architectures for a binary
classification of breast cytological images based fined tuned versions of seven deep learning techniques:
VGG16, VGG19, DenseNet201, InceptionResNetV2, InceptionV3, ResNet50 and MobileNetV2. The
empirical evaluations used: (1) four classification performance criteria (accuracy, recall, precision and F1-
score), (2) Scott Knott (SK) statistical test to select the best cluster of the outperforming architectures, and (3)
borda count voting system to rank the best performing architectures. All the evaluations were over the FNAC
dataset which contain 212 images. Results showed the potential of deep learning techniques to classify breast
cancer in malignant and benign, therefor the findings of this study recommend the use of MobileNetV2 for
the classification of the breast cancer cytological images since it gave the best results with an accuracy of
98.54%.
1 INTRODUCTION
Breast cancer (BC) is still the leading cause of death
among women worldwide (Metelko et al., 1995). It
remains a global challenge, causing over 1 million
deaths globally in 2019 (Bish et al., 2005). As the
number of patients infected by this disease increases,
it turns out to be increasingly hard for radiologists to
accurately deal with the diagnosis process in the
constrained accessible time (Zhang et al., 2011).
Medical images analysis is one of the most promising
research areas, it provides facilities for diagnosis and
making decisions of several diseases such as breast
cancer. Recently, more attention are paid to imaging
modalities and Deep Learning (DL) in BC
(Mendelson and Eb, 2019). Therefore, interpretation
of these images requires expertise and consequently
several algorithms have been developed and
evaluated to improve and help oncologist’s diagnosis.
a
https://orcid.org/0000-0001-7268-8404
b
https://orcid.org/0000-0002-4586-4158
In general, DL showed better performance in breast
cancer detection, and provided high accurate
classifications compared with classical Machine
Learning (ML) techniques (Saha, Mukherjee and
Chakraborty, 2016) (Sadoughi et al., 2018). For
instance, the study (Saha, Mukherjee and
Chakraborty, 2016) showed that the use of deep CNN
architectures is very powerful and efficient in the
domain of DL since it tested the InceptionRecurrent
Residual CNN in the dataset BreakHis and it gave
better results compared to existing techniques such as
CNN and SVM. The study (Xie et al., 2019) used
AlexNet and LeNet for the binary classification of the
BreakHis dataset and showed an improvement of the
accuracy compared to the traditional ML techniques.
However, the present study develops and evaluates
the performances measured in terms of accuracy,
sensitivity, recall, precision and F1-score of seven of
the most recent DL techniques in BC classification
Zerouaoui, H. and Idri, A.
Classifying Breast Cytological Images using Deep Learning Architectures.
DOI: 10.5220/0010850000003123
In Proceedings of the 15th Inter national Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 557-564
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
557

over the FNAC dataset. To the best of our knowledge,
this study is the first to evaluate and compare seven
DL techniques (VGG16, VGG19, DenseNet201,
InceptionResNetV2, InceptionV3, ResNet50 and
MobileNetV2) using the Scott Knott (SK) statistical
test and the borda count voting method in BC binary
classification. Note that the SK test has been widely
used to comparing, clustering and ranking multiple
machine learning models for parameters tunning
(Idri, Hosni and Abran, 2016; Ottoni et al., 2020) in
different fields such as software engineering (Ottoni
et al., 2020) and breast cancer (Idri et al., 2020) .
Therefore, we use the SK test since: (1) it shows high
performance compared to other statistical tests such
as Jollife (Jolliffe, Allen and Christie, 1989), Calinski
and Corsten (Calinski and Corsten, 1985), and Cox
and Spjotvoll (Worsley, 1986) and (2) its ability to
select the best non-overlapping groups of machine
learning techniques. Moreover, we use the Borda
Count voting method (García-Lapresta and Martínez-
Panero, 2002; Emerson, 2013) to rank the best SK
selected techniques based on the four performance
criterias.
The present study discusses two research
questions (RQs):
(RQ1): What is the overall performance of DL
techniques in BC classification?
(RQ2): Is there any DL techniques which
distinctly outperform the others?
The main contributions of this empirical study are the
following: (1) Designing seven DL architectures:
VGG16, VGG19, DenseNet201,
InceptionResNetV2, InceptionV3, ResNet50 and
MobileNetV2 in BC classification, (2) Avoiding
overfitting by using weight decay and L2
regularizers, (3) Comparing the performances of the
seven architectures using SK clustering test and borda
count voting method.
The remainder of this paper is organized as
follow. Section 2 describes the related work. In
Section 3, we present the configuration and
parametrization of the seven DL techniques, the
empirical methodology followed throughout the
research, the data preparation which includes data
acquisition and image processing and the
abbreviations. Section 4 presents and discusses the
empirical results. Section 5 outlines conclusions and
future works.
2 RELATED WORKS
This section presents the results and the main findings
of the related work as shown in Table 1, the results
are summarized as follow:
Accuracy is the most frequently criterion used to
evaluate the performance of the DL techniques in
BC when using balanced datasets (Zerouaoui and
Idri, 2021).
Most of the studies only compared two to three
DL techniques. Although the DL architectures
used in the selected studies were different, it is
worth notable that the most investigated
techniques were InceptionResNet, VGG16,
VGG19 and ResNet50
(Alom et al., 2019; Spanhol
et al., 2016; Xie et al., 2019)
Some studies (
Kassani et al., 2019; (Nahid, Mehrabi
and Kong, 2018; Zhu et al., 2019)
combined more
than two DL techniques in order to have better
Table 1: Summary of the literature review of the use of DL
techniques in BC classification.
Authors Findings and results
Alom et
al.
(Alom et
al.,
2019)
Conduct a study on the use of Inception
Recurrent Residual Convolutional Neural
Network (IRRCNN) which is a hybrid
DCNN architecture based on RCNN,
Residual Network and Inception tested on
two datasets: BreakHis and BC
classification challenge 2015.
The performance was evaluated on image
level, patch level, image based and patch-
based analysis. The results show an
improvement of 3,67% and 2.14% of
accuracy on the BreakHis dataset compared
to scientific results since 2016
Fabio et
al.
(Spanhol
et al.,
2016)
Investigate a deep learning approach, using
two DCNN architecture which are AlexNet
and LeNet, in order to avoid hand crafted
features. The results of the experiments
demonstrated an improved accuracy
compared to the experiments that used
traditional feature extractor techniques.
Xie et al.
(Xie et
al.,
2019)
Evaluate Inception_V3 and Inception_
ResNet_V2 for classification of Brekhis
using two types of learning: the supervised
and the unsupervised learning. The authors
used the transfer learning by pre training the
model on ImageNet, applying Data
augmentation and Fine tuning using the
BreakHis dataset. The experiment shows
that the results of the augmented dataset is
much better than the normal dataset, that
Inception_ ResNet_V2 has better results for
the feature extraction and the supervised
learning has a better accuracy than
clusterin
g
HEALTHINF 2022 - 15th International Conference on Health Informatics
558
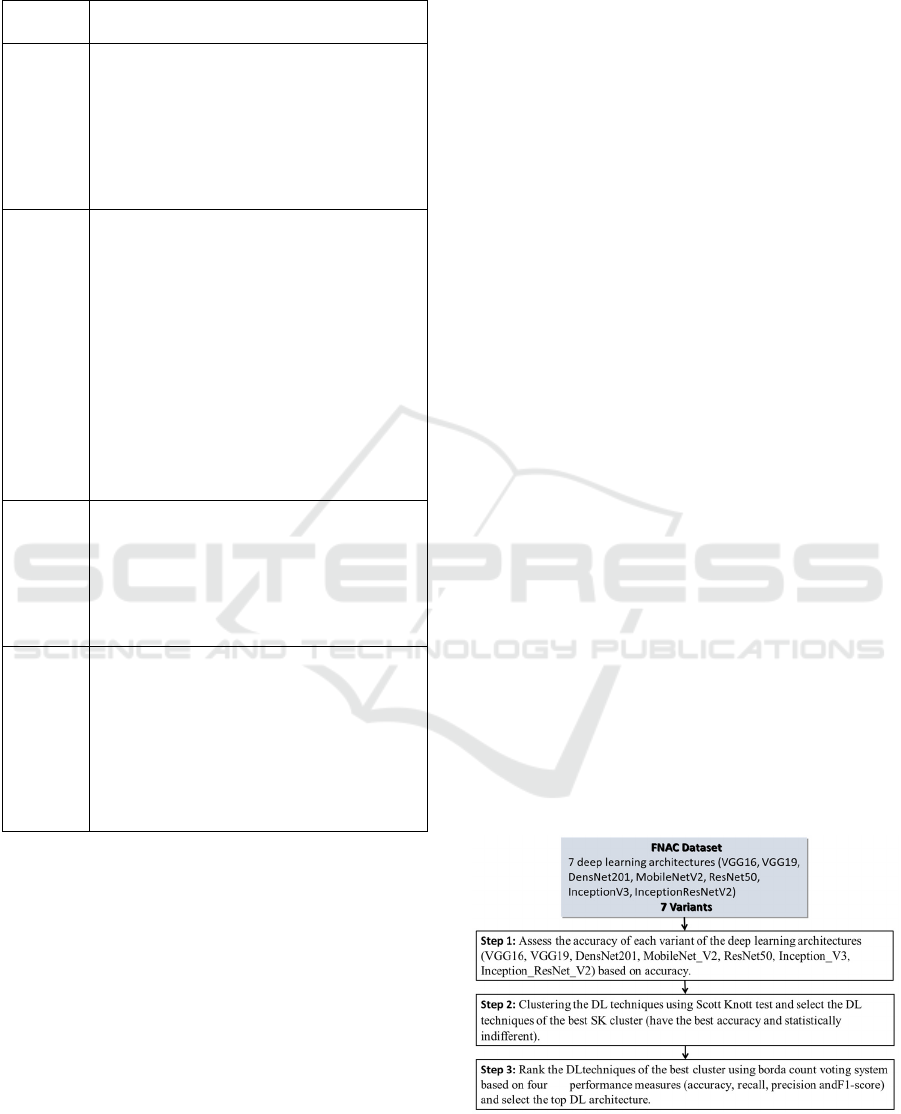
Table 1: Summary of the literature review of the use of DL
techniques in BC classification (cont.).
Authors
Findings and results
Al
Nahid et
al.
(Nahid,
Mehrabi
and
Kong,
2018)
Use three models: CNN techniques, LSTM
structure and the combination of CNN and
LSTM on the BreakHis Dataset. As results
91% of accuracy was obtained using 200x
images dataset, 96 of precision with 40x
images dataset, and the best F-Measure was
obtained using 40x and 100x datasets.
Kassani
et al.
(Kassani
et al.,
2019)
Use of a combination of CNNs deep
learning architectures such as VGG19,
MobileNet and Densenet for feature
extraction on 4 histopathological dataset
images: ICIAR, BreakHis, Densenet and
MobilNet. Then they compared the results
of the classification on traditional single
models and machine learning techniques
namely Decision tree, Random Forest,
XGBoost, AdaBoost and bagging classifier.
The main finding is that the proposed
ensemble method gives better results thant
the solo methods with an accuracy of:
98.13%,
Chuang
et al.
(Zhu et
al.,
2019)
Propose a hybrid architecture where they
assembled multiple CNN architectures
(Inception module, Residual Net and Batch
Normalization techniques) and tested it on
two datasets (BreakHis and BACH). The
proposed model shows a comparable and
b
etter performance.
Jiang et
al. (Jiang
et al.,
2019)
Design a new CNN that includes a
convolutional layer, a small SE-ResNet
module and a fully connected layer,
they tested their architecture on the
BreakHis dataset and the results
achieved 98,87% and 99,34% for the
binary class and 90.66% and 93.81% for
the multi class.
prediction results, since the ensemble methods in
general outperformed their single techniques.
3 EXPERIMENT
CONFIGURATION
This section presents the parameter tuning of the DL
models, the empirical design, the data preparation
followed and finally the abbreviations.
3.1 Experiment Configuration
Toward an automatic binary BC classification based
on publicly available image FNAC dataset, the
different DL architectures have been implemented
using several parameters tuning experiments. All the
images of the FNAC dataset were resized to 224x224
pixels except those of InceptionV3 and
InceptionResNetV2 models that were resized to
299x299 since it is the default input size in their
architectures. To train the models, we used the
transfer learning technique where we downloaded the
seven DL techniques pre-trained in the ImageNet
dataset (Fei-Fei, Deng and Li, 2010). For the
parameter tunning, we set the batch size to 32 and the
number of epochs to 300. As for the optimization, we
used Adam (adaptive moment estimation) (Kingma
and Ba, 2015) with β1=0.9, β2=0.999, and an initial
learning rate set to 0. 0001 and decrease exponentially
to 0.000001. Moreover, we used weight decay and
L2- regularizers to reduce the overfitting for different
models. A fully connected layer was trained with the
ReLU, followed by a dropout layer with a probability
of 0.5. We updated the last dense layer in all models
to output two classes corresponding to benign and
malignant instead of 1000 classes as was used for
ImageNet.
3.2 Empirical Design
Figure 1 shows the methodology followed to carry
out all the empirical evaluations. It consists of three
steps we describe hereafter. Note that similar
methodologies were used in (Worsley,
2009)(SHARMA et al., 2003)(Azzeh, Nassif and
Minku, 2015)(Idri, Abnane and Abran,
2018)(Zerouaoui et al., 2021)(Idri and Abnane,
2017).
Figure 1: Experimental process.
Classifying Breast Cytological Images using Deep Learning Architectures
559

3.3 Data Preparation
This section presents the data preparation process
followed for the FNAC as described in Figure 2,
which consists of Data pre-processing by using
intensity normalization and Contrast Limited
Adaptive Histogram Equalization (CLAHE) and Data
augmentation. The Images of the FNAC dataset were
captured by us using Leica ICC50 HD microscope
using 400 resolution and 24 bits color depth and with
5 megapixels camera associated with the
microscope(Saikia et al., 2019). Digitized images
captured were then reviewed by experienced certified
cyto-pathologists and selected a total of 212 images
(113 Malignant and 99 Benign). The database can be
downloaded from the link in (Saikia et al., 2019).
Figure 2: Data preparation process.
Data Processing: The next stage is to pre-process
input images using intensity normalization and
Contrast Limited Adaptive Histogram Equalization
(CLAHE). Intensity normalization is a pre-processing
step in image processing applications (Kassani et al.,
2019). We normalized input images to the standard
normal distribution using min-max normalization of
Equation 1. Furthermore, before feeding input images
into the proposed models, CLAHE is a necessary step
to improve the contrast in images as shown in Figure
3 (Makandar and Halalli, 2015; Kharel et al., 2017).
Figure 3: Original and transformed images.
minmax
min
xx
xx
X
norm
−
−
=
(1)
Data Augmentation: was used for the training
process after dataset pre-processing and has the goal
to avoid the risk of overfitting (Perez and Wang,
2017). Moreover, the strategies we used include
geometric transforms such as rescaling, rotations,
shifts, shears, zooms and flips.
3.4 Abbreviations
To assist the reader and shorten the names of the DL
techniques, we use the following naming rules in the
rest of this paper. We abbreviate the name of each
variant of DL techniques as shown in Table 2.
Table 2: Abbreviations used for the DL techniques for the
FNAC dataset.
D.L techniques with the image
magnification factor
Abbreviation
VGG 16 VGG16
VGG 19 VGG19
ResNet 50 Res50
Inception V3 INV3
Inception ResNet V2 INRES
DensNet201 DENS
MobilNetV2 MOB
4 RESULTS
This section presents and discusses the results of the
empirical evaluations of seven DL techniques:
VGG16, VGG19, InceptionV3, ResNet50,
InceptionResNetV2, DenseNet201, and
MobileNetV2, over the FNAC dataset. The
performances of the DL techniques were evaluated
using 5-fold cross validation and four criteria’s:
accuracy, recall, precision and F1-score. First the
performance is compared in terms of accuracy of each
DL technique (RQ1). Thereafter, we use the SK
statistical test to cluster the selected DL techniques,
and borda count to rank the DL techniques belonging
to the best SK cluster (RQ2).
4.1 Accuracy Evaluation and
Comparison of the Seven DL
Techniques
This section compares the accuracy values of the
seven DL techniques to each other over the FNAC
dataset. Note that the training and testing of the DL
techniques are implemented in Python using Keras
and Tensorflow DL frameworks and run on a TPU
processing unit of 8 cores with 35 GB in RAM and
Linux-based OS, provided by google in Colab
Notebook.
Figure 4 and Table 3 show the accuracy values of
the baseline VGG16, VGG19, DenseNet201,
InceptionResNetV2, InceptionV3, ResNet50 and
HEALTHINF 2022 - 15th International Conference on Health Informatics
560

MobileNetV2 vs the number of epochs over the
FNAC dataset. We observe that the best accuracy
values were achieved with the number of epochs 25
using VGG16, VGG19, InceptionV3, DenseNet201,
InceptionResNetV2 and MobileNetV2; and with the
number of epochs 100 when using ResNet 50 since.
The best accuracy value was achieved by MobileNet
V2 (98.54%) followed by DenseNet201, VGG16,
VGG19, InceptionV3 and InceptionResNetV2 with
an accuracy value of 98.07%, 97.65%, 95.72%,
93.51% and 91.34% respectively. The worst accuracy
value was achieved using ResNe 50 with a value of
91.34%
Figure 4: Accuracy values vs number of epochs of the seven
deep learning architectures over the FNAC dataset (The
abbreviation used in Figure 3 for the DL techniques for the
FNAC dataset are defined in Table 4 of section 6.5).
Table 3: Accuracy values of the seven deep learning
architectures over the FNAC dataset (The abbreviation used
in Table 3 for the DL techniques for the FNAC dataset are
defined in Table 1 of section 3.4).
DL technique Accuracy (%)
VGG16 97.65
VGG19 95.72
INV3 95.06
Res50 91.34
INRES 93.51
DENS 98.07
MOB 98.54
4.2 Clustering DL Techniques using
SK Test and Ranking Them using
Borda Count
This step uses the SK statistical test to evaluate the
predictive capabilities of the DL techniques evaluated
in step 4.1 and discusses the ranking results when
applying the borda count voting method based on
accuracy, recall, precision and F1-score on the best
SK clusters. Table 4 shows the values of the four
performance measures of all the DL techniques over
FNAC dataset. Note that the SK test consists of
grouping DL techniques with no significant
difference between their accuracy values. Since the
SK test requires that its inputs should be normally
distributed we verified the normality of the data by
the Kolmogorov-Smirnov test, and since the data is
normally distributed we didn’t use the Box Cox
transformation (Sakia, 2012). Afterwards, we
performed the SK test to cluster the selected DL
techniques into overlapping free groups and
identified the best group based on accuracy. The DL
techniques belonging to the same group have similar
predictive capability and the best group contains the
DL techniques that have the highest value of
accuracy.
Table 4: Best performance values of the DL techniques over
the FNAC Dataset (The abbreviation used in Table 4 for the
DL techniques for the FNAC dataset are defined in Table 1
of section 3.4).
DL Accur
acy
(%)
Recall
(%)
Precis
ion
(%)
F1 score
(%)
VGG16 97.65 98.15 97.49 97.8
VGG19 95.72 94.98 96.95 95.95
INV3 95.06 94.19 96.5 95.32
Res50 91.34 91.9 91.98 91.86
INRES 93.51 94.28 93.79 93.94
DENS 98.07 98.42 98.86 98.63
MOB 98.54 98.15 98.24 98.2
From the results of the SK test shown in Figure 5, it
is noticeable that the SK test results gives 4 clusters
which implies that the accuracy performances are
highly influenced by the DL model used for the
classification. The figure shows that the best SK
cluster contains 3 DL models including
DenseNet201, MobileNetV2 and VGG16 and that
last SK cluster contains the DL model ReseNet50.
Figure 5: Results of SK test for the DL techniques over the
FNAC dataset.
Table 5 shows the borda count ranking of the
architectures belonging to the best SK clusters for the
Classifying Breast Cytological Images using Deep Learning Architectures
561

FNAC dataset. The DL architecture MobileNetV2 is
ranked first followed by DenseNet201 and finally
VGG16.
Table 5: Best performance values.
Borda Count Ranking Deep Learning model
1 MOB
2 DENS
3 VGG16
As a summary, when using the cytological FNAC
dataset, it is recommended to use the DL architecture
MobileNetV2 since it was ranked first when using the
borda count voting method based on accuracy,
precision, recall and F1-score and achieved an
accuracy of 98.54%. On top of that MobileNetV2
remains the perfect DL architecture since it is light
weighted in terms of architecture and is designed for
mobile and web application. Therefore, we highly
recommend the use of MobileNetV2 for a binary
cytological classification.
5 CONCLUSIONS
The present paper presented and discussed the results
of an empirical comparative study of seven recent DL
techniques (VGG16, VGG19, DenseNet201,
InceptionResnetV2, InceptionV3, ResNet50 and
MobileNetV2) for BC binary imaging classification.
All the empirical evaluations used four performance
criteria’s, SK statistical test, and borda Count to
assess and rank these seven DL techniques over the
FNAC dataset. The findings of this study are:
(RQ1): What is the Overall Performance of DL
Techniques in BC Classification?
The accuracy results of the seven DL techniques were
highly influenced by the characteristics of the dataset.
Nevertheless, we observed that MobileNetV2,
DenseNet201, VGG16, VGG19 and InceptionV3
gave the best results. However, ReseNet50
underperformed compared to the others.
(RQ2): Is There Any DL Techniques, Which
Distinctly Outperform the Others?
MobileNetV2 technique gave the best results since it
belonged to the best SK clusters for the FNAC dataset
and was ranked first using the borda count voting test
based on accuracy, recall, precision and F1-score. As
results we recommend the use of MobileNet V2 to
develop DL computer assister diagnosis systems
since it gives good results when using cytological
images for binary BC classification.
Ongoing works investigate homogenous and
heterogeneous ensembles whose members are deep
learning techniques with different meta-learning
techniques such as bagging, boosting and stacking for
breast cancer imaging classification.
ACKNOWLEDGEMENTS
This work was conducted under the research project
“Machine Learning based Breast Cancer Diagnosis
and Treatment”, 2020-2023. The authors would like
to thank the Moroccan Ministry of Higher Education
and Scientific Research, Digital Development
Agency (ADD), CNRST, and UM6P for their
support.
This study was funded by Mohammed VI
polytechnic university at Ben Guerir Morocco.
Compliance with ethical standards.
Conflicts of interest/competing interests Not
applicable.
Code availability not applicable.
REFERENCES
Alom, M. Z. et al. (2019) ‘Breast Cancer Classification
from Histopathological Images with Inception
Recurrent Residual Convolutional Neural Network’,
Journal of Digital Imaging. Journal of Digital Imaging.
doi: 10.1007/s10278-019-00182-7.
Azzeh, M., Nassif, A. B. and Minku, L. L. (2015) ‘An
empirical evaluation of ensemble adjustment methods
for analogy-based effort estimation’, Journal of
Systems and Software. Elsevier Ltd., 103, pp. 36–52.
doi: 10.1016/j.jss.2015.01.028.
Bish, A. et al. (2005) ‘Understanding why women delay in
seeking help for breast cancer symptoms B’, 58, pp.
321–326. doi: 10.1016/j.jpsychores.2004.10.007.
Bony, S. et al. (2001) ‘The relationship between mycotoxin
synthesis and isolate morphology in fungal endophytes
of Lolium perenne’, New Phytologist, 152(1), pp. 125–
137. doi: 10.1046/j.0028-646X.2001.00231.x.
Calinski, T. and Corsten, L. C. A. (1985) ‘Clustering Means
in ANOVA by Simultaneous Testing’, Biometrics,
41(1), p. 39. doi: 10.2307/2530641.
Emerson, P. (2013) ‘The original Borda count and partial
voting’, Social Choice and Welfare, 40(2), pp. 353–
358. doi: 10.1007/s00355-011-0603-9.
Fei-Fei, L., Deng, J. and Li, K. (2010) ‘ImageNet:
Constructing a large-scale image database’, Journal of
Vision, 9(8), pp. 1037–1037. doi: 10.1167/9.8.1037.
García-Lapresta, J. L. and Martínez-Panero, M. (2002)
‘Borda count versus approval voting: A fuzzy
HEALTHINF 2022 - 15th International Conference on Health Informatics
562

approach’, Public Choice, 112(1), pp. 167–184. doi:
10.1023/A:1015609200117.
Hamza, M. and Larocque, D. (2005) ‘An empirical
comparison of ensemble methods based on
classification trees’, Journal of Statistical Computation
and Simulation, 75(8), pp. 629–643. doi: 10.1080/
00949650410001729472.
He, K. and Sun, J. (2016) ‘Deep Residual Learning for
Image Recognition’. doi: 10.1109/CVPR.2016.90.
Hosni, M. et al. (2019) ‘Reviewing ensemble classification
methods in breast cancer’, Computer Methods and
Programs in Biomedicine, 177, pp. 89–112. doi:
10.1016/j.cmpb.2019.05.019.
Huang, G. et al. (2017) ‘Densely Connected Convolutional
Networks’. doi: 10.1109/CVPR.2017.243.
Idri, A. et al. (2020) ‘Assessing the impact of parameters
tuning in ensemble based breast Cancer classification’,
Health and Technology. Health and Technology, 10(5),
pp. 1239–1255. doi: 10.1007/s12553-020-00453-2.
Idri, A. and Abnane, I. (2017) ‘Fuzzy Analogy Based Effort
Estimation: An Empirical Comparative Study’, IEEE
CIT 2017 - 17th IEEE International Conference on
Computer and Information Technology, (Ml), pp. 114–
121. doi: 10.1109/CIT.2017.29.
Idri, A., Abnane, I. and Abran, A. (2018) ‘Evaluating
Pred(p) and standardized accuracy criteria in software
development effort estimation’, Journal of Software:
Evolution and Process, 30(4), pp. 1–15. doi: 10.1002/
smr.1925.
Idri, A., Hosni, M. and Abran, A. (2016) ‘Improved
estimation of software development effort using
Classical and Fuzzy Analogy ensembles’, Applied Soft
Computing Journal. Elsevier B.V., 49, pp. 990–1019.
doi: 10.1016/j.asoc.2016.08.012.
Jiang, Y. et al. (2019) ‘Breast cancer histopathological
image classification using convolutional neural
networks with small SE-ResNet module’, PLoS ONE,
14(3), pp. 1–21. doi: 10.1371/journal.pone.0214587.
Jolliffe, I. T., Allen, O. B. and Christie, B. R. (1989)
‘COMPARISON OF VARIETY MEANS USING
advantage of this approach is that the divisions into
groups can be done at more’, 25, pp. 259–269.
K, H. T. (2013) ‘c r v i h o e f c r v i h o e f’, 4(2), pp. 627–
635.
Kassani, S. H. et al. (2019) ‘Classification of
Histopathological Biopsy Images Using Ensemble of
Deep Learning Networks’. Available at: http://
arxiv.org/abs/1909.11870.
Kharel, N. et al. (2017) ‘Early diagnosis of breast cancer
using contrast limited adaptive histogram equalization
(CLAHE) and Morphology methods’, 2017 8th
International Conference on Information and
Communication Systems, ICICS 2017, pp. 120–124.
doi: 10.1109/IACS.2017.7921957.
Kingma, D. P. and Ba, J. L. (2015) ‘Adam: A method for
stochastic optimization’, 3rd International Conference
on Learning Representations, ICLR 2015 - Conference
Track Proceedings, pp. 1–15.
Makandar, A. and Halalli, B. (2015) ‘Breast Cancer Image
Enhancement using Median Filter and CLAHE’,
International Journal of Scientific & Engineering
Research, 6(4), pp. 462–465. Available at: http://
www.ijser.org.
Mendelson, E. B. and Eb, M. (2019) ‘Imaging : Potentials
and Limitations’, American Journal of Roentgenology,
(February), pp. 1–7. doi: 10.2214/AJR.18.20532.
Metelko, Z. et al. (1995) ‘Pergamon THE WORLD
HEALTH ORGANIZATION QUALITY OF LIFE
ASSESSMENT ( WHOQOL ): POSITION PAPER
FROM THE WORLD HEALTH ORGANIZATION’,
41(10).
Mittas, N. and Angelis, L. (2013) ‘Ranking and clustering
software cost estimation models through a multiple
comparisons algorithm’, IEEE Transactions on
Software Engineering, 39(4), pp. 537–551. doi:
10.1109/TSE.2012.45.
Nahid, A. Al, Mehrabi, M. A. and Kong, Y. (2018)
‘Histopathological breast cancer image classification
by deep neural network techniques guided by local
clustering’, BioMed Research International, 2018. doi:
10.1155/2018/2362108.
Ottoni, A. L. C. et al. (2020) ‘Tuning of reinforcement
learning parameters applied to SOP using the Scott–
Knott method’, Soft Computing. Springer Berlin
Heidelberg, 24(6), pp. 4441–4453. doi: 10.1007/
s00500-019-04206-w.
Perez, L. and Wang, J. (2017) ‘The Effectiveness of Data
Augmentation in Image Classification using Deep
Learning’. Available at: http://arxiv.org/abs/
1712.04621.
Razzak, M. I., Naz, S. and Zaib, A. (no date) ‘Deep
Learning for Medical Image Processing : Overview ,
Challenges and the Future’.
Sadoughi, F. et al. (2018) ‘Artificial intelligence methods
for the diagnosis of breast cancer by image processing:
A review’, Breast Cancer: Targets and Therapy, 10,
pp. 219–230. doi: 10.2147/BCTT.S175311.
Sagi, O. and Rokach, L. (2018) ‘Ensemble learning: A
survey’, Wiley Interdisciplinary Reviews: Data Mining
and Knowledge Discovery, 8(4), pp. 1–18. doi:
10.1002/widm.1249.
Saha, M., Mukherjee, R. and Chakraborty, C. (2016)
‘Computer-aided diagnosis of breast cancer using
cytological images: A systematic review’, Tissue and
Cell. Elsevier Ltd, 48(5), pp. 461–474. doi:
10.1016/j.tice.2016.07.006.
Saikia, A. R. et al. (2019) ‘Comparative assessment of
CNN architectures for classification of breast FNAC
images’, Tissue and Cell. Elsevier Ltd, 57, pp. 8–14.
doi: 10.1016/j.tice.2019.02.001.
Sakia, A. R. M. (2012) ‘The Box-Cox transformation
technique : a review’, 41(2), pp. 169–178.
Sandler, M. et al. (2018) ‘MobileNetV2: Inverted Residuals
and Linear Bottlenecks’, Proceedings of the IEEE
Computer Society Conference on Computer Vision and
Pattern Recognition. IEEE, pp. 4510–4520. doi:
10.1109/CVPR.2018.00474.
SHARMA, J. et al. (2003) ‘Symbiotic Seed Germination
and Mycorrhizae of Federally Threatened Platanthera
praeclara (Orchidaceae)’, The American Midland
Classifying Breast Cytological Images using Deep Learning Architectures
563

Naturalist, 149(1), pp. 104–120. doi: 10.1674/0003-
0031(2003)149[0104:ssgamo]2.0.co;2.
Simonyan, K. and Zisserman, A. (2015) ‘Very deep
convolutional networks for large-scale image
recognition’, 3rd International Conference on Learning
Representations, ICLR 2015 - Conference Track
Proceedings, pp. 1–14.
Spanhol, F. A. et al. (2016) ‘A Dataset for Breast Cancer
Histopathological Image Classification’, IEEE
Transactions on Biomedical Engineering, 63(7), pp.
1455–1462. doi: 10.1109/TBME.2015.2496264.
Szegedy, C. et al. (2014) ‘Rethinking the Inception
Architecture for Computer Vision’.
Szegedy, C. et al. (no date) ‘the Impact of Residual
Connections on Learning’, pp. 4278–4284.
Worsley, A. K. J. (2009) ‘A Non-Parametric Extension of a
Cluster Analysis Method by Scott and Knott Published
by: International Biometric Society Stable URL: http://
www.jstor.org/stable/2529369’, 33(3), pp. 532–535.
Worsley, K. J. (1986) ‘Confidence regions and tests for a
change-point in a sequence of exponential family
random variables’, Biometrika, 73(1), pp. 91–104. doi:
10.1093/biomet/73.1.91.
Xie, J. et al. (2019) ‘Deep learning based analysis of
histopathological images of breast cancer’, Frontiers in
Genetics, 10(FEB), pp. 1–19. doi: 10.3389/
fgene.2019.00080.
Zerouaoui, H. et al. (2021) ‘Breast Fine Needle Cytological
Classification Using Deep Hybrid Architectures BT -
Computational Science and Its Applications – ICCSA
2021’, in Gervasi, O. et al. (eds). Cham: Springer
International Publishing, pp. 186–202.
Zerouaoui, H. and Idri, A. (2021) ‘Reviewing Machine
Learning and Image Processing Based Decision-
Making Systems for Breast Cancer Imaging’. Journal of
Medical Systems.
Zhang, G. et al. (2011) ‘A review of breast tissue
classification in mammograms’, Proceedings of the
2011 ACM Research in Applied Computation
Symposium, RACS 2011, pp. 232–237. doi: 10.1145/
2103380.2103426.
Zhu, C. et al. (2019) ‘Breast cancer histopathology image
classification through assembling multiple compact
CNNs’, BMC Medical Informatics and Decision
Making, 19(1), pp. 1–17. doi: 10.1186/s12911-019-
0913-x.
HEALTHINF 2022 - 15th International Conference on Health Informatics
564
