
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical
Language Models
Anastasios Lamproudis, Aron Henriksson and Hercules Dalianis
Department of Computer and System Sciences, Stockholm University, Stockholm, Sweden
Keywords:
Natural Language Processing, Language Models, Domain-adaptive Pretraining, Clinical Text, Swedish.
Abstract:
Research has shown that using generic language models – specifically, BERT models – in specialized domains
may be sub-optimal due to domain differences in language use and vocabulary. There are several techniques
for developing domain-specific language models that leverage the use of existing generic language models,
including continued and domain-adaptive pretraining with in-domain data. Here, we investigate a strategy
based on using a domain-specific vocabulary, while leveraging a generic language model for initialization.
The results demonstrate that domain-adaptive pretraining, in combination with a domain-specific vocabulary
– as opposed to a general-domain vocabulary – yields improvements on two downstream clinical NLP tasks
for Swedish. The results highlight the value of domain-adaptive pretraining when developing specialized
language models and indicate that it is beneficial to adapt the vocabulary of the language model to the target
domain prior to continued, domain-adaptive pretraining of a generic language model.
1 INTRODUCTION
The paradigm of pretraining and fine-tuning language
models has become a cornerstone of modern natural
language processing. By utilizing transfer learning
techniques, language models such as BERT (Devlin
et al., 2019) are pretrained using vast amounts of un-
labeled text data and subsequently adapted, or fine-
tuned, to carry out various downstream tasks using
labeled, task-specific data. While pretraining is gen-
erally computationally expensive – and therefore typi-
cally carried out by resource-rich organizations – fine-
tuning is computationally inexpensive. These fine-
tuned models have obtained state-of-the-art results in
many natural language processing tasks.
Language models are often pretrained using large,
readily available corpora in the general domain, e.g.
Wikipedia. However, the use of generic language
models in specialized domains may be sub-optimal
because of domain differences, in terms of, for in-
stance, language use and vocabulary (Lewis et al.,
2020, Gururangan et al., 2020). This has motivated
efforts to develop domain-specific language models,
e.g. SciBERT (Beltagy et al., 2019) and BioBERT
(Lee et al., 2020). Specialized language models have
been developed either by (i) pretraining a language
model with in-domain data from scratch, possibly in
combination with out-domain data, or by (ii) contin-
uing to pretrain an existing, generic language model
with in-domain data (domain-adaptive pretraining),
either by using large amounts of in-domain data, if
available, or by only using task-related unlabeled data
(task-adaptive pretraining).
However, the domain of the language model is
not only manifested in the pretraining or fine-tuning
sessions, but also in the model’s vocabulary. Every
language model requires a vocabulary for processing
the input text. For transformer-based language mod-
els, this vocabulary acts not only as a prepossessing
method but for connecting the input text to the model.
The vocabulary is used for creating the mapping of the
language to the learned – or to-be-learned – represen-
tations of the model at its lowest level: the embedding
table. This vocabulary can be built using different al-
gorithms, such as WordPiece (Wu et al., 2016), Sen-
tencePiece (Kudo and Richardson, 2018) and Byte-
Pair encoding (Sennrich et al., 2016). As such, the
vocabulary of a language model is determined by the
algorithm and the data which was used to generate it.
A drawback of continued, domain-adaptive pre-
training of an existing, generic language model is that
domain-specific words are often tokenized poorly.
For example, when tokenizing the common clinical
term r
¨
ontgenunders
¨
okning (English: x-ray investiga-
tion) with a general-domain language model (KB-
BERT), it is split into multiple subtokens: [‘ro’,
180
Lamproudis, A., Henriksson, A. and Dalianis, H.
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models.
DOI: 10.5220/0010893800003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 180-188
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

‘##nt’, ‘##gen’, ‘##under’, ‘##so’, ‘##kning’]. There
have therefore been efforts to adapt the vocabulary to
the target domain prior to domain-adaptive pretrain-
ing (Tai et al., 2020, Koto et al., 2021).
In this paper, a clinical language model for
Swedish is developed by adapting the vocabulary that
the model uses. More specifically, a clinical vocab-
ulary is constructed by applying the WordPiece al-
gorithm on large amounts of Swedish clinical text.
The clinical language model uses this vocabulary, but
also leverages an existing, generic language model
for Swedish for parameter initialization, both for the
shared vocabulary and for non-overlapping tokens.
An empirical investigation shows that this approach,
in addition to domain-adaptive pretraining, leads to
improved performance on two downstream clinical
natural language processing tasks.
2 RELATED WORK
There have been many efforts to develop domain-
specific, specialized language models by pretraining
with in-domain data, particularly for the biomedical
domain. A notable example is BioBERT (Lee et al.,
2020), which was initialized using a generic BERT
model while inheriting its vocabulary, after which
domain-adaptive pretraining was carried out. Another
example is BioMegatron (Shin et al., 2020) – a larger
model that obtained further improvements by train-
ing on even larger in-domain corpora and, in some
cases, using a domain-specific vocabulary. In another
study (Gu et al., 2021), it was shown that training
biomedical language models from scratch – as op-
posed to domain-adaptive pretraining of a generic lan-
guage model – may yield improved performance, al-
though requiring substantial computational resources.
Domain-specific language models have also been
developed for the clinical domain. Alsentzer et al.
(2019) pretrained BERT models using clinical notes
from MIMIC-III and showed that initializing the lan-
guage models with parameters from BioBERT, as op-
posed to BERT, yielded better downstream perfor-
mance. Lewis et al. (2020) developed clinical and
biomedical RoBERTa-based language models and
studied the impact of various training corpora and
model sizes, along with a domain-specific vocabulary.
Liu et al. (2019) conducted experiments which sug-
gest using a larger, more powerful generic language
model may be better than using a smaller, less power-
ful domain-specific language model. However, it was
also shown that using in-domain data may lead to im-
proved performance, in particular in the clinical do-
main. Learning a domain-specific vocabulary yielded
improvements on sequence labeling tasks, while the
impact was less clear for classification tasks. Further-
more, Gururangan et al. (2020) also investigated the
potential gains of domain-adaptive pretraining using
an existing BERT model and explored a number of
different settings: (i) domain-adaptive pretraining for
a limited amount of time, (ii) domain-adaptive pre-
training on the unlabeled training set of the intended
downstream task, and (iii) domain-adaptive pretrain-
ing on available unlabeled data directly related to the
future downstream task. A clinical language model
for Swedish was developed through domain-adaptive
pretraining of a generic language model while inherit-
ing its vocabulary, yielding improvements on several
downstream tasks (Lamproudis et al., 2021).
Some efforts have focused on adapting a generic
language model to a specialized domain by modify-
ing the vocabulary. In exBERT (Tai et al., 2020),
the vocabulary of a generic language model was aug-
mented with domain-specific terms, while adding ex-
tra parameters for the extended vocabulary. exBERT
was then pretrained using in-domain data and shown
to outperform its original counterpart on a number
of domain-specific tasks. In another relevant study
(Koto et al., 2021), albeit not in the biomedical do-
main, the general-domain vocabulary was replaced
with a domain-specific vocabulary, while using the
generic language model for initialization, both for
overlapping and non-overlapping tokens. The pro-
posed model yielded promising results without fur-
ther pretraining, while yielding improved results after
a further, domain-adaptive pretraining session.
3 DATA & METHODS
In this paper, we report on efforts to improve a clinical
language model for Swedish by adapting the model’s
vocabulary. A domain-specific, clinical vocabulary is
constructed by applying the WordPiece algorithm to
large amounts of clinical text. However, rather than
pretraining the model with a purely clinical vocab-
ulary from scratch, we leverage an existing, generic
language model for Swedish for parameter initializa-
tion. The approach is evaluated in two steps: (i)
through vocabulary adaptation and inheriting param-
eters from the generic language model alone (see sec-
tion 3.2), and (ii) followed by a session of domain-
adaptive pretraining (see section 3.3). The result-
ing language models are subsequently fine-tuned and
evaluated on two downstream clinical natural lan-
guage processing tasks: (i) detection of Protected
Health Information (PHI), i.e. a named entity recog-
nition task, and (ii) automatic assignment of ICD-
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models
181

10 codes to discharge summaries, i.e. a multi-class,
multi-label classification task. We save checkpoints
during the pretraining session in order to assess the
impact of different degrees of domain-adaptive pre-
training. The proposed models are compared to two
baselines: KB-BERT (Malmsten et al., 2020), which
is the generic language model for Swedish that is used
for initialization in the proposed approach, and Clin-
ical KB-BERT (Lamproudis et al., 2021), which car-
ries out domain-adaptive pretraining with the general-
domain vocabulary of KB-BERT.
The hypothesis is that pretraining a clinical lan-
guage model with a domain-specific vocabulary will
lead to improved performance on downstream tasks
in comparison to using a general-domain vocabulary.
We describe the evaluation setup in more detail in sec-
tion 3.4. Finally, differences between the vocabular-
ies are analyzed, in terms of term frequencies of the
shared vocabulary in the clinical domain, as well as
the impact of the two vocabularies on tokenization of
clinical text in terms of the resulting sample lengths.
3.1 Data
The clinical corpus used for domain-adaptive pre-
training contains 17.8 GB of clinical text
1
from the re-
search infrastructure Health Bank
2
– Swedish Health
Record Research Bank at DSV/Stockholm University
(Dalianis et al., 2015). The clinical text is written in
Swedish and encompasses a large number of clinical
units. The amount of text is comparable to the size
of the general-domain text used for pretraining KB-
BERT (Malmsten et al., 2020), a generic language
model for Swedish.
Two manually annotated data sets, corresponding
to two important downstream tasks, are used for fine-
tuning and evaluating the language models:
• Identifying Protected Health Information (PHI)
in clinical notes, which is a fundamental step in
de-identification and is typically approached as
a named entity recognition task. The Stockholm
EPR PHI Corpus comprises 21,653 sample sen-
tences, 380,000 tokens and contains 4,480 anno-
tated entities corresponding to 9 PHI classes: First
Name, Last Name, Age, Phone Number, Loca-
tion, Health Care Unit, Organization, Full Date,
and Date Part. Details about the dataset can be
found in (Velupillai et al., 2009).
• Assigning ICD-10 diagnosis codes to discharge
summaries, which is a document-level multi-
1
This research has been approved by the Swedish Ethi-
cal Review Authority under permission no. 2019-05679.
2
http://dsv.su.se/healthbank
class, multi-label classification task. The Stock-
holm EPR Gastro ICD-10 Corpus contains 6,062
discharge summaries (986,436 tokens) and their
assigned ICD-10 diagnosis codes that are gastro-
related and divided into 10 groups (classes) with a
more coarse granularity compared to the full ICD-
10 codes; the groups correspond to different body
parts and range from K00 to K99. There is, on av-
erage, 1.2 labels per sample. More details about
the dataset can be found in (Remmer et al., 2021).
3.2 Vocabulary Adaptation
The vocabulary of KB-BERT (Malmsten et al., 2020)
– an existing language model for Swedish, pretrained
using general-domain corpora encompassing newspa-
pers, government documents, e-books, social media
and Swedish Wikipedia – is adapted to the clinical do-
main by replacing its general-domain vocabulary with
a clinical vocabulary. The clinical vocabulary is con-
structed by applying the WordPiece algorithm to the
clinical corpus described in section 3.1. We set the
vocabulary size to be roughly equal to the vocabulary
size of KB-BERT, i.e. 50,325 words, and obtain a vo-
cabulary comprising 50,320 words plus the 5 special
tokens. A comparison of the general-domain vocab-
ulary of KB-BERT and the obtained clinical-domain
vocabulary shows that they have 16,519 words and
subwords in common (Figure 1). This means that
33,801 tokens are unique to the clinical domain and
do not exist in the vocabulary of KB-BERT.
Figure 1: The figure illustrates the two different vocabulary
sets. It shows the total amount of words present in each
vocabulary along with the number of words or tokens that
are shared between the two.
One option would be to pretrain a clinical lan-
guage model from scratch using the obtained clinical
vocabulary. However, in order to leverage the existing
language model for Swedish, KB-BERT, it is used for
HEALTHINF 2022 - 15th International Conference on Health Informatics
182

model initialization. More specifically, a new model
with the clinical-domain vocabulary is initialized in
which the learned parameters of KB-BERT are trans-
ferred. For shared tokens – i.e. tokens that exist in
both vocabularies – existing representations are in-
herited from KB-BERT. Clinical tokens that are not
present in KB-BERT are tokenized using the general-
domain tokenizer and initialized using the average of
the resulting subwords, as this has been shown to lead
to better performance compared to random initializa-
tion (Koto et al., 2021).
3.3 Domain-adaptive Pretraining
The language model obtained by using a clinical vo-
cabulary and leveraging KB-BERT for model initial-
ization is then further pretrained using clinical text,
i.e. it is subjected to domain-adaptive pretraining. To
that end, we use masked language modeling, omit-
ting the next sentence prediction task since it has
been shown that it is redundant in the development of
RoBERTA (Liu et al., 2019). Masked language mod-
eling can be described as randomly masking a per-
centage of the input tokens, using a special [MASK]
token. The model is then required to predict the
masked tokens based on the non-masked tokens, in
essence being forced to look for and build context in
its representations. The task is self-supervised, mean-
ing that it does not require manually annotated data.
When pretraining, the instructions and hyper-
parameters used in BERT (Devlin et al., 2019)
is followed with only two exceptions. Following
RoBERTA, instead of pretraining with a mix of se-
quence lengths, we only use sequences of maximum
length during our pretraining session. To create these
sequences, shorter sequences are concatenated using a
special [SEP] token to indicate the end and beginning
of each sequence. Furthermore, since the aim is to
adapt an already trained language model to a domain
of interest using limited resources, the pretraining is
carried out only for a limited amount of steps, corre-
sponding to one epoch of the data. The pretraining
hyperparameters are shown in Table 1.
To evaluate the domain-adaptive pretraining ses-
sion, the experiments do not rely on the loss computed
by a validation set; instead, the resulting models are
fine-tuned and evaluated on downstream tasks.
3.4 Fine-tuning & Evaluation
The models are fine-tuned following the general prac-
tice introduced by BERT and followed since. This
means using each language model as the core of each
task-specific model and only changing the last layer
Table 1: Hyperparameters for domain-adaptive pretraining.
hyperparameters values
learning rate 10
−4
batch size 256
Adam optimizer 4
β
1
0.9
β
2
0.999
L2 weight decay 0.01
warm up steps 10,000
linear learning rate decay 4
dropout probability 10%
update steps ≈ 40,000 equal to 1 epoch
training sequence length 512
Masked language modeling probability 15%
to match the requirements of each task. As input to
these task-specific layers, the [CLS] token represen-
tations are used for the classification task (ICD-10),
while, for the named entity recognition task (PHI), all
of the individual token representations are used.
An extensive hyperparameter search is not per-
formed as the goal is not to obtain state-of-the-art re-
sults on the downstream tasks, but rather to evaluate
and compare the underlying language models. How-
ever, in a setup where the goal is to achieve the best
possible performance, an extensive hyperparameter
search should be conducted. Instead, we perform a
narrow hyperparameter search to obtain the ones used
in the experiments in this work. These hyperparame-
ters are shown in Table 2.
Table 2: Fine-tuning hyperparameters.
hyperparameters PHI ICD-10 Uncertainty
learning rate 3 · 10
−5
2 · 10
−5
3 · 10
−5
batch size 64 32 64
For each experiment, 10% of the dataset is used as
a validation or development set and 10% as a held-out
test set; the remaining 80% is used for training. In or-
der to obtain more reliable performance estimates, ten
experiments for each model and downstream task are
performed, in which the held-out test set is randomly
selected; we then report the mean performance.
4 RESULTS
The results after fine-tuning and evaluating the lan-
guage models on the two downstream tasks are shown
in Table 3. As can be seen, the best performance is ob-
tained when using a clinical vocabulary in conjunc-
tion with domain-adaptive pretraining. On the PHI
task, this model obtains an F
1
-score of 0.942, com-
pared to 0.920 when fine-tuning a generic language
model without adapting the vocabulary or carrying
out domain-adaptive pretraining. Clinical KB-BERT
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models
183

Table 3: A comparison of pretrained models using different vocabularies and with or without domain-adaptive pretraining
with Swedish clinical text for two downstream tasks (F
1
-score).
Model
Vocabulary
adaptation
Domain-adaptive
pretraining
PHI ICD-10
General Vocabulary (KB-BERT) No No 0.920 0.799
General Vocabulary (Clinical KB-BERT) No Yes 0.934 0.831
Clinical Vocabulary (ours) Yes No 0.911 0.796
Clinical Vocabulary (ours) Yes Yes 0.942 0.835
obtains a F
1
-score of 0.934 on this task, showing that
domain-adaptive pretraining is useful even without
vocabulary modifications; however, the performance
is improved further when also adapting the vocabu-
lary to the target domain. There is a similar pattern
for the ICD-10 task, even if the improvement in per-
formance by adapting the vocabulary prior to domain-
adaptive pretraining is rather small (F
1
-score of 0.835
vs. 0.831). Compared to KB-BERT, however, the dif-
ference is substantial (F
1
-score of 0.835 vs. 0.799).
Furthermore, it is evident from the results that
modifying the vocabulary and inheriting model pa-
rameters from a generic language model alone – with-
out any domain-adaptive pretraining – is not suffi-
cient. In fact, doing so leads to worse downstream
performance compared to the generic language model
(PHI: 0.911 vs. 0.920; ICD-10: 0.796 vs 0.799 F
1
-
score).
In Figure 2, downstream performance of the lan-
guage models at various checkpoints of the domain-
adaptive pretraining session is shown. The purpose of
this evaluation is to track and detect any possible dif-
ferences between the two domain-adaptive pretrain-
ing sessions, i.e. when using a general vocabulary
(Clinical KB-BERT) vs. using a clinical vocabulary
with parameters inherited from a generic language
model (ours). For reference, the two models without
domain-adaptive pretraining are also included, using
a general vocabulary (KB-BERT) vs. a clinical vo-
cabulary (ours). As already observed from the final
results, the vocabulary-adapted (clinical vocabulary)
model starts off with a worse performance compared
to Clinical KB-BERT (general vocabulary). How-
ever, already after 10% of a single epoch of domain-
adaptive pretraining, the results have been overturned.
Overall, for 9 (ICD-10) and 7 (PHI) out of 10 check-
points of domain-adaptive pretraining, respectively,
the vocabulary-adapted language model outperforms
the general-vocabulary alternative.
In order to gain further insights into domain differ-
ences w.r.t. vocabulary and, in particular, the nature of
the shared vocabulary, we investigate the distribution
of the intersection of the general and clinical vocabu-
laries according to term frequency in the clinical do-
main (Figure 3). This distribution is obtained by sam-
pling 10% of the clinical corpus used for pretraining;
in this analysis, we only consider whole words, i.e.
not the subwords in the vocabularies, denoted by the
‘##token’. The whole words in the clinical vocabulary
(∼ 36.000) are ranked according to term frequency in
the clinical corpus; the figure shows how the whole
words in the shared vocabulary are distributed accord-
ing to their ranked term frequency in the clinical cor-
pus. Each bin corresponds to 100 words and is ranked
from top to bottom, meaning that on the left we have
the most frequent words and on the right the least fre-
quent. As each bin corresponds to 100 words, the
count value can be interpreted as the percentage of the
shared words that belong to the corresponding rank-
ing. The figure shows that that the shared vocabulary
is not over-represented among the most frequent clin-
ical terms; if anything, it appears that many of the in-
tersecting terms are among the less frequent clinical
terms. The figure also shows that, among the top 100
most frequent clinical terms, only around a third are
present in the general-domain vocabulary.
Finally, the impact of using a tokenizer based on a
general vs. a clinical vocabulary is investigated. Fig-
ure 4 shows this impact in terms of sequence length
before and after tokenization on one of the datasets
for the downstream tasks (ICD-10). The length of se-
quences before tokenization corresponds to the num-
ber of space-separated tokens, while after tokeniza-
tion, words may, to different degrees, be split into
subwords. The more words are split into subwords,
the longer the resulting sequences will be. The fig-
ure shows that, in both cases and as expected, the se-
quences become longer after tokenization. However,
when tokenizing clinical text with a general vocabu-
lary the sequences become longer – i.e. more words
are split into subwords – compared to when using a
clinical vocabulary.
Below are some examples of terms in the clinical
vocabulary and how they are split by the general vo-
cabulary tokenizer of KB-BERT:
• rituximab → [‘ritu’, ‘##xi’, ‘##ma’, ‘##b’]
(English: rituximab)
HEALTHINF 2022 - 15th International Conference on Health Informatics
184
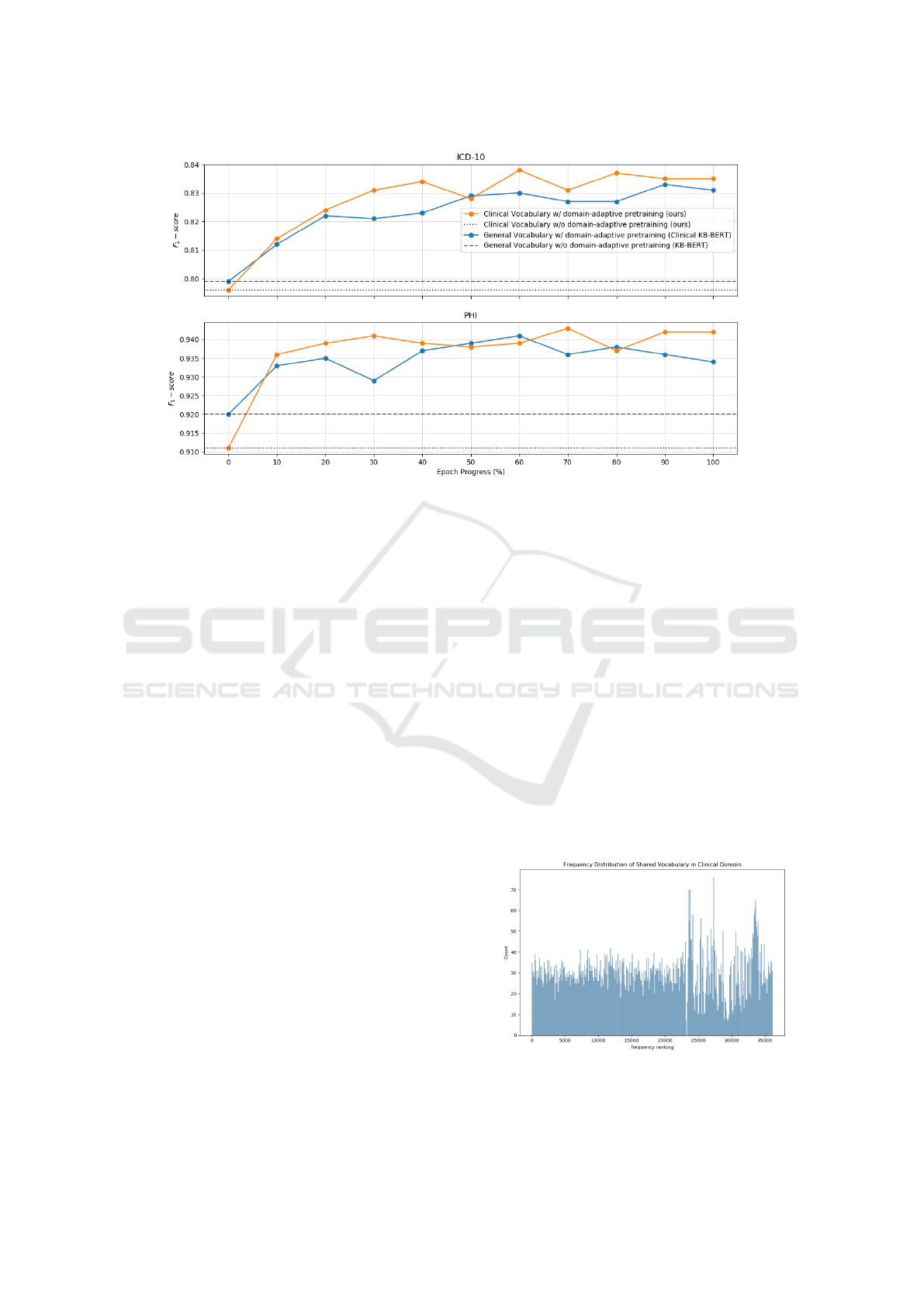
Figure 2: Downstream performance (F
1
-score) of various checkpoints during the pretraining process.
• somatiskt → [‘som’, ‘##atiskt’]
(English: somatic)
• mna → [‘mn’, ‘##a’]
(English: mna, mini nutritional assessment)
• parkinsson → [‘park’, ‘##ins’, ‘##son’]
(English: parkinson)
• lymfk
¨
ortelstationer → [‘lym’, ‘##f’, ‘##kort’,
‘##els’, ‘##tat’, ‘##ioner’]
(English: lymph node stations)
• lungf
¨
or
¨
andringarna → [‘lung’, ‘##for’, ‘##and’,
‘##ringarna’]
(English: the lung changes)
• r
¨
ontgenunders
¨
okning → [‘ro’, ‘##nt’, ‘##gen’,
‘##under’, ‘##so’, ‘##kning’]
(English: x-ray examination)
5 DISCUSSION
In this paper, we have investigated two factors of
potential importance when developing a clinical lan-
guage model based on exploiting the existence of a
generic language model: (i) vocabulary adaptation
and (ii) domain-adaptive pretraining, i.e. continued
pretraining with in-domain data. This situation is
important as it potentially allows for developing a
specialized language model with limited resources:
by adapting the vocabulary to the target domain in
conjunction with only one epoch of domain-adaptive
pretraining, the performance on downstream tasks is
clearly improved. This approach benefits from an
existing, albeit generic, language model, which has
been trained for many more epochs. However, the
results indicate that vocabulary adaptation alone, i.e.
without domain-adaptive pretraining, is not beneficial
and, in contrast, degrades performance. This might
indicate that the parameter transfer and approxima-
tion of the unknown representations disrupt the cal-
ibration of the model. However, with only a little
bit of domain-adaptive pretraining – as little as 10%
of an epoch – the performance of this model outper-
forms Clinical KB-BERT (Lamproudis et al., 2021),
i.e. the alternative model that does not involve any
vocabulary modifications. In future work, we plan to
compare these two approaches to pretraining a clin-
ical language model from scratch, which would cor-
respond to adapting the vocabulary and carrying out
domain-adaptive pretraining, but without resorting to
parameter transfer from a generic language model.
Furthermore, an alternative to replacing entirely the
general vocabulary with a clinical vocabulary would
Figure 3: The distribution of the intersection of the general
and clinical vocabularies according to term frequency in the
clinical domain. Each bin corresponds to 100 words and is
ranked from left (most frequent) to right (least frequent).
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models
185
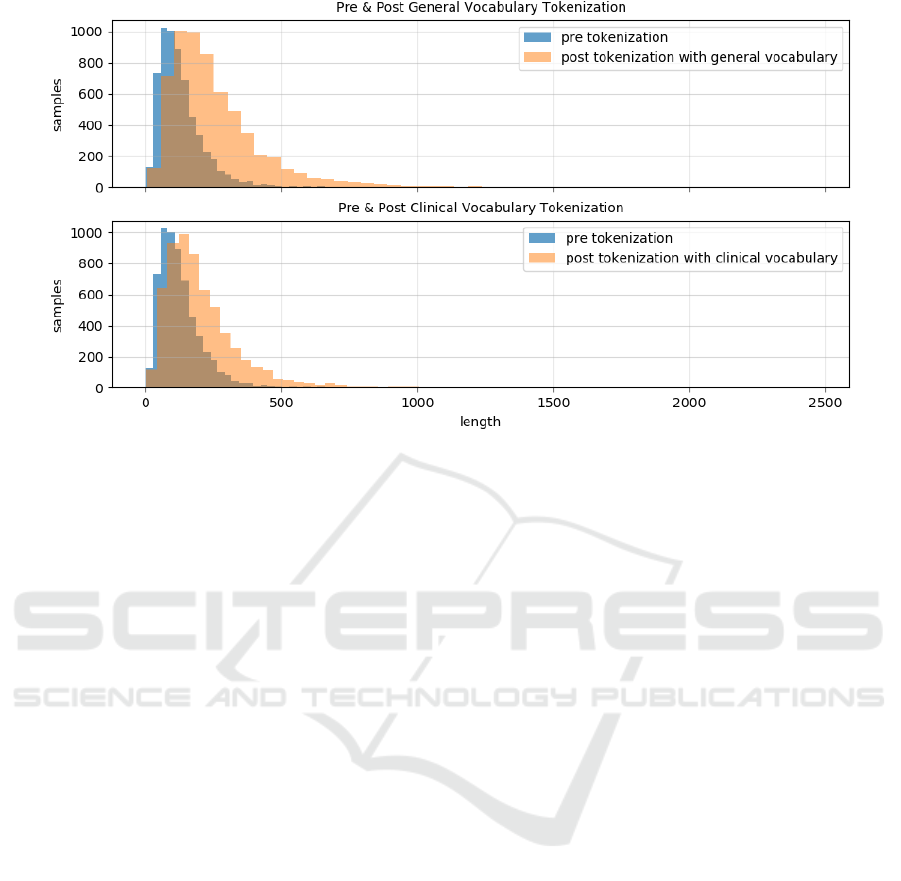
Figure 4: The distribution of sample lengths in terms of number of tokens from the Stockholm EPR Gastro ICD-10 Corpus.
be instead to extend the general vocabulary to in-
clude also clinical terms, e.g. by adding the non-
overlapping part of the clinical vocabulary, following
exBERT (Tai et al., 2020).
The fact that the size of the intersection between
the general-domain and clinical-domain vocabularies
is only around a third of each respective vocabulary
size indicates that there are substantial domain dif-
ferences between the general domain and the clini-
cal domain in terms of terminology. When looking
deeper at the shared vocabulary and how these terms
are distributed w.r.t to term frequency in the clinical
domain, we observe that they are not over-represented
among the most frequent clinical terms. However, it
should be noted that both vocabularies are limited to
approximately 50,000 tokens, which means that rela-
tively rare words and subwords in both domains have
already been excluded. When using a general-domain
tokenizer, such as the one in KB-BERT and Clini-
cal KB-BERT, these terms would be split into sub-
words by the tokenizer. The impact of this is shown
in Figure 4. It is, moreover, illustrated by the ex-
amples given in section 4, where important clinical
terms are split into subwords, often in such a way
that the meaning of the term cannot be derived from
the subwords. This applies also to abbreviations and
acronyms, which are prevalent in clinical text, and il-
lustrated by mna, which stands for mini nutritional
assessment and is split into two subwords: ‘mn’ and
‘##a’. For domain-specific tasks, it would be sub-
optimal for the tokenizer to split important domain-
specific terms into subwords, especially when done
in a manner that is not semantically decomposable.
Instead, by using a domain-specific vocabulary prior
to domain-adaptive pretraining, representations for
common clinical terms would be learned: it makes
sense that this would be beneficial when fine-tuning
language models to perform domain-specific tasks.
6 CONCLUSIONS
In this paper, we report on the development of a
clinical language model for Swedish that leverages
an existing generic language model and adapts it
to the clinical domain through vocabulary adapta-
tion and domain-adaptive pretraining. The results on
two downstream natural language processing tasks in
the clinical domain demonstrate that applying both
of these two strategies yields improved performance
compared to applying only one or neither. Most of
the improvement seems to stem from the domain-
adaptive pretraining, while vocabulary adaptation
without any domain-adaptive pretraining is counter-
productive. However, very little domain-adaptive pre-
training – as little as 10% of an epoch – is needed
for vocabulary adaptation to be effective and outper-
form the same amount of domain-adaptive pretrain-
ing with the general vocabulary of the existing lan-
guage model. Furthermore, we have provided some
insights into vocabulary-based domain differences in
an effort to motivate the results, which demonstrate
that the proposed approach outperforms the baselines.
In future work, we plan to compare the approach
proposed in this paper, as well as Clinical KB-
BERT, with pretraining a clinical language model
from scratch, i.e. without relying on KB-BERT for
HEALTHINF 2022 - 15th International Conference on Health Informatics
186

tokenization or model initialization. We also plan to
carry out longer domain-adaptive pretraining sessions
and study the impact of this on the respective ap-
proaches. Finally, another approach we plan to inves-
tigate is to extend and augment the general-domain
vocabulary with clinical terms instead of completely
replacing the general vocabulary with a clinical vo-
cabulary, as was done here.
ACKNOWLEDGEMENTS
This work was partially funded by Region Stockholm
through the project Improving Prediction Models for
Diagnosis and Prognosis of COVID-19 and Sepsis
with Natural Language Processing of Clinical Text.
REFERENCES
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jindi,
D., Naumann, T., and McDermott, M. (2019). Pub-
licly available clinical BERT embeddings. In Pro-
ceedings of the 2nd Clinical Natural Language Pro-
cessing Workshop, pages 72–78, Minneapolis, Min-
nesota, USA. Association for Computational Linguis-
tics.
Beltagy, I., Lo, K., and Cohan, A. (2019). SciBERT: A Pre-
trained Language Model for Scientific Text. In Pro-
ceedings of the 2019 Conference on Empirical Meth-
ods in Natural Language Processing and the 9th Inter-
national Joint Conference on Natural Language Pro-
cessing (EMNLP-IJCNLP), pages 3606–3611.
Dalianis, H., Henriksson, A., Kvist, M., Velupillai, S.,
and Weegar, R. (2015). HEALTH BANK- A Work-
bench for Data Science Applications in Healthcare.
CEUR Workshop Proceedings Industry Track Work-
shop, pages 1–18.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). BERT: Pre-training of Deep Bidirectional
Transformers for Language Understanding. In Pro-
ceedings of the 2019 Conference of the North Amer-
ican Chapter of the Association for Computational
Linguistics: Human Language Technologies, Volume
1 (Long and Short Papers), pages 4171–4186, Min-
neapolis, Minnesota. Association for Computational
Linguistics.
Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama,
N., Liu, X., Naumann, T., Gao, J., and Poon,
H. (2021). Domain-specific language model pre-
training for biomedical natural language process-
ing. ACM Transactions on Computing for Healthcare
(HEALTH), 3(1):1–23.
Gururangan, S., Marasovi
´
c, A., Swayamdipta, S., Lo, K.,
Beltagy, I., Downey, D., and Smith, N. A. (2020).
Don’t Stop Pretraining: Adapt Language Models to
Domains and Tasks. In Proceedings of the 58th An-
nual Meeting of the Association for Computational
Linguistics, pages 8342–8360, Online. Association
for Computational Linguistics.
Koto, F., Lau, J. H., and Baldwin, T. (2021). IndoBER-
Tweet: A Pretrained Language Model for Indone-
sian Twitter with Effective Domain-Specific Vocabu-
lary Initialization. In Proceedings of the 2021 Con-
ference on Empirical Methods in Natural Language
Processing, pages 10660–10668.
Kudo, T. and Richardson, J. (2018). SentencePiece: A sim-
ple and language independent subword tokenizer and
detokenizer for Neural Text Processing. In Proceed-
ings of the 2018 Conference on Empirical Methods
in Natural Language Processing: System Demonstra-
tions, pages 66–71.
Lamproudis, A., Henriksson, A., and Dalianis, H. (2021).
Developing a Clinical Language Model for Swedish:
Continued Pretraining of Generic BERT with In-
Domain Data. In Proceedings of RANLP 2021: Re-
cent Advances in Natural Language Processing, 1-3
Sept 2021, Varna, Bulgaria, pages 790–797.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). BioBERT: a pre-trained biomed-
ical language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Lewis, P., Ott, M., Du, J., and Stoyanov, V. (2020). Pre-
trained Language Models for Biomedical and Clinical
Tasks: Understanding and Extending the State-of-the-
Art. In Proceedings of the 3rd Clinical Natural Lan-
guage Processing Workshop, pages 146–157.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen,
D., Levy, O., Lewis, M., Zettlemoyer, L., and Stoy-
anov, V. (2019). RoBERTa: A Robustly Opti-
mized BERT Pretraining Approach. arXiv preprint
arXiv:1907.11692.
Malmsten, M., B
¨
orjeson, L., and Haffenden, C. (2020).
Playing with Words at the National Library of
Sweden–Making a Swedish BERT. arXiv preprint
arXiv:2007.01658.
Remmer, S., Lamproudis, A., and Dalianis, H. (2021).
Multi-label Diagnosis Classification of Swedish Dis-
charge Summaries – ICD-10 Code Assignment Using
KB-BERT. In Proceedings of RANLP 2021: Recent
Advances in Natural Language Processing, RANLP
2021, 1-3 Sept 2021, Varna, Bulgaria, pages 1158–
1166.
Sennrich, R., Haddow, B., and Birch, A. (2016). Neu-
ral Machine Translation of Rare Words with Subword
Units. In Proceedings of the 54th Annual Meeting
of the Association for Computational Linguistics (Vol-
ume 1: Long Papers), pages 1715–1725, Berlin, Ger-
many. Association for Computational Linguistics.
Shin, H.-C., Zhang, Y., Bakhturina, E., Puri, R., Patwary,
M., Shoeybi, M., and Mani, R. (2020). BioMegatron:
Larger Biomedical Domain Language Model. In Pro-
ceedings of the 2020 Conference on Empirical Meth-
ods in Natural Language Processing (EMNLP), pages
4700–4706.
Tai, W., Kung, H., Dong, X. L., Comiter, M., and Kuo,
C.-F. (2020). exBERT: Extending Pre-trained Models
with Domain-specific Vocabulary Under Constrained
Training Resources. In Proceedings of the 2020 Con-
Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models
187

ference on Empirical Methods in Natural Language
Processing: Findings, pages 1433–1439.
Velupillai, S., Dalianis, H., Hassel, M., and Nilsson, G. H.
(2009). Developing a standard for de-identifying elec-
tronic patient records written in Swedish: precision,
recall and F-measure in a manual and computerized
annotation trial. International Journal of Medical In-
formatics, 78(12):e19–e26.
Wu, Y., Schuster, M., Chen, Z., Le, Q. V., Norouzi,
M., Macherey, W., Krikun, M., Cao, Y., Gao, Q.,
Macherey, K., et al. (2016). Google’s neural ma-
chine translation system: Bridging the gap between
human and machine translation. arXiv preprint
arXiv:1609.08144.
HEALTHINF 2022 - 15th International Conference on Health Informatics
188
