
DeIDNER Model: A Neural Network Named Entity Recognition
Model for Use in the De-identification of Clinical Notes
Mahanazuddin Syed
1a
, Kevin Sexton
1b
, Melody Greer
1
, Shorabuddin Syed
1c
, Joseph VanScoy
2
,
Farhan Kawsar
2
, Erica Olson
2
, Karan Patel
2
, Jake Erwin
2
, Sudeepa Bhattacharyya
3
,
Meredith Zozus
4d
and Fred Prior
1e
1
Department of Biomedical Informatics, University of Arkansas for Medical Sciences, Little Rock, AR, U.S.A.
2
College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR, U.S.A.
3
Department of Biological Sciences and Arkansas Biosciences Institute, Arkansas State University, Jonesboro, AR, U.S.A.
4
Department of Population Health Sciences, University of Texas Health Science Center at San Antonio,
San Antonio, TX, U.S.A.
Keywords: Clinical Named Entity Recognition, Deep Learning, De-identification, Word Embeddings, and Natural
Language Processing.
Abstract: Clinical named entity recognition (NER) is an essential building block for many downstream natural language
processing (NLP) applications such as information extraction and de-identification. Recently, deep learning
(DL) methods that utilize word embeddings have become popular in clinical NLP tasks. However, there has
been little work on evaluating and combining the word embeddings trained from different domains. The goal
of this study is to improve the performance of NER in clinical discharge summaries by developing a DL
model that combines different embeddings and investigate the combination of standard and contextual
embeddings from the general and clinical domains. We developed: 1) A human-annotated high-quality
internal corpus with discharge summaries and 2) A NER model with an input embedding layer that combines
different embeddings: standard word embeddings, context-based word embeddings, a character-level word
embedding using a convolutional neural network (CNN), and an external knowledge sources along with word
features as one-hot vectors. Embedding was followed by bidirectional long short-term memory (Bi-LSTM)
and conditional random field (CRF) layers. The proposed model reaches or overcomes state-of-the-art
performance on two publicly available data sets and an F1 score of 94.31% on an internal corpus. After
incorporating mixed-domain clinically pre-trained contextual embeddings, the F1 score further improved to
95.36% on the internal corpus. This study demonstrated an efficient way of combining different embeddings
that will improve the recognition performance aiding the downstream de-identification of clinical notes.
1 INTRODUCTION
Named entity recognition (NER) is an important step
in any natural language processing (NLP) task.
Clinical NER, i.e., NER applied to unstructured data
in medical records, has received much more attention
recently (Catelli et al., 2020). Clinical NER allows
recognizing and labeling entities in the clinical text,
which is an essential building block for many
downstream NLP applications such as information
a
https://orcid.org/0000-0002-8978-1565
b
https://orcid.org/0000-0002-1460-9867
c
https://orcid.org/0000-0002-4761-5972
d
https://orcid.org/0000-0002-9332-1684
e
https://orcid.org/0000-0002-6314-5683
extraction from and de-identification of entities in
clinical narratives.
Early clinical NER systems often relied on rule-
based techniques, which heavily depend on
dictionaries or ontologies (Liu et al., 2017). Later, a
number of machine learning (ML), deep learning
(DL), and hybrid models emerged and provided
improved performance (Syeda et al., 2021; S. Wu et
al., 2020). A major challenge with these later models
is access to a corpus of labeled data for training,
640
Syed, M., Sexton, K., Greer, M., Syed, S., VanScoy, J., Kawsar, F., Olson, E., Patel, K., Erwin, J., Bhattacharyya, S., Zozus, M. and Prior, F.
DeIDNER Model: A Neural Network Named Entity Recognition Model for Use in the De-identification of Clinical Notes.
DOI: 10.5220/0010884500003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 640-647
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

testing, and evaluating the models (Syed, Syed, et al.,
2021). To promote the development and evaluation of
clinical NER systems, workshops like Informatics for
Integrating Biology and the Bedside (I2B2) have
released open access labeled corpora (Stubbs &
Uzuner, 2015). In addition, several studies devoted a
significant amount of resources to developing corpora
for domain-specific clinical NER tasks (Syed, Al-
Shukri, et al., 2021). Models trained on such domain-
specific corpora demonstrate improved accuracy in
named entity recognition and model peformance
(Habibi, Weber, Neves, Wiegandt, & Leser, 2017).
However, because of privacy concerns, these data sets
are either not publicly released or contain synthetic
identifiers affecting the generalizability of the
models.
Recently, DL-based NER algorithms have been
proposed for solving a wide range of text mining and
NLP tasks (Luo et al., 2018; Y. Wu, Jiang, Xu, Zhi,
& Xu, 2018). They have a significant advantage over
traditional ML methods as DL techniques do not
require extensive feature engineering (Burns, Li, &
Peng, 2019). The core concept of such DL methods is
to compute distributed representations of words, i.e.,
word embeddings, in the form of vectors (Li, Sun,
Han, & Li, 2020). Early word embedding models
such as Word2Vec (Mikolov, Chen, Corrado, &
Dean, 2013), GloVe (Pennington, Socher, &
Manning, 2014), and fastText (Bojanowski, Grave,
Joulin, & Mikolov, 2017) have demonstrated good
performance in many NLP tasks. However, these
models are context-free generating a single word
embedding representation for each word not adjusted
to the surrounding context. More recent word
embedding models such as Bidirectional Encoder
Representations from Transformers (BERT) (Devlin,
2019), embeddings from language models (ELMo)
(Peters et al., 2018), and FLAIR (Akbik, Blythe, &
Vollgraf, 2018) addresses this deficiency by adjusting
the context of the word based on the surroundings
(Khattak et al., 2019). The major difference between
above contextual word embedding models is that
BERT uses vocabulary that contains words and
subwords extracted from general domain and
generally needs to be updated when pre-trained to
specific domain, whereas FLAIR and ELMo uses
sequence of characters to build word-level
embeddings and independent of such vocabulary
(Tai, Kung, Dong, Comiter, & Kuo, 2020).
In addition to word embeddings, combining
character-level word embeddings have proven to be
critical for NER tasks. In the context of combining
word embeddings along with character-level word
embeddings, Lample et al.(Lample, Ballesteros,
Subramanian, Kawakami, & Dyer, 2016) used long
short-term memory (LSTM) character-level word
embeddings whereas Ma and Hovy (Ma & Hovy,
2016) used convolutional neural networks (CNN).
Zhai et al. (Zhai, Nguyen, & Verspoor, 2018) further
confirmed the choice of LSTM or CNN character-
level word embeddings in Bi-LSTM CRF model did
not have clear positive effect on the pefromance,
however CNN shows advantage in reducing training
complexity and was recommended.
To date, the most successful DL model for NER
is a bidirectional long short-term memory (Bi-LSTM)
with conditional random field (CRF) architecture first
proposed by Huang et al. (Huang, Xu, & Yu, 2015)
Several recent studies have applied off-the-shelf
general domain, pre-trained clinical embeddings, and
mixed-domain embeddings methods as input to NER
models (Alsentzer et al., 2019; Catelli, Casola, De
Pietro, Fujita, & Esposito, 2021). However, very few
studies have explored the full potential of combining
these embeddings, especially in identifying the
entities related to de-identification from clinical
narratives. Jiang et al. (Jiang, Sanger, & Liu, 2019)
demonstrated the combination of the standard word
embeddings (Word2Vec), contextual embeddings
(FLAIR and ELMo), and semantic embeddings to
identify clinical entities such as treatments, problems,
and tests. However, above study utilized Word2Vec
standard word embeddings, but not GloVe, which is
a superior standard word embeddings method and
uses word co-occurrence matrix in generating quality
word representations (Min, Zeng, Chen, Chen, &
Jiang, 2017). In addition, above study did not utilize
additional character-level word embedding and
instead rely on FLAIR and ELMo character-based
architecture. Augmenting the embeddings with an
additional character-level word embedding using a
CNN architecture could play a complementary role in
fully exploiting the potential of different embeddings
and improve the overall performance.
This study describes our NER model, which
addresses the limitation of earlier studies in
combining different embeddings, and its potential to
generate accurate named entities that will aid in the
de-identification of medical texts and other
downstream clinical NLP tasks. Our main
contributions include the following:
Carefully preparing a human-annotated corpus
with hospital discharge summaries to train and
evaluate the model for generalizability.
We present an innovative DL NER model,
DeIDNER, specifically useful for generating
named entities that will be used in a larger model
to de-identify clinical texts (e.g., discharge
DeIDNER Model: A Neural Network Named Entity Recognition Model for Use in the De-identification of Clinical Notes
641

summaries). The model comprises of an input
embedding layer that combines: standard word
embeddings (GloVe), context based word
embeddings (FLAIR) , a character-level word
embedding using a CNN, and external knowledge
bases along with word features as one-hot vectors
followed by Bi-LSTM CRF layers.
A detailed systematic analysis is performed on the
effect of mixed-domain clinical pre-trained
contextual embeddings on the internal corpus.
2 MATERIALS AND METHODS
2.1 Data Sets
This study was conducted with University of
Arkansas for Medical Sciences (UAMS) institutional
review board approval (IRB #228649). This study
used three datasets, two of which are publicly
available (CoNLL-2003 data and 2014 I2B2
challenge datasets), and an internally generated
corpus. The internal corpus was developed from our
previous work, and consists of 500 discharge
summary notes. The named entity labels in the
internal corpus include: PERSON, LOCATION,
ORGANIZATION, DATE, AGE, ID, and PHONE
which are in BIO (Begin, Inside, Outside) notation
format.
2.2 Neural Network Model
Architecture
Figure 1 gives the main structure of our model that
consists of 1) input embeddings layer with all
different word embeddings, and 2) a recurrent neural
network (RNN) layer that takes concatenated
embeddings from the input layer, and finally 3) a CRF
layer that obtains a globally optimal chain of labels
for a given sequence considering the correlations
between adjacent tags.
2.2.1 Input Embeddings Layer
This layer is used to generate word embeddings in
order to fit the text stream to our neural network
model. The word embeddings capture semantic and
syntactic meanings of words based on their
surrounding words, and are widely used in many NLP
tasks. This layer consists of four different
embeddings: 1) Standard word embeddings using
GloVe, 2) Contextualized word embeddings using
FLAIR, 3) Character level word embeddings using
CNN, and 4) Semantic embeddings as one-hot
vectors.
Figure 1: Details of the model architecture with input
embeddings layer, Bi-LSTM layer, and followed by output
CRF layer.
1. Standard Word Embeddings: For standard
word embeddings, we have used Stanford’s pre-
trained GloVe (Pennington et al., 2014) with 100-
dimensional word embeddings to generate vector
representation of the input words. GloVe pre-
trained embeddings on an out-of-domain corpus
provide broad vocabulary coverage. In addition,
GloVe embeddings tend to perform better where
context is not important.
2. Context-based Word Embeddings: FLAIR
embeddings, a context-dependent word
representation, were used to generate vector
representations of a word based on its context in a
sentence. The experiment performed by Habibi et
al. (Habibi et al., 2017) showed that word
embeddings trained on biomedical corpora
improve the model’s performance. Thus, we have
experimented with two different FLAIR
embeddings in our model based on the data-set:
• A base version of FLAIR embeddings for two
public data-sets evaluation and
• A pre-trained version of FLAIR trained on
12,000 discharge summaries from the UAMS
Electronic Health Record (EHR) system
separate from 500 discharge summaries used
for evaluation. We followed the method Akbik
et al. (Akbik et al., 2018) introduced to train
the FLAIR base model with similar parameter
settings: learning rate 0.1, batch size 32,
dropout 0.5, patience number 6, and 250
epochs. This pre-trained version was used
only against the internal corpus evaluation.
HEALTHINF 2022 - 15th International Conference on Health Informatics
642

3. Character-level Word Embeddings: Apart from
the word-level embeddings, character-level word
embeddings contain rich structural information of
an entity and are widely used in many NLP tasks.
To provide character-level morphological
information and alleviate out of vocabulary
problems, we generated character-level word
embeddings using a CNN adopted from Zhang et
al. (Zhang, Zhao, & LeCun, 2015) with all the
default settings.
4. Semantic One-hot Vector Embeddings: Due to
the complexity of the clinical domain, integrating
external knowledge sources into deep learning
models has shown improved results in clinical
NLP tasks. Here, we employed a one-hot vector
representation for each type of external
knowledge sources and lexical word feature. The
external knowledge sources are adopted from the
Neamatullah et al. (Neamatullah et al., 2008)
study including common first names, last names,
geo locations, and selected list of medical terms
compiled from Unified Medical Language System
(UMLS) thesaurus.
Finally, all the different embeddings are combined
with a simple concatenation method before feeding it
to the deep neural network model.
2.2.2 Neural Network Model and CRF
Layer
To date, most state-of-the-art sequential text data
algorithms have used LSTM (Yu, Si, Hu, & Zhang,
2019). However, conventional LSTMs are only able
to make use of a previous context. To overcome this
limitation, we adopted the Bi-LSTM proposed by
Huang et al. (Huang et al., 2015), which can leverage
the context information in both forward and backward
directions.
i
t
= σ (W
xi
x
t
+ W
hi
h
t−1
+ W
ci
c
t−1
+ b
i
) (1
)
f
t
= σ(W
xf
x
t
+ W
hf
h
t−1
+ W
cf
c
t−1
+ b
f
) (2
)
c
t
= f
t
c
t−1
+ i
t
tanh(W
xc
x
t
+ W
hc
h
t−1
+ b
c
) (3
)
o
t
= σ(W
xo
x
t
+ W
ho
h
t−1
+ W
co
c
t
+ b
o
) (4
)
h
t
= o
t
tanh(c
t
) (5
)
where σ is the logistic sigmoid function, x
t
is the input
vector at time t and h
t
is the hidden state vector.
Weights W
i
, W
f
, W
c
, W
o
and bias constants b
i
, b
f
, b
c
,
b
o
are the parameters to be learned. x=(x
1
, x
2
……,
x
n
) is the input sequence that can be fed into a
classification layer for many classification tasks. As
shown in Figure 1, the output from the input
embeddings layer which is a concatenation of the
different types of embeddings described in the
previous sections is given as input to the Bi-LSTM
layer.
Finally, the last layer is a CRF layer that utilizes
hidden states from the word-level Bi-LSTM and uses
neighbor tag information in predicting the current tag.
Again, it has been shown that CRFs can produce
better tagging accuracy.
3 EXPERIMENTS AND
EVALUATION
This section outlines the experiments performed to
validate and evaluate the proposed model separately
against internal corpus and public data-sets. We used
the following configuration for all the experiments:
learning rate 0.1, batch size 32, dropout 0.5, patience
number 5, and the number of epochs was 150.
3.1 Experiments on Public Data-sets
In this experimental setup, we evaluate our proposed
model, DeIDNER by combining all four different
embeddings described earlier; namely (1) standard
word embeddings using GloVe, (2) base version of
FLAIR contextual embeddings, (3) character-level
word embeddings using a CNN, and (4) semantic
one-hot vector embeddings.
This experiment was to verify and demonstrate
the effectiveness of the developed model using
domain agnostic or general embeddings. Specifically,
against two publicly available data sets, CoNLL-2003
and I2B2 2014, to benchmark and compare the
performance with state-of-the-art models that used
above public datasets.
3.2 Experiments on Internal Corpus
In this experimental setup, we evaluate our proposed
model, DeIDNER, by first using base version of
FLAIR and then mixed-domain clinically pre-trained
FLAIR with other three different embeddings
described earlier.
In all the experiments, we used the training set
(70%) to learn model parameters, the validation set
(10%) to select optimal hyper-parameters, and the test
set (20%) to report the final results.
DeIDNER Model: A Neural Network Named Entity Recognition Model for Use in the De-identification of Clinical Notes
643

3.3 Evaluation
For evaluating the model, we used the most common
evaluation metrics for multi-class classification tasks:
precision, recall, and F1 score. In addition, the
performance of the model was assessed by a five-fold
cross-validation approach. In order to obtain
comparable and reproducible results, we followed the
CoNLL-2003 model evaluation methodology at the
entity level exact match and reporting the metrics
after repeating the procedure five times and averaging
the results.
Precision = P = C/M (6
)
Recall = R = C/N (7
)
F1 Score = 2PR / (P+R) (8
)
M = total number of predicted entities in the
sequence.
N = total number of ground truth entities in the
sequence.
C = total number of correct entities.
A Tesla (NVIDIA Corporation) graphics
processing unit was used to conduct the experiment.
The source code was written in Pytorch 1.8 (GPU-
enabled version) for Python 3.6.
4 RESULTS
4.1 Public Data-Set
Table 1&2 summarizes the performance of the
proposed model against the public data-sets. Our
proposed model reaches or overcomes state-of-the-art
performance on two publicly available data sets,
CoNLL-2003 and I2B2 2014.
The proposed model DeIDNER using general
domain embeddings achieved an F1 score of 93.25%
on CoNLL-2003 corpus and 94.89% on I2B2 2014
test data sets.
4.2 Internal Corpus
As shown in Table 3, the proposed model with
general domain embeddings achieved an F1 score of
94.31%. The experimental setup with mixed-domain
clinically pre-trained FLAIR embeddings yielded an
improved F1 score of 95.36%. This clearly
demonstrates the importance of mixed-domain
clinically pre-trained word embeddings. The 5-fold
cross validation of aforementioned best model against
the internal gold corpus reported F1 score range of
94.85% - 95.36% with mean confidence interval (CI
= 95%) was 95.19 ± 0.36.
Figures 2, show confusion matrix of our best
performing model that combines all four embeddings
on the internal corpus. The performance for DATE
entity was best, and the worst for LOCATION
followed by ORGANIZATION.
5 DISCUSSION
In this study, we investigated a DL based approach
for clinical NER specifically focused on entities
important for a clinical free de-identification task. We
systematically analyzed the contributions of different
embeddings in the input embeddings layer and the
effect of mixed-domain clinically pre-trained
Table 1: DeIDNER model performance on CoNLL-2003 data set along with best published method in literature as baseline.
Model F1 Score (%) Precision (%) Recall (%)
Akbik et al. (Akbik et al., 2018) 93.09 - -
DeIDNER 93.25 92.99 93.50
Table 2: DeIDNER model performance on I2B2 2014 data set along with best published method in literature as baseline.
Model F1 Score (%) Precision (%) Recall (%)
Catelli et al.
(
Catelli et al., 2021
)
94.80 95.52 94.09
DeIDNER 94.89 95.96 93.84
Table 3: DeIDNER model performance on internal corpus with and without mixed-domain pre-training.
Model F1 Score (%) Precision (%) Recall (%)
DeIDNER (without mixe
d
-domain pre-trainin
g
)
94.31 93.86 93.37
DeIDNER (with mixed-domain pre-training)
95.36 96.23 94.51
HEALTHINF 2022 - 15th International Conference on Health Informatics
644
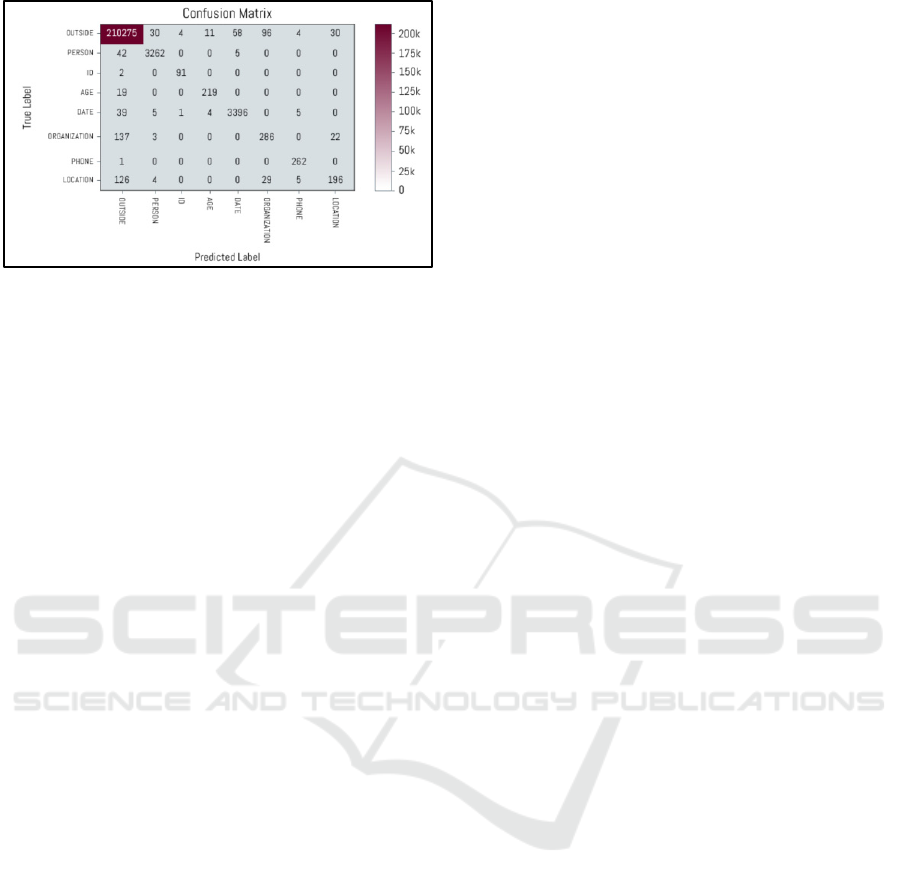
Figure 2: Confusion matrix of our best performing model
that combines all four embeddings on the internal corpus.
embeddings. We also demonstrated the
generalizability of the model on the clinical corpus
that we generated from discharge summary
documents. Our methodology and findings are
significant for future work in clinical NLP research.
The success of a NER model relies on how the text
is represented and integrated in the input layer with
different embeddings. Analysis by Akbik et al.
(Akbik et al., 2018) demonstrated that combining
standard word embeddings with contextual
embeddings improves the performance of the NER
models. In addition, Luo et al. (Luo et al., 2018)
combined word embeddings with character-level
embeddings and produced promising results. In our
study, we demonstrated an effective way to
incorporate different embeddings each representing
different characteristics of knowledge contained in
the corpora being analyzed. The results from our
experiment against the two publicly available data-
sets showed that our model is competitive compared
to other methods in literature that used above data.
Many previous studies have demonstrated
improved performance in the clinical NER models
when using domain-specific embeddings, but not in
entities that are non-clinical in nature like de-
identification (Alsentzer et al., 2019; Catelli et al.,
2021). However, in our experiments against the
internal corpus, we have achieved an F1 score of
94.31% using general embeddings and improved F1
score of 95.36% when mixed-domain clinically pre-
trained embeddings are used. This was an interesting
finding and we wanted to further validate the F1 score
improvement is statistically significant between the
proposed models with and without mixed-domain
clinically pre-trained embeddings, we performed a
5x2cv paired t-test (Dietterich, 1998). The results
showed that the improvement of F-measure between
DeIDNER model without pre-training and DeIDNER
with mixed-domain clinical pre-training were
statistically significant (P value < 0.05).
We conducted an analysis of errors in our
proposed system. We observed that most errors
occurred in entities that are long combined texts. For
instance, in a LOCATION entity “4301 W Markham
Street”, only part of it “W Markham Street” was
predicted as a LOCATION. Another problem was
ambiguities in the text. For instance, “St Louis” was
predicted as ORGANIZATION instead of
LOCATION. These errors are likely due to a lack of
training data.
This research study has some inherent limitations.
First, although we focused on different combinations
of embeddings, we have not exhausted all individual
embedding methods. For instance, standard word
embeddings can be accomplished using many
approaches, e.g., word2vec, FastText. We only used
GloVe embeddings, as we believe this approach is
more robust for generating standard embeddings.
Similar to standard embeddings, we also did not
exhaustively research the more recent context-based
embeddings models but chose FLAIR which is the
current state-of-the-art model and do not depend on
vocabulary like BERT based variants. Finally, we
used concatenation method to combine all the
embeddings, which was simple and straightforward
way of combining knowledge from different corpora.
However, in our future work, we plan to explore
ensemble method of combining the embeddings and
evaluate the performance of downstream de-
identification algorithms using the entities from our
work.
6 CONCLUSIONS
In this paper, we proposed a unique deep learning
method, combining multiple word embeddings that
include both standard word embeddings and
contextual word embeddings trained on clinical
discharge summaries to improve recognition of
named entities. The mixed-domain clinically pre-
trained model achieved a best F1 score of 95.36%,
which offers potential for further utilization in NLP
research. The knowledge generated here may
contribute to the downstream tasks, especially the de-
identification of clinical narratives and has generated
new insights relevant to the pre-processing stage of
biomedical NLP end models.
DeIDNER Model: A Neural Network Named Entity Recognition Model for Use in the De-identification of Clinical Notes
645

ACKNOWLEDGEMENTS
This study was supported in part by the Translational
Research Institute (TRI), grant UL1 TR003107
received from the National Center for Advancing
Translational Sciences of the National Institutes of
Health (NIH) and award AWD00053499, Supporting
High Performance Computing in Clinical
Informatics. The content of this manuscript is solely
the responsibility of the authors and does not
necessarily represent the official views of the NIH.
REFERENCES
Akbik, A., Blythe, D., & Vollgraf, R. (2018). Contextual
String Embeddings for Sequence Labeling.
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jin, D.,
Naumann, T., & McDermott, M. (2019). Publicly
Available Clinical BERT Embeddings.
Bojanowski, P., Grave, E., Joulin, A., & Mikolov, T.
(2017). Enriching word vectors with subword
information. Transactions of the Association for
Computational Linguistics, 5, 135-146.
Burns, G. A., Li, X., & Peng, N. (2019). Building deep
learning models for evidence classification from the
open access biomedical literature. Database : the
journal of biological databases and curation, 2019,
baz034. doi: 10.1093/database/baz034
Catelli, R., Casola, V., De Pietro, G., Fujita, H., & Esposito,
M. (2021). Combining contextualized word
representation and sub-document level analysis through
Bi-LSTM+CRF architecture for clinical de-
identification. Knowledge-Based Systems, 213, 106649.
doi: https://doi.org/10.1016/j.knosys.2020.106649
Catelli, R., Gargiulo, F., Casola, V., De Pietro, G., Fujita,
H., & Esposito, M. (2020). Crosslingual named entity
recognition for clinical de-identification applied to a
COVID-19 Italian data set. Applied soft computing, 97,
106779-106779. doi: 10.1016/j.asoc.2020.106779
Devlin, J. e. a. (2019). Bert: pre-training of deep
bidirectional transformers for language understanding.
In: Proceedings of the 2019 Conference of the North
American Chapter of the Association for
Computational Linguistics: Human Language
Technologies, Volume 1 (Long and Short Papers),
Minneapolis, MN, USA. pp. 4171–4186. Association
for Computational Linguistics. https://
www.aclweb.org/anthology/N19-1423.
Dietterich, T. G. (1998). Approximate statistical tests for
comparing supervised classification learning
algorithms. Neural Comput., 10(7), 1895–1923. doi:
10.1162/089976698300017197
Habibi, M., Weber, L., Neves, M., Wiegandt, D. L., &
Leser, U. (2017). Deep learning with word embeddings
improves biomedical named entity recognition.
Bioinformatics, 33(14), i37-i48. doi: 10.1093/
bioinformatics/btx228
Huang, Z., Xu, W., & Yu, K. (2015). Bidirectional LSTM-
CRF Models for Sequence Tagging.
Jiang, M., Sanger, T., & Liu, X. (2019). Combining
Contextualized Embeddings and Prior Knowledge for
Clinical Named Entity Recognition: Evaluation Study.
JMIR Med Inform, 7(4), e14850. doi: 10.2196/14850
Khattak, F. K., Jeblee, S., Pou-Prom, C., Abdalla, M.,
Meaney, C., & Rudzicz, F. (2019). A survey of word
embeddings for clinical text. Journal of Biomedical
Informatics: X, 4, 100057. doi: https://doi.org/10.1016/
j.yjbinx.2019.100057
Lample, G., Ballesteros, M., Subramanian, S., Kawakami,
K., & Dyer, C. (2016). Neural Architectures for Named
Entity Recognition.
Li, J., Sun, A., Han, R., & Li, C. (2020). A Survey on Deep
Learning for Named Entity Recognition. IEEE
Transactions on Knowledge and Data Engineering, PP,
1-1. doi: 10.1109/TKDE.2020.2981314
Liu, Z., Yang, M., Wang, X., Chen, Q., Tang, B., Wang, Z.,
& Xu, H. (2017). Entity recognition from clinical texts
via recurrent neural network. BMC Med Inform Decis
Mak, 17(Suppl 2), 67-67. doi: 10.1186/s12911-017-
0468-7
Luo, L., Yang, Z., Yang, P., Zhang, Y., Wang, L., Wang, J.,
& Lin, H. (2018). A neural network approach to
chemical and gene/protein entity recognition in patents.
Journal of Cheminformatics, 10(1), 65. doi: 10.1186/
s13321-018-0318-3
Ma, X., & Hovy, E. (2016). End-to-end Sequence Labeling
via Bi-directional LSTM-CNNs-CRF.
Mikolov, T., Chen, K., Corrado, G., & Dean, J. (2013).
Efficient Estimation of Word Representations in Vector
Space. http://arxiv.org/abs/1301.3781
Min, X., Zeng, W., Chen, N., Chen, T., & Jiang, R. (2017).
Chromatin accessibility prediction via convolutional
long short-term memory networks with k-mer
embedding. Bioinformatics, 33(14), i92-i101. doi:
10.1093/bioinformatics/btx234
Neamatullah, I., Douglass, M. M., Lehman, L.-w. H.,
Reisner, A., Villarroel, M., Long, W. J., . . . Clifford, G.
D. (2008). Automated de-identification of free-text
medical records. BMC Med Inform Decis Mak, 8(1), 32.
doi: 10.1186/1472-6947-8-32
Pennington, J., Socher, R., & Manning, C. (2014). Glove:
Global Vectors for Word Representation (Vol. 14).
Peters, M., Neumann, M., Iyyer, M., Gardner, M., Clark,
C., Lee, K., & Zettlemoyer, L. (2018). Deep
contextualized word representations.
Stubbs, A., & Uzuner, Ö. (2015). Annotating longitudinal
clinical narratives for de-identification: The 2014
i2b2/UTHealth corpus. J Biomed Inform, 58
Suppl(Suppl), S20-S29. doi: 10.1016/j.jbi.2015.07.020
Syed, M., Al-Shukri, S., Syed, S., Sexton, K., Greer, M. L.,
Zozus, M., Prior, F. (2021). DeIDNER Corpus:
Annotation of Clinical Discharge Summary Notes for
Named Entity Recognition Using BRAT Tool. Stud
Health Technol Inform, 281, 432-436. doi: 10.3233/
shti210195
Syed, M., Syed, S., Sexton, K., Syeda, H. B., Garza, M.,
Zozus, M., Prior, F. (2021). Application of Machine
HEALTHINF 2022 - 15th International Conference on Health Informatics
646

Learning in Intensive Care Unit (ICU) Settings Using
MIMIC Dataset: Systematic Review. Informatics
(MDPI), 8(1). doi: 10.3390/informatics8010016
Syeda, H. B., Syed, M., Sexton, K. W., Syed, S., Begum,
S., Syed, F., Yu, F., Jr. (2021). Role of Machine
Learning Techniques to Tackle the COVID-19 Crisis:
Systematic Review. JMIR Med Inform, 9(1), e23811.
doi: 10.2196/23811
Tai, W., Kung, H., Dong, X., Comiter, M., & Kuo, C.-F.
(2020). exBERT: Extending Pre-trained Models with
Domain-specific Vocabulary Under Constrained
Training Resources.
Wu, S., Roberts, K., Datta, S., Du, J., Ji, Z., Si, Y., . . . Xu,
H. (2020). Deep learning in clinical natural language
processing: a methodical review. J Am Med Inform
Assoc, 27(3), 457-470. doi: 10.1093/jamia/ocz200
Wu, Y., Jiang, M., Xu, J., Zhi, D., & Xu, H. (2018). Clinical
Named Entity Recognition Using Deep Learning
Models. AMIA Annu Symp Proc, 2017, 1812-1819.
Retrieved from https://pubmed.ncbi.nlm.nih.gov/
29854252
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5977567/
Yu, Y., Si, X., Hu, C., & Zhang, J. (2019). A Review of
Recurrent Neural Networks: LSTM Cells and Network
Architectures. Neural Comput, 31(7), 1235-1270. doi:
10.1162/neco_a_01199
Zhai, Z., Nguyen, D., & Verspoor, K. (2018). Comparing
CNN and LSTM character-level embeddings in
BiLSTM-CRF models for chemical and disease named
entity recognition.
Zhang, X., Zhao, J., & LeCun, Y. (2015). Character-level
convolutional networks for text classification. Paper
presented at the Proceedings of the 28th International
Conference on Neural Information Processing Systems
- Volume 1, Montreal, Canada.
DeIDNER Model: A Neural Network Named Entity Recognition Model for Use in the De-identification of Clinical Notes
647
