
The h-ANN Model: Comprehensive Colonoscopy Concept
Compilation using Combined Contextual Embeddings
Shorabuddin Syed
1a
, Adam Jackson Angel
2
, Hafsa Bareen Syeda
3b
, Carole France Jennings
4
,
Joseph VanScoy
5
, Mahanazuddin Syed
1c
, Melody Greer
1
, Sudeepa Bhattacharyya
6
,
Meredith Zozus
7d
, Benjamin Tharian
8
and Fred Prior
1e
1
Department of Biomedical Informatics, University of Arkansas for Medical Sciences, U.S.A.
2
Department of Internal Medicine, Washington University, U.S.A.
3
Department of Neurology, University of Arkansas for Medical Sciences, U.S.A.
4
Department of Internal Medicine, Tulane University, U.S.A.
5
College of Medicine, University of Arkansas for Medical Sciences, U.S.A.
6
Department of Biological Sciences, Arkansas State University, U.S.A.
7
Department of Population Health Sciences, University of Texas Health Science Centre at San Antonio, U.S.A.
8
Division of Gastroenterology and Hepatology, University of Arkansas for Medical Sciences, U.S.A.
Keywords: Colonoscopy, Natural Language Processing, Deep Learning, Word Embeddings, Clinical Concept Extraction.
Abstract: Colonoscopy is a screening and diagnostic procedure for detection of colorectal carcinomas with specific
quality metrics that monitor and improve adenoma detection rates. These quality metrics are stored in
disparate documents i.e., colonoscopy, pathology, and radiology reports. The lack of integrated standardized
documentation is impeding colorectal cancer research. Clinical concept extraction using Natural Language
Processing (NLP) and Machine Learning (ML) techniques is an alternative to manual data abstraction.
Contextual word embedding models such as BERT (Bidirectional Encoder Representations from
Transformers) and FLAIR have enhanced performance of NLP tasks. Combining multiple clinically-trained
embeddings can improve word representations and boost the performance of the clinical NLP systems. The
objective of this study is to extract comprehensive clinical concepts from the consolidated colonoscopy
documents using concatenated clinical embeddings. We built high-quality annotated corpora for three report
types. BERT and FLAIR embeddings were trained on unlabeled colonoscopy related documents. We built a
hybrid Artificial Neural Network (h-ANN) to concatenate and fine-tune BERT and FLAIR embeddings. To
extract concepts of interest from three report types, 3 models were initialized from the h-ANN and fine-tuned
using the annotated corpora. The models achieved best F1-scores of 91.76%, 92.25%, and 88.55% for
colonoscopy, pathology, and radiology reports respectively.
1 INTRODUCTION
Colonoscopy plays a critical role in screening of
colorectal carcinomas (CC) (Kim et al., 2020).
Although it is a most frequently performed procedure,
the lack of standardized reporting is impeding clinical
and translational research. Vital details related to the
procedure are stored in disparate documents,
colonoscopy, pathology, and radiology reports
a
https://orcid.org/0000-0002-4761-5972
b
https://orcid.org/0000-0001-9752-4983
c
https://orcid.org/0000-0002-8978-1565
d
https://orcid.org/0000-0002-9332-1684
e
https://orcid.org/0000-0002-6314-5683
respectively. The established quality metrics such as
adenoma detection rates, bowel preparation, and
cecal intubation rate are documented in endoscopy
and pathology reports (Anderson & Butterly, 2015;
Rex et al., 2015). Procedure indicators, medical
history require review of clinical history and
radiology reports. A comprehensive study of quality
metrics often involves labour-intensive chart review,
thereby limiting the ability to report, monitor, and
Syed, S., Angel, A., Syeda, H., Jennings, C., VanScoy, J., Syed, M., Greer, M., Bhattacharyya, S., Zozus, M., Tharian, B. and Prior, F.
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings.
DOI: 10.5220/0010903300003123
In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022) - Volume 5: HEALTHINF, pages 189-200
ISBN: 978-989-758-552-4; ISSN: 2184-4305
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
189

ultimately improve procedure quality (Syed et al.,
2021).
Natural language processing (NLP) has been used
as an alternative to manual data abstraction (Syeda et
al., 2021). Most studies to date, built NLP based
solutions to extract limited clinical concepts from
unconsolidated colonoscopy documents, with limited
data extraction, which is inadequate to provide a
complete clinical picture (Harkema et al., 2011; J. K.
Lee et al., 2019; Nayor, Borges, Goryachev, Gainer,
& Saltzman, 2018; Patterson, Forbush, Saini, Moser,
& DuVall, 2015; Raju et al., 2015). Manual chart
review is often still required to collect other
procedure metrics embedded as free text in disparate
colonoscopy related documents. Early studies
adopted rule-based NLP algorithms to extract
procedure metrics (Mehrotra et al., 2012; Raju et al.,
2015), but these algorithms have limited applicability
to diverse health care settings. A recent study by Lee
et al. (J. K. Lee et al., 2019) addressed the
generalization problem by employing traditional ML
technique to extract procedure findings from varying
colonoscopy report formats. To improve model
performance a dictionary of terms and phrases that
identify procedure metrics was created in addition to
annotations. The application of their proposed
solution is subject to the availability of a large
annotated clinical corpus and semantic and lexical
features manually crafted by domain experts.
With the emergence of deep learning (DL)
techniques, research on clinical concept extraction
has shifted from traditional ML to DL as DL
techniques eliminate the need for feature
representation (i.e. word embeddings) by domain
experts (H. Liang, Sun, Sun, & Gao, 2018; Yang,
Bian, Hogan, & Wu, 2020). These algorithms are
trained and evaluated in the general English domain
and later applied to cross-domain settings (X. Jiang,
Pan, Jiang, & Long, 2018; Malte & Ratadiya, 2019;
Schmidt, Marques, Botti, & Marques, 2019). Such
off-the-shelf models perform poorly when
identifying clinical concepts due to the presence of
domain specific abbreviations and terminologies
(Griffis, Shivade, Fosler-Lussier, & Lai, 2016; K.
Huang, Altosaar, & Ranganath, 2019; J. Lee et al.,
2019). Training the ML models on large annotated
clinical corpora can improve performance, but
availability of such corpora is rare due to legal and
institutional concerns arising from the sensitivity of
clinical data (Abdalla, Abdalla, Rudzicz, & Hirst,
2020; Caufield et al., 2018)
.
Contextual language representation models such
as Embeddings from Language Models (ELMO)
(Peters et al., 2018), Bidirectional Encoder
Representations from Transformers (BERT) (Devlin,
Chang, Lee, & Toutanova, 2018), and Flair (Akbik,
Blythe, & Vollgraf, 2018), can mitigate the
bottleneck of requiring a large annotated clinical
corpus (K. Huang et al., 2019; M. Jiang, Sanger, &
Liu, 2019; Si, Wang, Xu, & Roberts, 2019). These
LMs adopt semi-supervised learning, where the
models are trained to learn domain linguistics (i.e.,
clinical context-sensitive embeddings or clinical
embeddings) using a large volume of Unlabelled
clinical texts, commonly referred as “pre-training”
(M. Jiang et al., 2019; Sharma & Daniel, 2019). The
LMs need to be pre-trained on clinical texts only
once, then they can be adapted to various NLP tasks
using small, labelled corpora (referred as fine-
tuning). Thus, the time-consuming task of expert-
annotation to create large training datasets is
significantly decreased.
Using clinical embeddings, several studies
reported performance improvement across all NLP
tasks (Alsentzer et al., 2019; K. Huang et al., 2019; Si
et al., 2019; Yang et al., 2020). However, very few
studies have explored the full potential of combining
the clinical embeddings from multiple language
representation models. Jiang et al. (M. Jiang et al.,
2019) investigated the effects of combining
contextualized word embeddings (ELMO + FLAIR)
with classic word embeddings, Word2Vec (Mikolov,
Chen, Corrado, & Dean, 2013). Similarly, Boukkouri
et al. (El Boukkouri, Ferret, Lavergne, &
Zweigenbaum, 2019) studied the combination of
ELMO and Word2Vec. Both studies either pre-
trained or fine-tuned embeddings on clinical
narratives, and the trained-concatenated embeddings
were used to enhance downstream Name Entity
Recognition (NER) accuracies. However, compared
to ELMO, BERT has been found to have superior
performance on various NLP tasks due to its deep
bidirectional architecture (Alsentzer et al., 2019; Si et
al., 2019). The self-attention mechanism of BERT
efficiently models long-term dependencies, but
clinical feature representation is curtailed by its fixed
vocabulary (Bressem et al., 2021; J. Lee et al., 2019).
In contrast, FLAIR generates strong character-level
features and is independent of tokenization and
vocabulary (Akbik et al., 2018). Combining BERT
and FLAIR embeddings can improve word
representations and further boost the performance of
the clinical NLP systems. The objective of this study
is to extract comprehensive clinical concepts from the
consolidated colonoscopy documents using
concatenated clinical embeddings. In our previous
work, we built an automated algorithm that links
colonoscopy related documents (Syed et al., 2021).
HEALTHINF 2022 - 15th International Conference on Health Informatics
190

Leveraging this work done, main contributions of this
study are as follows, 1) Built high-quality annotated
corpora for the three document types (~ 425 reports
each). 2) We present a hybrid Artificial Neural
Network (h-ANN) architecture with concatenated GI
domain-trained BERT and FLAIR embeddings as the
input layer followed by BiLSTM and CRF layers. 3)
Using fine-tuned h-ANN models, we extracted
comprehensive clinical concepts from the three
colonoscopy document types with relatively small
annotated corpora. We evaluated the model’s
performance against manual chart review. 4) We
conducted a systematic evaluation of the effects of
combining clinical embeddings from multiple word-
based LMs on the downstream NLP tasks. 5) We
compared model performance across the three
document types.
2 METHODS
2.1 Dataset - Annotation
For this study, we used colonoscopy related
documents of patients undergoing the procedure at
the University of Arkansas for Medical Sciences
(UAMS) from May 2014 to September 2020. The
original dataset included 16,900 colonoscopy, 11,182
pathology, and 7,364 radiology reports respectively.
From the dataset, a random sample of 1,281 reports
were selected for annotation. The unlabeled corpus
contains 34,165 notes from the three document types,
and was used to pre-train LMs. We will refer to the
unlabeled corpus as “Un-GIC”.
To identify clinical entities that are essential to
improve procedure quality and to facilitate
colonoscopy research, we did an extensive literature
review and interviewed a panel of domain experts.
We identified 74 unique entities from colonoscopy
report, this includes scope times, quality of bowel
preparation, size and location of polyps, and findings
etc. From pathology reports, we identified 61 entities
including specimen type, type of polyp, location, and
pathological classifications (benign and malignant)
etc. Similarly, from radiology reports 47 entities were
identified, this includes diverticulosis, inflammation,
mass, haemorrhage, and stricture etc.
Several studies have been done to understand
factors effecting the annotation time and the quality
of clinical corpora (Fan et al., 2019; Roberts et al.,
2007; Wei, Franklin, Cohen, & Xu, 2018). Roberts et
al. (Roberts et al., 2007) and Wei et al. (Wei et al.,
2018) identified number of entities to annotate and
long term dependencies between the entities as the
key factors hindering clinical text annotations. Use of
standard terminologies to annotate clinical narratives
reduces entity identification ambiguities and
improves syntactical relation accuracies, allowing for
high inter-annotator agreement (Fan et al., 2019).
Taxonomies facilitate injecting domain knowledge
into ML models and improve clinical concept
extraction accuracy (M. Jiang et al., 2019; Wu et al.,
2018). However, colonoscopy documents are
annotated to identify specific procedure metrics and
employing generic terminologies will not be
beneficial. To address this problem, for each
document type, we built taxonomies by classifying
the identified entities into various classes, as shown
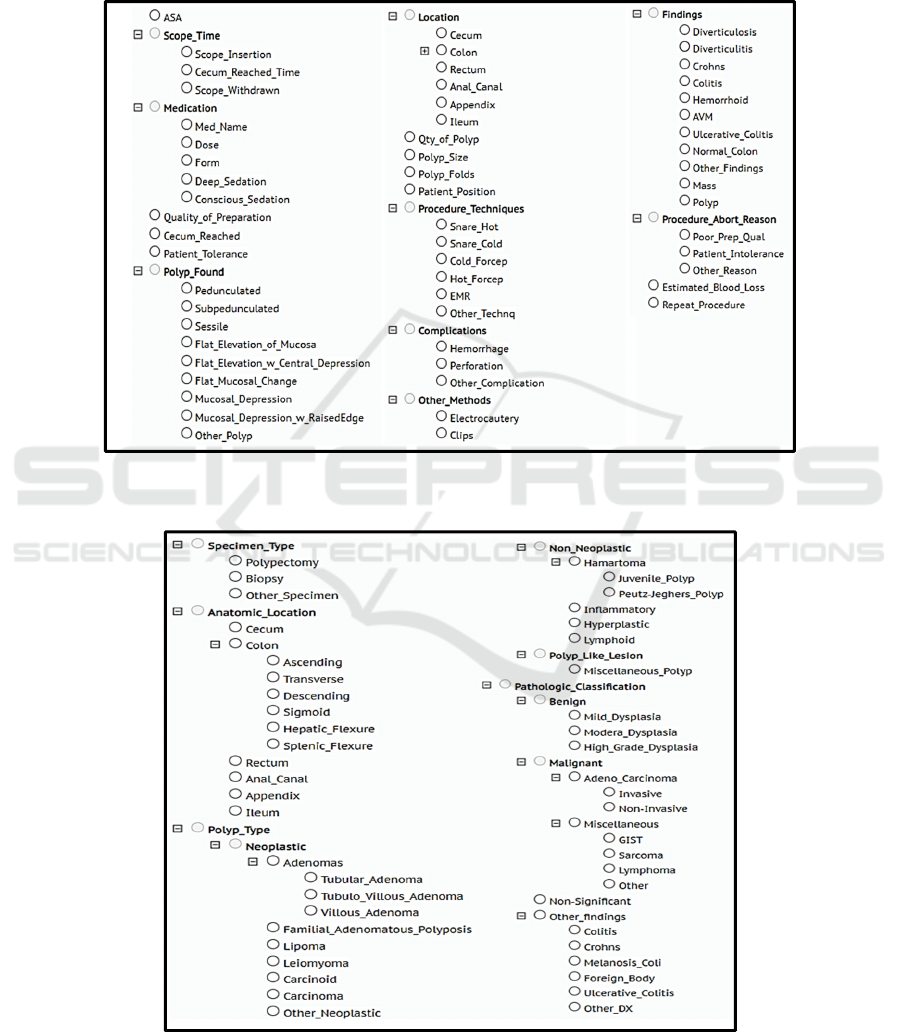
in Figure 1, 2, and 3 respectively. Using the domain
specific taxonomies and adopting standard annotation
guidelines we built three high-quality annotated
corpora, 1) Colonoscopy Corpus (CC): containing
442 labelled colonoscopy reports, 2) Pathology
Corpus (CP): containing 426 labelled pathology
reports, and 3) Radiology Corpus (CR): containing
413 labelled radiology reports that are associated with
the colonoscopy procedure. The CC, CP, and CR
contain a total of 10,672, 4,136, and 3,071
annotations respectively. As shown in Table 1, for
downstream clinical concept extraction tasks, the
annotated corpora were split into train, test, and
validation sets (70%-20%-10% respectively) for each
of the three document types.
2.2 Concept Extraction Architecture
To extract clinical concepts, we followed the
following procedure: 1) Clinical Embedding
Generation: pre-train LMs BERT and FLAIR on Un-
GIC; 2) Hybrid Artificial Neural Network (h-ANN)
creation: build a h-ANN network to concatenate and
fine-tune clinical embeddings; 3) Concept Extraction:
to extract concepts from the 3 report types, initialize
three models with the same h-ANN architecture and
fine-tune each model with CC, CP, and CR
respectively. The overall process of training LMs,
concatenating embeddings, and initializing and fine-
tuning downstream h-ANN models is shown in
Figure 4.
2.3 Clinical Embedding Generation
To generate clinical embeddings, we pre-trained
BERT and FLAIR models on Un-GIC.
Devlin et al. (Devlin et al., 2018) introduced the
language representational model BERT, based on a
Transformer architecture. BERT learns contextual
representations using two unsupervised tasks, masked
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings
191
language model (MLM) and next sequence prediction
(NSP). During pre-training, the MLM randomly
masks some of the tokens from the input sequence
and then predicts the original masked word based on
its surrounding context. To learn relationships
between sentences, NSP predicts whether the second
sentence is likely to follow the first. Devlin et al.
(Devlin et al., 2018) pre-trained BERT on English
text, BooksCorpus (800M words) and Wikipedia
(2,500M words) and open sourced the model
(BERT
Base
). To generate GI domain specific
Figure 1: Colonoscopy taxonomy depicting clinical entities and their classifications. Colonoscopy reports were annotated for
entities mentioned in the taxonomy.
Figure 2: Pathology taxonomy depicting clinical entities and their classifications. Pathology reports were annotated for entities
mentioned in the taxonomy.
HEALTHINF 2022 - 15th International Conference on Health Informatics
192

embeddings, we initialized the general-purpose
language representation model BERTBase and pre-
trained the model on Un-GIC. As clinical narratives
are not always well formatted, we performed sentence
segmentation on the entire corpus and delimited
documents by empty lines. About 12% of the
sentences in the corpus were longer than 128 tokens.
To limit the input sentence length to 128 tokens, we
split longer sentences.
As clinical documents contain
numerous domain-specific words which were not
present in the vocabulary files of the BERT
Base
, we
replaced 80 unused tokens from the vocabulary with
GI specific concepts. These concepts included
various disease names, pathological classifications,
and procedure names found in the colonoscopy
corpus. The vocabulary size remained the same
(28,996 tokens) to match the original configuration
file of BERT
Base.
For the hyperparameters, we used
the recommended settings and pre-training was
carried out using the TensorFlow library (Abadi et al.,
2016).
Figure 3: Radiology imaging taxonomy depicting clinical
entities and their classifications. Radiology reports were
annotated for entities mentioned in the taxonomy.
Akbik et al. (Akbik et al., 2018) proposed novel
contextual string embeddings also known as FLAIR
embeddings. During pre-training, each sentence is
passed as a sequence of characters to a bidirectional
character-level neural network language model. The
internal states of the forward and backward character
LMs are concatenated to generate contextualized
word-level embeddings. To train FLAIR embeddings
on the GI domain, we leveraged ‘pubmed-X’
embeddings (Sharma & Daniel, 2019). The pubmed-
X embeddings were generated by instantiating a
character-level LM (trained on the general English
domain), and further training the model using 5% of
PubMed abstracts (~ 1.2 million abstracts published
on or before 2015). To learn GI linguistics, we
initialized the pubmed-X LM and bi-directionally
trained the model using Un-GIC. Documents from the
Un-GIC were randomly split into train (80%),
validation (10%) and test (10%) sets. For the
hyperparameters, we used the recommended settings
and training was carried out using the Pytorch
framework (Paszke et al., 2019).
2.4 Hybrid Artificial Neural Network
(h-ANN) Architecture
To concatenate and fine-tune embeddings, we
designed our h-ANN model based on a BiLSTM-CRF
(Bidirectional long-short-memory-conditional
random fields) sequence labelling architecture
proposed by Huang et al. (Z. Huang, Xu, & Yu,
2015). The BiLSTM-CRF model has been found to
have superior performance on part of speech tagging
(POS), chunking and NER tasks (M. Jiang et al.,
2019). Figure 5 shows the architecture of the h-ANN;
the input embedding layer combines GI domain
trained BERT and contextual string embeddings. The
concatenated embeddings are used as input features
to the BiLSTM layers. Based on the forward and
backward output states, the CRF layers compute the
final sequence probability. The h-ANN was
implemented using the FLAIR framework described
in Akbik et al. (Akbik et al., 2019).
2.5 Concept Extraction
To extract clinical entities from colonoscopy reports,
we trained the h-ANN model on CC and named the
model as h-ANNcol. Similarly, we fine-tuned the
other two h-ANN models on CP and CR respectively
and named them as h-ANNpath and h-ANNrad. For
fine-tuning the models, we used the recommended
FLAIR framework hyperparameter settings, learning
rate as 0.1, and mini batch size as 32. The maximum
epoch was set to 250. We integrated the 3 fine-tuned
models (h-ANNcol, h-ANNpath, and h- ANNrad)
into one toolkit and named it GIN (Gastroenterology
NLP toolkit).
2.6 Evaluation
The three models (h-ANNcol, h-ANNpath, and
h-ANNrad) were trained on 80% of the annotated
corpus and evaluated on the remaining 20%. To avoid
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings
193
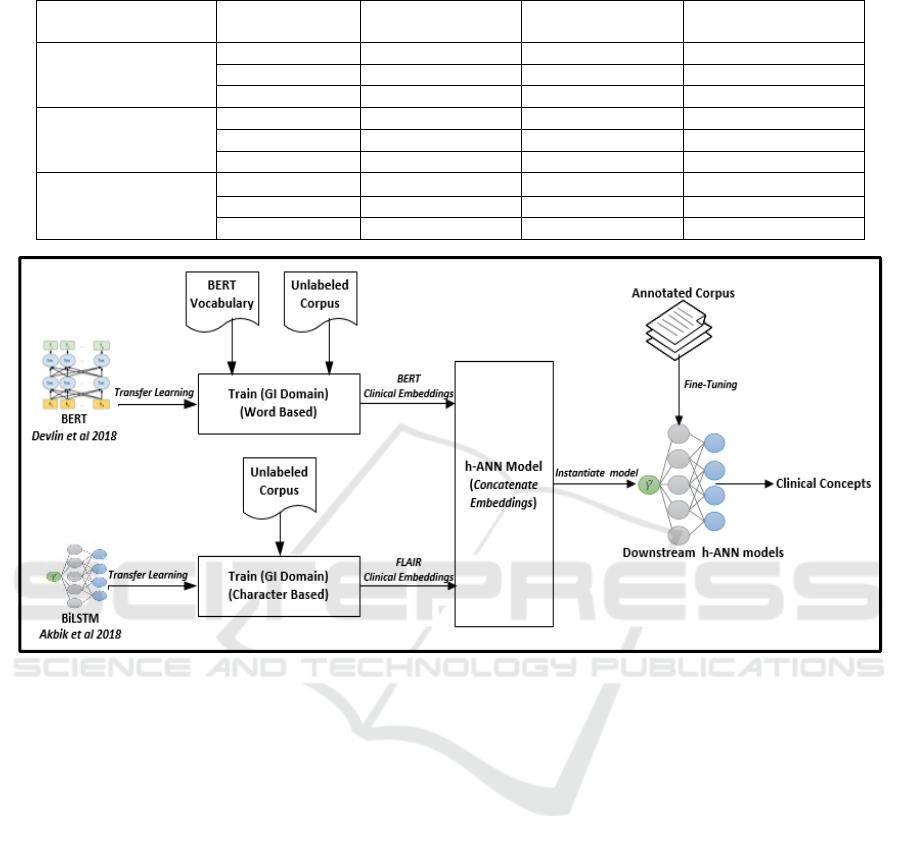
Table 1: Distribution of documents, sentences, and clinical entities in train, test, and validation sets across the three corpora.
Document Type Subset Avg. Number of
Notes
Avg. Number of
Sentences
Avg. Number of
Clinical Entities
Colonoscopy Train 309 13,862 7,467
Test 89 3,967 2,145
Validation 44 1,998 1,060
Pathology Train 299 2,896 2,900
Test 85 851 825
Validation 42 412 411
Radiology Train 289 3,387 2,192
Test 83 952 606
Validation 41 461 273
Figure 4: Workflow depicting training of language models, concatenating embeddings, instantiating and fine-tuning h-ANN
models to extract clinical concepts from colonoscopy related documents. GI: Gastroenterology, h-ANN: Hybrid artificial
neural network.
sample bias, we used a 5-fold cross-validation
technique and each document was used only once in
the test set. Performance of these models was
measured by the following metrics: precision, recall
and F1 scores.
3 RESULTS
For Pre-training BERT and FLAIR embeddings on
the Un-GIC (34,165 unlabeled notes) took
approximately 8 and 14 days respectively using an
NVidia Tesla V100 GPU (32GB) (NVIDIA, Santa
Clara, CA). The 14 days training for FLAIR is the
sum of the time taken to forward and backward train
the RNN based LM on the Un-GIC. It took
approximately 4, 5, and 7 hours to fine-tune the
models h-ANNpath, h-ANNcol, and h-ANNrad
respectively. For the three models, Figure 6 shows the
test F1 scores computed after completion of each
training epoch. The range of accuracies achieved by
the 3 models during 5-fold cross validation on the test
set is shown in Table 2. The h-ANNpath achieved the
best overall F1-score of 92.25%, followed by h-
ANNcol (91.76%), and h-ANNrad (88.55%). For the
best performing h-ANNcol model on the colonoscopy
narratives, Table 3 lists precision, recall, and F1 score
of each entity. Similarly, the results from the best
performing models on pathology and radiology notes
are shown in Table 4 and Table 5 respectively. The h-
ANNpath achieved F1 scores of 0.950 and 0.937 for
identifying neoplastic and malignant polyps which
are the confirmatory findings for colorectal cancer in
pathology reports. The best performing h-ANNcol
model achieved over 95% accuracy to identify scope
times and 92.63% accuracy in extracting polyp
findings from the colonoscopy reports. Similarly, h-
ANNrad achieved F1 score of over 95% for
identifying entities like hemorrhage, abscess,
steatosis, and stones from the radiology reports.
HEALTHINF 2022 - 15th International Conference on Health Informatics
194
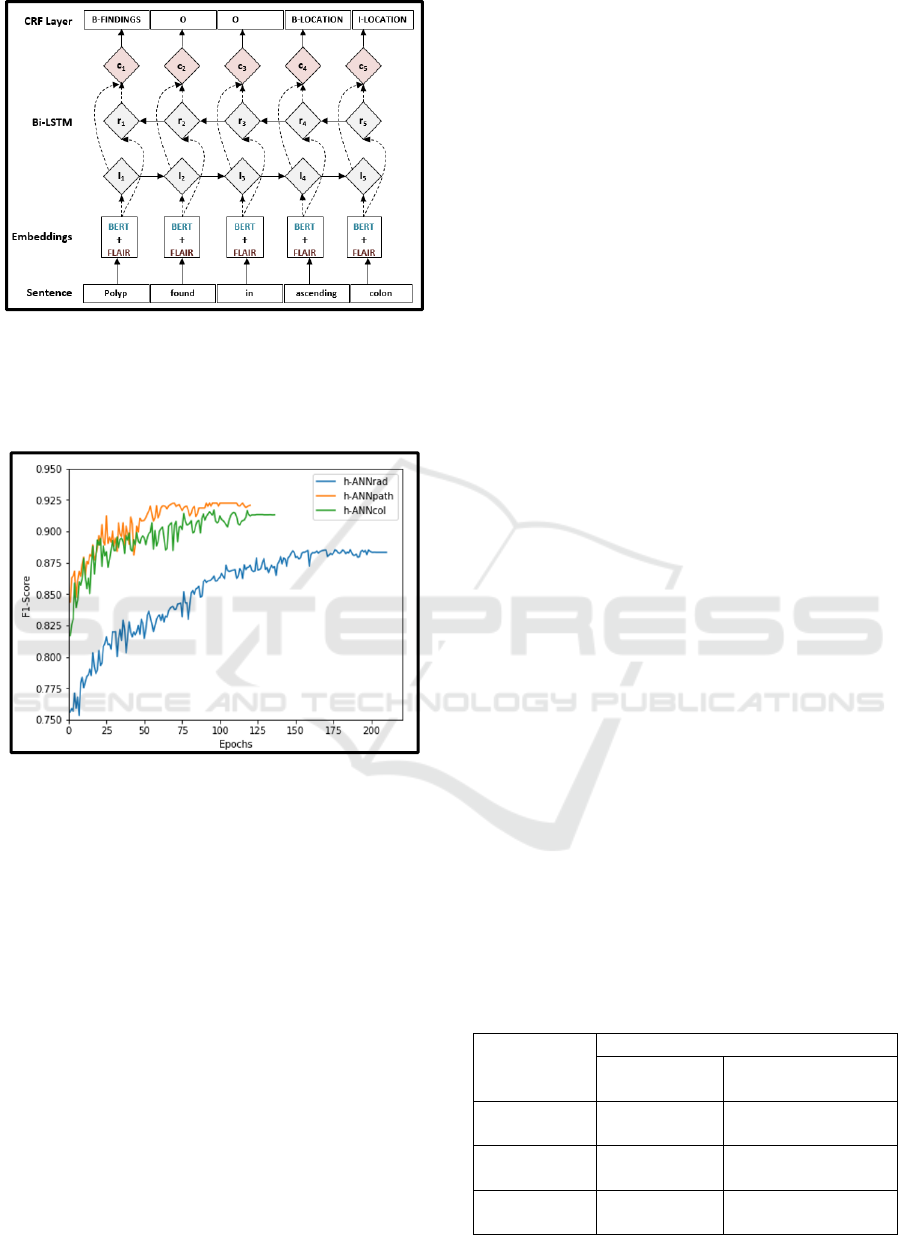
Figure 5: The h-ANN architecture depicting embedding,
Bi-LSTM, and CRF layers. Concatenated BERT and
FLAIR embeddings are given as input features to the Bi-
LSTM layer. BERT: Bidirectional Encoder
Representations from Transformers.
Figure 6: Training curves for h-ANNpath, h-ANNcol, and
h-ANNrad models. F1 score on the test set was measured
after completion of each epochs.
We further validated GIN’s accuracy to extract
clinical concepts from the three document types using
manual medical record abstraction. We randomly
selected 300 (N=16,900, confidence interval = 90%,
ε = 5%) colonoscopy procedures for chart review. For
the 300 procedures, 219 associated pathology, and
123 radiology notes were identified respectively. The
three document types (642 total) were chart reviewed
for 15 entities as shown in Table 6. These variables
were selected based on the quality metrics published
by the American College of Gastroenterology and
recommendations from a panel of gastroenterologists
(lead by BT). The entities include type of polyp
(neoplastic and non-neoplastic) and location,
pathological classification (benign and malignant
carcinomas), scope times, quality of bowel
preparation, and abnormalities found in radiology
reports (obstruction, tumor, and perforation). These
concepts are vital to colonoscopy quality
improvement, care management, and colorectal
cancers research. The chart review was done by 4
reviewers (1 medical student and 3 trained data
warehouse analysts) under the guidance of domain
expert (BT). Discrepancies between the chart
reviewers were resolved by the domain expert. We
extracted the same entities from the evaluation
sample using GIN. Using findings from manual data
abstraction as the gold standard, we evaluated
extraction accuracy of GIN and report the results in
Table 6. Overall GIN achieved an accuracy of
91.05%, and the accuracy for extracting entities from
colonoscopy reports was 94.69%. Similarly, for
identifying concepts from the pathology and
radiology reports, the toolkit achieved accuracies of
92.40% and 86.05% respectively.
4 DISCUSSION
In this study, we extracted comprehensive clinical
concepts from consolidated colonoscopy documents
using a unique DL model that combines GI-domain
trained BERT and FLAIR embeddings. Pre-training
and concatenating embeddings has two main
advantages: 1) better representation of clinical
concepts and 2) minimizing annotated corpus size
required for training. Using relatively smaller
annotated corpora (~ 430 notes per document type),
the GIN achieved competitive accuracy (91.05%) in
extracting an exhaustive list of clinical entities from
the three document types. The h-ANNcol model
extracted polyp findings from colonoscopy reports
with an accuracy of 92.63% which is comparable to
the results presented by Lee et al. (J. K. Lee et al.,
2019), who trained a traditional ML model on
approximately 800 annotated documents and
achieved 92.50% accuracy. Most studies to date have
extracted few clinical predictors from colonoscopy or
Table 2: Five-fold cross validation results of the three fine-
tuned models on respective documents.
Fine-tuned
Models
Model Performance - F1 Score
Range (%) Mean Confidence
Interval (95%)
Pathology (h-
ANN
path
)
89.66 – 92.25 91.03 ± 1.53
Colonoscopy
(h-ANN
col
)
89.42 – 91.76 90.60 ± 1.19
Radiology (h-
ANN
rad
)
85.91 – 88.55 87.22 ± 1.45
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings
195
pathology reports (Nayor et al., 2018; Patterson et al.,
2015; Raju et al., 2015). Moreover, imaging reports
were not integrated to gather procedure indications
and other related findings. Leveraging our previous
work (Syed et al., 2021), in this study, we extracted
74, 61, and 47 unique entities from consolidated
colonoscopy, pathology, and radiology reports
respectively. Integrating these vital concepts with
discrete EHR data has the potential to decrease or
altogether eliminate manual data abstraction and
facilitate colonoscopy quality assessment, treatment
plan, and colorectal cancer research.
Table 3: Performance results of the h-ANNcol model on
identifying clinical entities from colonoscopy reports.
Colonoscopy Entity Precision Recall F1
Score
Polyp_Found 0.9178 0.9350 0.9263
Polyp_Size 0.8942 0.9171 0.9055
Qty_of_Polyp 0.8906 0.8706 0.8805
Location 0.9111 0.8937 0.9023
Findings 0.8760 0.8834 0.8797
Scope_Insertion 0.9470 0.9620 0.9544
Cecum_Reached_Time 0.9520 0.9510 0.9515
Scope_Withdrawn 0.9670 0.9350 0.9507
ASA 0.9780 0.9550 0.9664
Med_Name 0.9650 0.9580 0.9615
Form 0.9710 0.9540 0.9624
Dose 0.9450 0.9510 0.948
Conscious_Sedation 0.8570 0.8710 0.8639
Deep_Sedation 0.8950 0.8743 0.8845
Cecum_Reached 0.9368 0.9500 0.9434
Estimated_Blood_Loss 0.8667 0.8890 0.8777
Patient_Position 0.8387 0.8667 0.8525
Patient_Tolerance 0.8989 0.8999 0.8994
Procedure_Techniques 0.8876 0.8650 0.8762
Quality_of_Preparation 0.9764 0.9550 0.9656
Several studies built in-house NLP solutions to
extract concepts of interests from colonoscopy
documents (J. K. Lee et al., 2019; Mehrotra et al.,
2012; Raju et al., 2015). But, these solutions used
either rule based algorithms or proprietary software,
lacking generalization and applicability to diverse
health care settings. Pre-training contextual LMs on
domain-specific corpora and sharing pre-trained
weights can solve these problems (Alsentzer et al.,
2019; J. Lee et al., 2019). In our study, we pre-trained
BERT and FLAIR on the Un-GIC (~34,165 notes) to
learn domain linguistic. The trained LMs can be
utilized by any healthcare institution with minimal to
no pre-training efforts. Using institution-specific
annotated corpora, the models can be fine-tuned for
various downstream NLP tasks. Moreover, compared
to pre-training, fine-tuning is relatively less resource
intensive and can be done in few hours. This can
eliminate the need for high performance computing
and the associated technical expertise.
Table 4: Performance results of the h-ANNpath model on
identifying clinical entities from pathology reports.
Pathology Entity Precision Recall F1
Score
Location 0.920 0.972 0.945
Specimen Type 0.911 0.967 0.938
Neoplastic Polyp 0.971 0.930 0.950
Non Neoplastic Polyp 0.915 0.870 0.892
Polyp Like Lesion 0.887 0.875 0.881
Pathological
Classification Benign
0.887 0.964 0.924
Pathological
Classification Malignant
0.924 0.950 0.937
Table 5: Performance results of the h-ANNrad model on
identifying clinical entities from radiology reports.
Imaging Entity Precision Recall F1 Score
Abscess 0.963 0.911 0.936
Cirrhosis 0.964 0.958 0.961
Colitis 0.956 0.917 0.936
Crohns 0.800 0.812 0.806
Diverticulosis 0.803 0.837 0.819
Edema 0.835 0.818 0.826
Hemorrhage 0.954 0.983 0.968
Inflammation 0.833 0.821 0.827
Ischemia 0.944 0.962 0.953
Location 0.862 0.807 0.833
Mass or Tumor 0.83 0.882 0.855
Obstruction 0.843 0.865 0.853
Perforation 0.863 0.838 0.850
Tumor Size 0.842 0.862 0.851
Steatosis 0.964 0.944 0.953
Stones 0.971 0.954 0.962
Stranding 0.913 0.921 0.917
Liver 0.851 0.890 0.870
Thickening 0.832 0.860 0.845
Both BERT and FLAIR models have shown
improved performance on various NLP tasks when
trained on domain-specific corpora (Alsentzer et al.,
2019; M. Jiang et al., 2019; Sharma & Daniel, 2019).
During pre-training, BERT learns semantics at both
word and sentence levels (Kalyan & Sangeetha,
2021). Moreover, its multi-head self-attention
mechanism enables the model to capture long-range
dependencies, often found in clinical narratives
(K. Huang et al., 2019; Kalyan & Sangeetha, 2021).
HEALTHINF 2022 - 15th International Conference on Health Informatics
196

Table 6: Results of the GIN’s accuracy when compared to chart review based on a list of 15 entities selected from the
colonoscopy, pathology, and radiology reports. GIN: Gastroenterology NLP toolkit.
Report Type
(Sample Size)
Entity No. of Documents
in which the Entity
was Found During
Chart Review (%)
GIN Accuracy based
on Chart Review (%) -
95% confidence
Interval
Colonoscopy (n=300)
Presence of Polyp 156 (52.00) 91.82 (87.16 – 96.04)
Polyp Location 156 (52.00) 89.13 (84.11 – 94.09)
Scope Insertion Time 300 (100.00) 96.82 (94.51 – 98.69)
Cecum Reached Time 296 (98.78) 96.50 (94.08 – 98.48)
Scope Withdrawn Time 276 (92.00) 96.13 (93.65 – 98.37)
Adequacy of Bowel
Preparation
298 (99.40) 97.75 (95.89 – 99.41)
Pathology (n=219)
Specimen Type 219 (100.00) 92.80 (89.17 – 96.21)
Neoplastic Polyps 45 (20.55) 95.10 (86.00 – 1.00)
Non Neoplastic Polyps 31 (14.16) 89.11 (75.06 – 99.14)
Malignant 17 (7.76) 93.22 (73.02 – 98.95)
Benign 46 (21.00) 91.76 (82.99 – 99.61)
Imaging (n=123)
Mass or Tumor 31 (25.20) 85.83 (70.66 – 97.08)
Obstruction 25 (20.30) 86.20 (65.35 – 93.60)
Perforation 7 (5.80) 85.14 (48.68 – 97.43)
Liver Abnormality 27 (22.00) 86.85 (67.52 – 94.08)
But, the clinical feature representation of BERT is
curtailed by its fixed vocabulary (Bressem et al.,
2021; J. Lee et al., 2019). Flair models words and
context as sequences of characters to form word-level
embeddings, this has the advantages of generating
strong character-level features, being independent of
tokenization and vocabulary, and efficiently handling
rare and misspelled words (Akbik et al., 2018). But,
character-level representation performs poorly when
processing long sentences (D. Liang, Xu, & Zhao,
2017). Due to these characteristics, we specifically
chose to combine BERT and FLAIR embeddings, this
generated strong word representations for the
downstream NLP tasks. Using the best performing
models on the three document types and the
associated annotated corpora respectively, we tested
the performance of 3 model configurations” 1) BERT
embeddings alone, 2) FLAIR embeddings alone, and
3) concatenated BERT and FLAIR embeddings. As
shown in Table 7, for the three document types, the
models with concatenated embeddings performed
best compared to models with either BERT or FLAIR
embeddings alone.
To validate if the F1-score improvement is
statistically significant for the three models (h-
ANN
col
, h-ANN
path
, and h-ANN
rad
) with concatenated
embeddings, we conducted a 5x2cv paired t-test
(Dietterich, 1998). For each report type (colonoscopy,
pathology, and radiology), we did a pairwise
comparison between the model with the concatenated
embeddings (M
BERT+FLAIR
) and models with
individual BERT (M
BERT
) and FLAIR (M
FLAIR
)
embeddings respectively. Resulting in 6 pair
comparisons, 2 for each report type (M
BERT+FLAIR
vs
M
BERT
and M
BERT+FLAIR
vs M
FLAIR
). The results show
Table 7: Performance of models with 1) BERT, 2) FLAIR,
and 3) concatenated BERT and FLAIR embeddings on
imaging, colonoscopy, and pathology documents
respectively.
Model Precision
(%)
Recall
(%)
F1 Score
(%)
Imaging
M
BERT
84.21 82.53 83.36
M
FLAIR
83.32 80.56 81.92
M
BERT+FLAIR
89.18 87.94 88.55
Colonoscopy
M
BERT
89.79 90.11 89.95
M
FLAIR
91.12 89.86 90.48
M
BERT+FLAIR
91.22 92.32 91.76
Pathology
M
BERT
89.59 90.41 90.00
M
FLAIR
90.27 92.89 91.56
M
BERT+FLAIR
91.38 93.14 92.25
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings
197

that the improvement of F-measures for all six pairs
were statistically significant (P value < 0.05). For
identifying individual entities using concatenated
embeddings from the three document types, we
noticed F1score improvement between 3.4% - 7.2%
compared to using models with individual
embedding. The most F1-score improvement
wasseen on radiology concept extraction.
Of the three report types from which we extracted
data, colonoscopy and pathology were semi-
structured and radiology reports were unstructured.
Unsurprisingly, the h-ANN
col
and h-ANN
path
model
accuracies were higher than h-ANN
rad
. Moreover,
imaging reports are known to be complex, lack
clarity, and often omit a definitive conclusion (Brady,
2018). These could be the reasons that the h-ANN
rad
model took relatively more epochs (~215) to
converge during fine-tuning, as shown in Figure 6.
Further study is needed to assimilate key information
from radiology reports and improve information
extraction accuracy.
5 CONCLUSIONS
In Domain-trained contextualized embeddings are
powerful word representations. Using concatenated
embeddings, we extracted comprehensive clinical
concepts from consolidated colonoscopy documents
with a high degree of confidence (F1 score 91.05%)
and relatively smaller annotated corpora (~50%).
Integrating these vital concepts with discrete EHR
data can eliminate manual data abstraction and
increase secondary use of information in narrative
colonoscopy-related reports for colonoscopy quality
assessment and colorectal cancer research. The NLP
framework demonstrated here is generalizable and
can be applied to diverse clinical narratives and
potentially beyond healthcare to improve NLP
performance in specialty domains, we extracted
comprehensive clinical.
ACKNOWLEDGEMENTS
Patients’ data used for this study were obtained under
IRB approval (IRB# 262202) at the University of
Arkansas for Medical Sciences (UAMS). The study
was supported in part by the Translational Research
Institute (TRI), grant UL1 TR003107 received from
the National Center for Advancing Translational
Sciences of the National Institutes of Health (NIH)
and award AWD00053499, Supporting High
Performance Computing in Clinical Informatics.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Zheng, X. J. A. (2016). TensorFlow: Large-
Scale Machine Learning on Heterogeneous Distributed
Systems. abs/1603.04467.
Abdalla, M., Abdalla, M., Rudzicz, F., & Hirst, G. (2020).
Using word embeddings to improve the privacy of
clinical notes. Journal of the American Medical
Informatics Association, 27(6), 901-907. doi:10.1093/
jamia/ocaa038
Akbik, A., Bergmann, T., Blythe, D., Rasul, K., Schweter,
S., & Vollgraf, R. (2019). FLAIR: An Easy-to-Use
Framework for State-of-the-Art NLP. https://doi.org/
10.18653/v1/n19-4010
Akbik, A., Blythe, D., & Vollgraf, R. (2018). Contextual
String Embeddings for Sequence Labeling. Santa Fe,
New Mexico, USA: Association for Computational
Linguistics.
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jindi,
D., Naumann, T., & McDermott, M. (2019). Publicly
Available Clinical BERT Embeddings. Minneapolis,
Minnesota, USA: Association for Computational
Linguistics.
Anderson, J. C., & Butterly, L. F. (2015). Colonoscopy:
quality indicators. Clinical and translational
gastroenterology, 6(2), e77-e77. doi:10.1038/
ctg.2015.5
Brady, A. P. (2018). Radiology reporting-from Hemingway
to HAL? Insights into imaging, 9(2), 237-246.
doi:10.1007/s13244-018-0596-3
Bressem, K. K., Adams, L. C., Gaudin, R. A., Tröltzsch, D.,
Hamm, B., Makowski, M. R., Niehues, S. M. (2021).
Highly accurate classification of chest radiographic
reports using a deep learning natural language model
pre-trained on 3.8 million text reports. Bioinformatics
(Oxford, England), 36(21), 5255-5261. doi:10.1093/
bioinformatics/btaa668
Caufield, J. H., Zhou, Y., Garlid, A. O., Setty, S. P., Liem,
D. A., Cao, Q., Ping, P. (2018). A reference set of
curated biomedical data and metadata from clinical case
reports. Scientific data, 5(1), 180258. doi:10.1038/
sdata.2018.258
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2018).
BERT: Pre-training of Deep Bidirectional
Transformers for Language Understanding.
Dietterich, T. G. (1998). Approximate Statistical Tests for
Comparing Supervised Classification Learning
Algorithms. Neural Computation, 10(7), 1895-1923.
doi:10.1162/089976698300017197 %J Neural
Computation
El Boukkouri, H., Ferret, O., Lavergne, T., &
Zweigenbaum, P. (2019). Embedding Strategies for
Specialized Domains: Application to Clinical Entity
HEALTHINF 2022 - 15th International Conference on Health Informatics
198

Recognition. Florence, Italy: Association for
Computational Linguistics.
Fan, Y., Wen, A., Shen, F., Sohn, S., Liu, H., & Wang, L.
(2019). Evaluating the Impact of Dictionary Updates on
Automatic Annotations Based on Clinical NLP
Systems. AMIA Jt Summits Transl Sci Proc, 2019, 714-
721.
Griffis, D., Shivade, C., Fosler-Lussier, E., & Lai, A. M.
(2016). A Quantitative and Qualitative Evaluation of
Sentence Boundary Detection for the Clinical Domain.
AMIA Jt Summits Transl Sci Proc, 2016, 88-97.
Harkema, H., Chapman, W. W., Saul, M., Dellon, E. S.,
Schoen, R. E., & Mehrotra, A. (2011). Developing a
natural language processing application for measuring
the quality of colonoscopy procedures. J Am Med
Inform Assoc, 18 Suppl 1(Suppl 1), i150-156.
doi:10.1136/amiajnl-2011-000431
Huang, K., Altosaar, J., & Ranganath, R. (2019).
ClinicalBERT: Modeling Clinical Notes and Predicting
Hospital Readmission.
Huang, Z., Xu, W., & Yu, K. J. A. (2015). Bidirectional
LSTM-CRF Models for Sequence Tagging.
abs/1508.01991.
Jiang, M., Sanger, T., & Liu, X. (2019). Combining
Contextualized Embeddings and Prior Knowledge for
Clinical Named Entity Recognition: Evaluation Study.
JMIR Med Inform, 7(4), e14850. doi:10.2196/14850
Jiang, X., Pan, S., Jiang, J., & Long, G. (2018, 8-13 July
2018). Cross-Domain Deep Learning Approach For
Multiple Financial Market Prediction. Paper presented
at the 2018 International Joint Conference on Neural
Networks (IJCNN).
Kalyan, K. S., & Sangeetha, S. (2021). BertMCN: Mapping
colloquial phrases to standard medical concepts using
BERT and highway network. Artif Intell Med, 112,
102008. doi:https://doi.org/10.1016/j.artmed.2021.10
2008
Kim, K., Polite, B., Hedeker, D., Liebovitz, D., Randal, F.,
Jayaprakash, M., Lam, H. (2020). Implementing a
multilevel intervention to accelerate colorectal cancer
screening and follow-up in federally qualified health
centers using a stepped wedge design: a study protocol.
Implementation Science, 15(1), 96. doi:10.1186/
s13012-020-01045-4
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., &
Kang, J. (2019). BioBERT: a pre-trained biomedical
language representation model for biomedical text
mining. Bioinformatics (Oxford, England).
doi:10.1093/bioinformatics/btz682
Lee, J. K., Jensen, C. D., Levin, T. R., Zauber, A. G.,
Doubeni, C. A., Zhao, W. K., & Corley, D. A. (2019).
Accurate Identification of Colonoscopy Quality and
Polyp Findings Using Natural Language Processing. J
Clin Gastroenterol, 53(1), e25-e30. doi:10.1097/
mcg.0000000000000929
Liang, D., Xu, W., & Zhao, Y. (2017). Combining Word-
Level and Character-Level Representations for
Relation Classification of Informal Text. Paper
presented at the Rep4NLP@ACL.
Liang, H., Sun, X., Sun, Y., & Gao, Y. (2018). Correction
to: Text feature extraction based on deep learning: a
review. EURASIP Journal on Wireless
Communications and Networking, 2018(1), 42.
doi:10.1186/s13638-018-1056-y
Malte, A., & Ratadiya, P. (2019). Evolution of transfer
learning in natural language processing. CoRR,
abs/1910.07370.
Mehrotra, A., Dellon, E. S., Schoen, R. E., Saul, M.,
Bishehsari, F., Farmer, C., & Harkema, H. (2012).
Applying a natural language processing tool to
electronic health records to assess performance on
colonoscopy quality measures. Gastrointest Endosc,
75(6), 1233-1239.e1214. doi:10.1016/j.gie.2012.01.0
45
Mikolov, T., Chen, K., Corrado, G., & Dean, J. (2013).
Efficient Estimation of Word Representations in Vector
Space. Paper presented at the ICLR.
Nayor, J., Borges, L. F., Goryachev, S., Gainer, V. S., &
Saltzman, J. R. (2018). Natural Language Processing
Accurately Calculates Adenoma and Sessile Serrated
Polyp Detection Rates. Dig Dis Sci, 63(7), 1794-1800.
doi:10.1007/s10620-018-5078-4
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J.,
Chanan, G., Chintala, S. (2019). PyTorch: An
Imperative Style, High-Performance Deep Learning
Library. Paper presented at the NeurIPS.
Patterson, O. V., Forbush, T. B., Saini, S. D., Moser, S. E.,
& DuVall, S. L. (2015). Classifying the Indication for
Colonoscopy Procedures: A Comparison of NLP
Approaches in a Diverse National Healthcare System.
Stud Health Technol Inform, 216, 614-618.
Peters, M. E., Neumann, M., Iyyer, M., Gardner, M., Clark,
C., Lee, K., & Zettlemoyer, L. (2018, jun). Deep
Contextualized Word Representations, New Orleans,
Louisiana.
Raju, G. S., Lum, P. J., Slack, R. S., Thirumurthi, S., Lynch,
P. M., Miller, E., Ross, W. A. (2015). Natural language
processing as an alternative to manual reporting of
colonoscopy quality metrics. Gastrointest Endosc,
82(3), 512-519. doi:10.1016/j.gie.2015.01.049
Rex, D. K., Schoenfeld, P. S., Cohen, J., Pike, I. M., Adler,
D. G., Fennerty, M. B., Weinberg, D. S. (2015). Quality
indicators for colonoscopy. Gastrointest Endosc, 81(1),
31-53. doi:10.1016/j.gie.2014.07.058
Roberts, A., Gaizauskas, R., Hepple, M., Davis, N.,
Demetriou, G., Guo, Y., Wheeldin, B. (2007). The
CLEF corpus: semantic annotation of clinical text.
AMIA ... Annual Symposium proceedings. AMIA
Symposium, 2007, 625-629.
Schmidt, J., Marques, M. R. G., Botti, S., & Marques, M.
A. L. (2019). Recent advances and applications of
machine learning in solid-state materials science. npj
Computational Materials, 5(1), 83.
doi:10.1038/s41524-019-0221-0
Sharma, S., & Daniel, R., Jr. (2019). BioFLAIR: Pretrained
Pooled Contextualized Embeddings for Biomedical
Sequence Labeling Tasks. arXiv e-prints,
arXiv:1908.05760.
The h-ANN Model: Comprehensive Colonoscopy Concept Compilation using Combined Contextual Embeddings
199

Si, Y., Wang, J., Xu, H., & Roberts, K. (2019). Enhancing
clinical concept extraction with contextual embeddings.
Journal of the American Medical Informatics
Association, 26(11), 1297-1304. doi:10.1093/jamia/
ocz096 %J Journal of the American Medical
Informatics Association
Syed, S., Tharian, B., Syeda, H. B., Zozus, M., Greer, M.
L., Bhattacharyya, S., Prior, F. (2021). Consolidated
EHR Workflow for Endoscopy Quality Reporting. Stud
Health Technol Inform, 281, 427-431.
doi:10.3233/shti210194
Syeda, H. B., Syed, M., Sexton, K. W., Syed, S., Begum,
S., Syed, F., Yu, F., Jr. (2021). Role of Machine
Learning Techniques to Tackle the COVID-19 Crisis:
Systematic Review. JMIR Med Inform, 9(1), e23811.
doi:10.2196/23811
Wei, Q., Franklin, A., Cohen, T., & Xu, H. (2018). Clinical
text annotation - what factors are associated with the
cost of time? AMIA Annu Symp Proc, 2018, 1552-1560.
Wu, Y., Yang, X., Bian, J., Guo, Y., Xu, H., & Hogan, W.
(2018). Combine Factual Medical Knowledge and
Distributed Word Representation to Improve Clinical
Named Entity Recognition. AMIA Annu Symp Proc,
2018, 1110-1117.
Yang, X., Bian, J., Hogan, W. R., & Wu, Y. (2020). Clinical
concept extraction using transformers. Journal of the
American Medical Informatics Association, 27(12),
1935-1942. doi:10.1093/jamia/ocaa189
HEALTHINF 2022 - 15th International Conference on Health Informatics
200
