
Named Entity Recognition for the Extraction of
Emerging Technological Knowledge from Medical Literature
Sabrina Lamberth-Cocca
1a
, Bernhard Maier
1
, Christian Nawroth
1
,
Paul Mc Kevitt
2b
and Matthias Hemmje
1c
1
Faculty of Mathematics and Computer Science, University of Hagen, Germany
2
Academy for International Science & Research (AISR), Derry/Londonderry, Northern Ireland
p.mckevitt@aisr.org.uk
Keywords: Named Entity Recognition, Natural Language Processing, Information Retrieval, Knowledge Extraction,
Machine Learning, Emerging Medical Technology, Clinical Argumentation Support.
Abstract: In this paper, we show the results of an experimental Information Retrieval System (IRS) prototype to support
the detection of emerging medical technology using the method of Named-Entity Recognition (NER). The
overall goal is to automatically identify and classify entities and structures in scientific medical articles, which
represent the concept of Medical Technologies (MedTech) with high topicality. As a first approach, we
combine learning-based NER with rule-based emerging Named-Entity Recognition (eNER). We train a
machine-learning model on manually annotated NER candidates representing medical devices. We then
match the results with entries from vocabularies containing medical devices according to our definition, using
a handcrafted rule-based approach and fuzzy functions. The main outcome is an experimental prototype which
we call, MedTech-eNER-IRS, which shows that such an approach works in general, including pointers for
further research and prototype improvements.
1 INTRODUCTION
The work presented in this paper is part of the
RecomRatio (Recommendation Rationalization)
project (cf. University of Bielefeld, 2017). The main
objective here is to support decision making in
various medical areas by developing Information
Retrieval (IR) systems for clinical Virtual Research
Environments (VREs). RecomRatio is a VRE to
support argumentation processes of medical staff in
determining clinical decisions.
Medical experts need to conduct research based
on knowledge sources such as scientific publications
for various reasons. One purpose is to gather
information about the state of the art, and in particular
new technologies in relevant biomedical fields.
Databases such as PubMed (NCBI, 2022) support
document research in relevant domains, for instance
specific diagnostic areas like gene expression
analysis. The general problem of information
a
https://orcid.org/0000-0002-8092-5219
b
https://orcid.org/0000-0001-9715-1590
c
https://orcid.org/0000-0001-8293-2802
explosion does not end at the medical domain and
leads to a growing volume of literature.
In the case of this study and our experimental
prototype, called MedTech-eNER-IRS, we observe a
problem that is unsolved to the best of our knowledge:
detecting Named Entities (NEs) that represent the
concept of emerging Medical Technology
(MedTech). Whilst the automatic recognition of
emerging NEs (eNEs) has already been addressed by
Nawroth et al. (2018), current IR systems are not
capable of recognizing and classifying MedTech
entities. Additionally, existing vocabularies such as
MeSH (Medical Subject Headings) (NLM, 2021) and
SNOMED CT (Systematized Nomenclature of
Medicine – Clinical Terms) (SNOMED International,
2021) can be generally used for medical NER, but do
not contain explicit classes for distinguishing
MedTech entities, which adds another degree of
complexity to the recognition task.
Lamberth-Cocca, S., Maier, B., Nawroth, C., Kevitt, P. and Hemmje, M.
Named Entity Recognition for the Extraction of Emerging Technological Knowledge from Medical Literature.
DOI: 10.5220/0011369300003335
In Proceedings of the 14th International Joint Conference on Knowledge Discovery, Knowledge Engineer ing and Knowledge Management (IC3K 2022) - Volume 2: KEOD, pages 101-108
ISBN: 978-989-758-614-9; ISSN: 2184-3228
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
101

In the following Section 2, we give an overview
of the state of the art and related work, followed by a
description of how we prepared the experimental data
and conducted the preparatory study in Section 3. In
Section 4, we present the design and implementation
of our experimental prototype, MedTech-eNER-IRS,
including use cases, architecture and experimental
functions. Section 5 summarizes the evaluation
results of the MedTech-eNER-IRS’s components.
We close this article with a discussion (Section 6),
and conclusion and future research (Section 7).
2 STATE OF THE ART AND
RELATED WORK
Detecting medical terms from collections of textual
documents that are relevant to a specific information
need is a problem of Information Retrieval (IR) and
Natural Language Processing (NLP). More precisely,
the problem can be subsumed under the NLP task of
Named Entity Recognition (NER). NER denotes the
method of automatically detecting and classifying
Named Entities, i.e. named objects from the real
world such as persons, organizations, locations, in
unstructured text data. In the medical domain, NER
focuses on the detection of specific medical terms.
For instance, biomedical information extraction
applies biomedical NER (BioNER) to detect relevant
entities representing genes and diseases, in order to
infer relations from text-based publications, e.g.,
Perera et al. (2020).
2.1 Definition of Medical Technology
In order to define entities for recognition in a NER
model, we first need to define the underlying concept.
Thus, we first answer the question, what exactly
MedTech-eNER-IRS is meant to recognize and
classify. In general, medical technology (MedTech)
means the, “application of science to develop
solutions to health problems or issues such as the
prevention or delay of onset of diseases or the
promotion and monitoring of good health” (National
Center for Health Statistics, 2010, p. 4). The World
Health Organization (WHO) defines health
technology as the “application of organized
knowledge and skills in the form of devices,
medicines, vaccines, procedures, and systems
developed to solve a health problem and improve
quality of lives” (WHO, 2022, para. 2). A similar
definition of health technology can be found in the
Health Technology Assessment (HTA) glossary: “An
intervention developed to prevent, diagnose or treat
medical conditions; promote health; provide
rehabilitation; or organize healthcare delivery. The
intervention can be a test, device, medicine, vaccine,
procedure, program or system” (International
Network of Agencies for Health Technology
Assessment, 2022).
The European medical devices directive,
Regulation (EU) 2017/745 of the European
Parliament and of the Council of 5 April 2017 on
medical devices, applies the following definition for
medical devices: “any instrument, apparatus,
appliance, software, implant, reagent, material or
other article intended by the manufacturer to be used,
alone or in combination, for human beings for one or
more of the following specific medical purposes:
diagnosis, prevention, monitoring, prediction,
prognosis, treatment or alleviation of disease,
diagnosis, monitoring, treatment, alleviation of, or
compensation for, an injury or disability,
investigation, replacement or modification of the
anatomy or of a physiological or pathological
process or state,
providing information by means of in-
vitro examination of specimens derived from the
human body, including organ, blood and tissue
donations,
and which does not achieve its principal intended
action by pharmacological, immunological or
metabolic means, in or on the human body, but which
may be assisted in its function by such means.
The following products shall also be deemed to be
medical devices:
devices for the control or support of conception;
products specifically intended for the cleaning,
disinfection or sterilisation of devices […]”
(EU, 2020, p. 15).
In-vitro diagnostic devices (IVD) are not part of
this directive, but the IVD directive also defines them
as medical devices: “in vitro diagnostic medical
device means any medical device which is a reagent,
reagent product, calibrator, control material, kit,
instrument, apparatus, equipment, or system, whether
used alone or in combination, intended by the
manufacturer to be used in vitro for the examination
of specimens, including blood and tissue donations,
derived from the human body, solely or principally
for the purpose of providing information:
concerning a physiological or pathological state, or
concerning a congenital abnormality, or
to determine the safety and compatibility with
potential recipients, or
to monitor therapeutic measures”
(EU, 2012, p. 5).
KEOD 2022 - 14th International Conference on Knowledge Engineering and Ontology Development
102

The United States of America (U.S.) Food and
Drug Administration (FDA) uses a similar definition
for medical devices:
“an instrument, apparatus, implement, machine,
contrivance, implant, in vitro reagent, or other
similar or related article, including any component,
part, or accessory, which is
(1) recognized in the official National Formulary,
or the United States Pharmacopeia, or any
supplement to them,
(2) intended for use in the diagnosis of disease or
other conditions, or in the cure, mitigation,
treatment, or prevention of disease, in man or
other animals, or
(3) intended to affect the structure or any function
of the body of man or other animals, and
which does not achieve its primary intended
purposes through chemical action within or on the
body of man or other animals and which is not
dependent upon being metabolized for the
achievement of its primary intended purposes”
(FDA, 2017, p. 5).
Based on the regulatory definitions of the
European Union (EU) and the U.S., we use the
following shortened and summarized definition of
medical technology for all following analyses and
MedTech-eNER-IRS.
Instrument, apparatus, device, software, machine,
appliance, implant, in-vitro reagent for the
Diagnosis, prevention, monitoring, prognosis or
treatment of diseases with its
Main effect not through in-vivo biochemical action
(no drugs).
2.2 Characteristics of Emerging
Technological Knowledge
The concept of technology has both instrumental and
processual components and is strongly tied to the
concept of knowledge. A systemic approach
explaining the emergence of new technology
distinguishes between knowledge (technological
know-how), activities (problem solving by applying
technology), and artifacts (problem solution;
machines, devices, products; technical systems) as
constituents (Bullinger, 1994, p. 32 ff.).
Technological knowledge “derives from, and finds
meaning, in activity”, is strongly tied to practical
applications; tacit knowledge as a form of
technological knowledge is embedded in
technological activity, and “isolated from activity and
removed from the implementing context, much of
technological knowledge loses its meaning and
identity“ (Herschbach, 1995, p. 38).
The difficulty with emerging knowledge and its
systematic detection is that it, “arises suddenly and
unexpectedly and it cannot be planned and predicted”
(Patel and Ghonheim, 2011, p. 425). Thus, the
challenging task is to detect entities representing
emerging (technological) knowledge that are yet
unknown, which we define as non-existent in a
relevant vocabulary or knowledge base.
We limited our definition of MedTech in the
previous section to its instrumental dimension, and
thus are focusing on MedTech artifacts for MedTech-
eNER-IRS discussed here. However, for further
refinement, procedural information and application
context will be included.
2.3 Emerging Named Entity
Recognition (eNER)
Emerging Named Entity Recognition (eNER) is a
relatively new research area that deals with the
automatic detection of NEs that are useful to
automatically extract emerging knowledge according
to the definition in the previous section. Nawroth et
al. (2018) introduced the concept of emerging Named
Entities (eNE) and eNER in order to recognize and
classify characteristic NEs in the context of
arguments in clinical decisions. According to their
definition, eNEs are terms in use that are not
acknowledged yet, i.e., not listed in controlled
vocabularies or databases. We apply the following
formal definition (1) to determine if an NE is an eNE:
If Y
D
< Y
NE
=> eNE (1)
Y
D
: publication year of documents from text
corpus
Y
NE
: entry year of each entity in a controlled
vocabulary or database.
3 EXPERIMENTAL DATA AND
PREPARATORY STUDY
In order to discover the patterns of NEs that represent
medical technology as well as their appearance in
relevant text documents, we conducted a manual
corpus analysis and annotation of such NEs.
We analyzed eleven research papers in the field
of medical diagnostics from different journals, which
had been selected by a medical expert from the field
of laboratory diagnostics (see Table 1). Medical
devices in the text corpus are referenced to mostly not
by using general descriptive terms, but brand names.
Additionally, product- or manufacturer names are
often incomplete or imprecise. We observed some
Named Entity Recognition for the Extraction of Emerging Technological Knowledge from Medical Literature
103
patterns, such as <product name> followed by
<manufacturer name> helpful for rule-based NER.
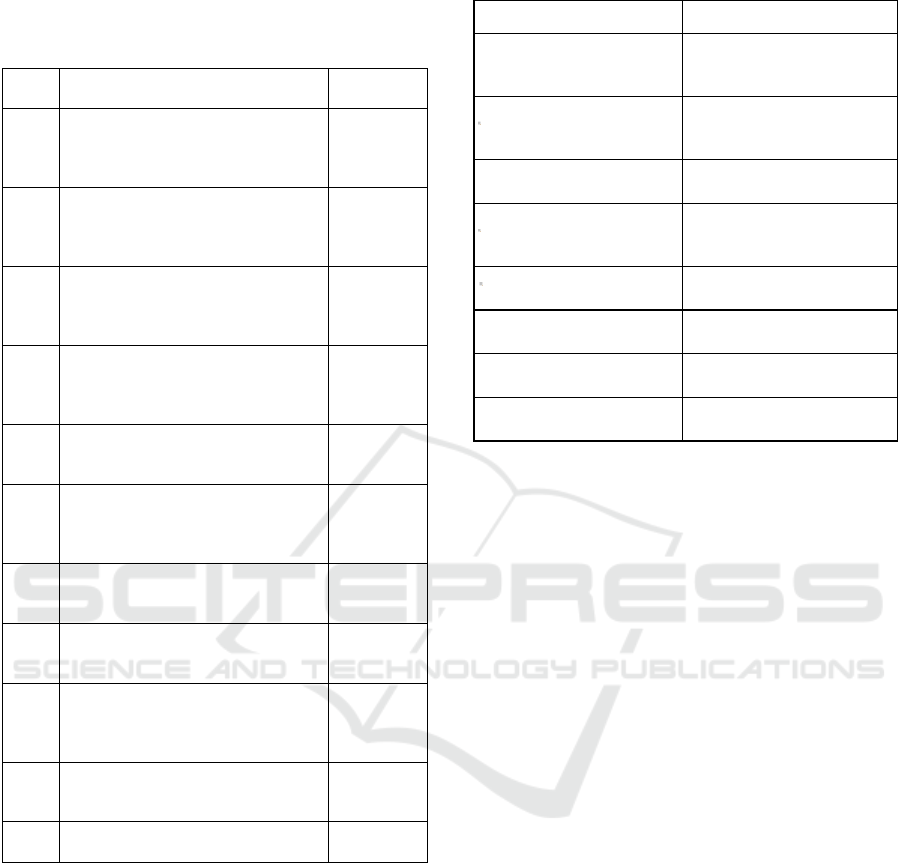
Table 1: Text corpus of papers in the field of diagnostics.
Text
No.
Paper Title Reference
1 The clinical significance of EBV
DNA in the plasma and peripheral
blood mononuclear cells of patients
with or without EBV diseases
Kanakry et
al. (2016)
2 Cell-Free DNA in blood reveals
significant 1 cell, tissue and organ
specific injury and predicts
COVID-19 severity
Cheng et
al. (2020)
3 Assessment of cell free
mitochondrial DNA as a biomarker
of disease severity in different viral
infections
Ali et al.
(2020)
4 Absolute measurement of the tissue
origins of cell-free DNA in the
healthy state and following
p
aracetamol overdose
Laurent et
al. (2020)
5 Clinical utility of circulating cell-
free Epstein–Barr virus DNA in
p
atients with gastric cance
r
Katsutoshi
et al.
(2017)
6 Analytical and clinical validation of
a microbial cell-free DNA
sequencing test for infectious
disease
Blauwkam
p et al.
(2018)
7 Detection of cell-free Epstein-Barr
virus DNA in serum during acute
infectious mononucleosis
Gan et al.
(1993)
8 Circulating cell-free nucleic acids:
main characteristics and clinical
application
Szilágyi et
al. (2020)
9 Detection and quantification of
virus DNA in plasma of patients
with Epstein-Barr virus-associated
diseases
Yamamoto
et al.
(1994)
10 Monitoring of cell-free viral DNA
in primary Epstein-Barr virus
infection
Kimura et
al. (1999)
11 A powerful, non-invasive test to
rule out infection
O’Grady
(2019)
However, the patterns occur in an inconsistent
manner. Table 2 shows a list of different patterns and
corresponding examples observed in the analyzed
text corpus. The results were discussed and validated
with the medical expert based on a questionnaire
which contained the manually annotated NE
candidates that we assumed to represent medical
technologies according to the definition above.
Table 2: Different patterns of medical-device naming.
Pattern Example from text corpus
Device (product name;
manufacturer, city, US
federal state)
Automated counts (Sysmex
KX-21N; Sysmex,
Lincolnshire, IL)
Product name (manufacturer,
city, US federal state)
QIAmp DNA blood mini
reagents (Qiagen,
Gaithersburg, MD)
Product name (manufacturer)
Qiagen/Artus EBV analyte
specific reagents (Qiagen)
Product name (manufacturer,
reference #[Nr])
DNA cryostorage vials
(Eppendorf, reference
#0030079400)
Product name (manufacturer
reference #[Nr])
DNA cryostorage vials
(Thermo Scientific #363401)
Device (manufacturer,
country of origin)
kit (Machery-Nagel, Germany)
Product name (manufacturer,
country of origin)
Eva green qPCR Master mix
(Solis Biodyne, Estonia)
Product name contains
manufacturer name
Qiagen Circulating Nucleic
Acid Ki
t
4 DESIGN AND
IMPLEMENTATION
Our model extends the eNER-IRS (emerging Named
Entity Recognition System) by Nawroth et al. (2018)
for the recognition of medical technology terms,
which we abbreviate as MedTech-NEs and MedTech-
eNEs for emerging terms respectively. We first
discuss the use cases of the MedTech-eNER-IRS.
Then we discuss preparation of the test and evaluation
data, the algorithmic constituents of MedTech-eNER-
IRS as well as its overall architecture.
4.1 MedTech-eNER-IRS Use Cases
Based on the principles of User-Centered System
Design (UCD) (Norman and Draper, 1986), we define
first the user context and requirements of the
MedTech-eNER-IRS. Overall context is given by the
RecomRatio project which aims to support medical
experts in decision making by providing them with
relevant content from large volumes of text
documents such as scientific articles. In particular, the
pipeline of MedTech-eNER-IRS is intended to accept
unknown texts and present the identified NEs/eNEs
as output. In brief, MedTech-eNER-IRS is to fulfill
the following two key requirements: (1) Perform
NER/eNER in unknown texts; (2) Presentation of the
annotated text, i.e. the MedTech-NERs/eNERs.
Derived from this, the use cases supported by
MedTech-eNER-IRS are as follows (see Figure 1):
KEOD 2022 - 14th International Conference on Knowledge Engineering and Ontology Development
104
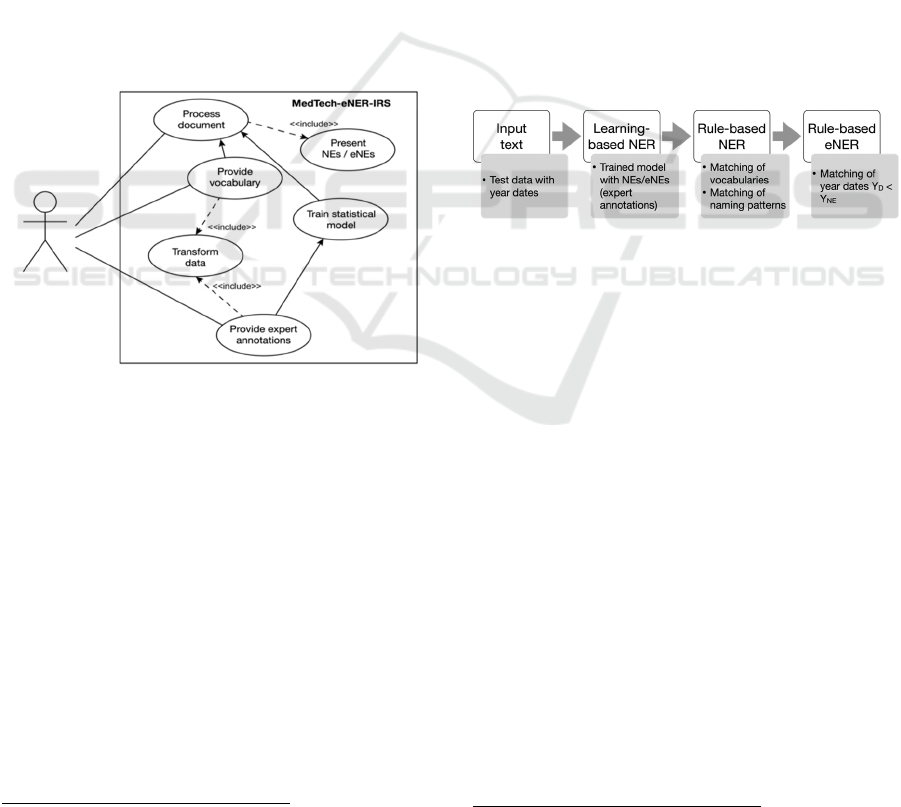
Transform data: Transforms manually prepared
data, as well as raw data from medical vocabularies
in a data schema suitable for machines-based
processing.
Provide expert annotations: Provides MedTech-
eNER-IRS with the data from the preparatory study,
i.e. the manually annotated, validated MedTech-
NEs (expert annotations).
Train statistical model: Trains the learning-based
NER/eNER model with the expert annotations
using spaCy models.
Provide vocabulary: Provides MedTech-eNER-IRS
with the data for rule-based NER/eNER, including
year dates from medical vocabularies for the
identification of eNEs.
Process document: Processes a document of choice
by the user from its original raw format through the
whole NER/eNER system’s pipeline, including
learning-based NER, rule-based NER, and rule-
based eNER (see Figure 2).
Present NEs/eNEs: Presents the results of the
MedTech-NER/eNER task to the user.
Figure 1: MedTech-eNER-IRS use cases.
Learning-Based NER: MedTech-eNER-IRS
consists of a combination of learning-based and rule-
based NER in order to detect emerging MedTech
terms. The training and evaluation data were obtained
as follows:
Training data: We used the manually annotated NE
candidates representing MedTech.
Evaluation data: We chose a 5-fold cross-
validation, since the training corpus was limited (11
documents).
Rule-Based NER and eNER: Medical
vocabularies such as MeSH and SNOMED CT
contain terms that can be used for medical NER.
However, they are limited to rather general terms
related to medical technology, whilst relevant text
corpora often contain medical devices and reagents
4
Archive files for historical and research purposes
with specific brand names or manufacturer-specific
product names. Thus, we additionally used
manufacturer-specific databases for training the
detection of MedTech-NEs classified as product
names and manufacturers: Premarket Approval
(PMA) (FDA, 2021a), 510(k) (FDA, 2021b) as well
as Device Registration and Listing (FDA, 2021c) of
the U.S. Food and Drug Administration (FDA).
We identified and extracted the relevant entries of
MedTech terms from MeSH and SNOMED CT
together with the year date. We used the following
versions: MeSH XML Descriptors 2021; SNOMED
CT International 20210131
4
; Premarket Approval
PMA 202109; 501(k): PMN since 1996 (as per 13
September 2021), PMN 1991-1995, PMN 1986-1990,
PMN 1981-1985, PMN 1976-1980; Device
Registration & Listing (as per 06 November 2021).
The year dates are required for the automatic
matching of the identified NEs with the vocabulary or
database entries, in order to determine, if an NE is an
eNE or not. In order to deal with the inconsistent use
of naming patterns, we applied fuzzy functions using
the RapidFuzz library (Bachmann, 2021).
Figure 2: MedTech-eNER-IRS pipeline.
4.2 Architecture and Experimental
NER/eNER Functions
The overall architecture of MedTech-eNER-IRS
consists of three components according to the Model-
View-Control (MVC) design pattern (see Figure 3).
We used the Python programming language in
Jupyter Notebook (Kluyver et al., 2016) as framework
for the realization of the experimental MedTech-
eNER-IRS, as well as pretrained models from the
NLP library spaCy (Honnibal and Montani, 2017):
en_core-sci_lg, en_core_web_lg. From the corpus of
11 documents (323,118 words), 202 annotations
(NEs/eNEs) were extracted and used for training. We
used the open-source tool Doccano
5
for annotation.
The following code lines illustrate the function to
match the year dates for eNE classification.
# eNER function to match year dates
def ener(entities, year):
for entity in entities:
if int(year)<int(entity._.year):
entity._.emerging=True
return entities
5
https://github.com/doccano/doccano
Named Entity Recognition for the Extraction of Emerging Technological Knowledge from Medical Literature
105
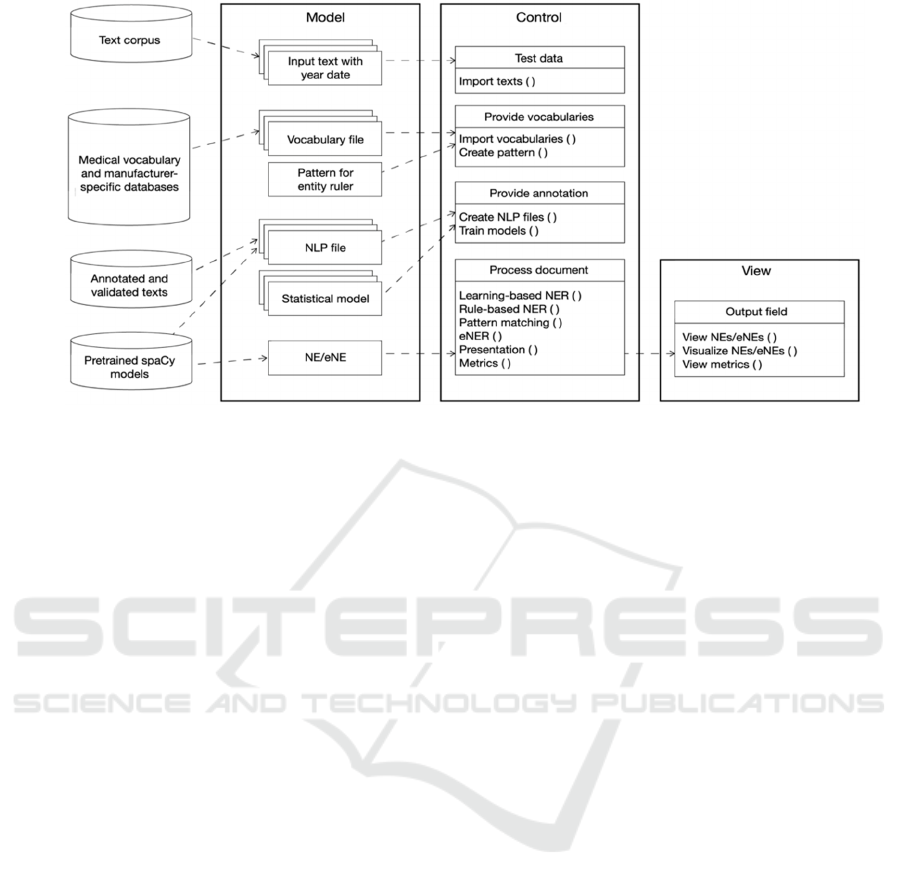
Figure 3: Overall architecture of MedTech-eNER-IRS according to the MVC paradigm.
5 EVALUATION
We evaluated the results of MedTech-eNER-IRS in
respect of its three key components: (1) preparatory
study, (2) learning-based NER, and (3) rule-based
NER/eNER. In this section, we also describe possible
improvements to further develop MedTech-eNER-
IRS.
Preparatory study: We conducted a qualitative
study, in order to generate a training data set as well
as a gold standard for testing the results of MedTech-
eNER-IRS. This preparatory step was based on a
questionnaire with the manually annotated MedTech-
NE/eNE candidates that were presented to a medical
expert (professor in the field of laboratory
diagnostics), who had to choose between “Named
Entity”, “emerging Named Entity”, and “No Medical
Technology”. The expert had difficulties in
classifying many of the cases presented because the
context was missing. The unclear cases and the
reasons for the difficulties were clarified in a second
in-depth interview with the same expert. One key
result was that even if a term represents a medical
technology, it might be irrelevant in the context of an
expert’s specific information need, and thus would
not be a relevant NE/eNE to be presented to the user
of MedTech-eNER-IRS. Additionally, some terms
were borrowed from a domain not known to the
expert, e.g., molecular biology, and it was not clear if
they were relevant for a specific MedTech
application. To improve these deficiencies, methods
to automatically determine the descriptivity of
identified NE/eNE candidates such as TF-IDF and
Word2Vec can be applied.
Learning-based NER: For evaluation of the
learning-based model we used the Scorer method of
spaCy to calculate the metrics Precision, Recall and
F
1
. Test data were created from the corpus by
performing standard text cleaning such as removal of
empty lines, irrelevant head- or footnotes or line
numbers. Since the training corpus was limited, the
spaCy output during training showed an error and the
quality of the statistical model was low (F
1
: 0.39,
Precision: 0.42, Recall: 0.35), which was accepted for
the experimental setup, but would be solved in future
setups by increasing the corpus size.
Rule-based NER/eNER: Both on the basis of our
vocabulary file of MedTech terms, including the
naming patterns we found in the manufacturer-
specific databases, MedTech-eNER-IRS returned
relevant hits. False-positive results occurred several
times, mostly due to homonyms or the naming pattern
<product name> contains <manufacturer name>.
Evaluation was not metrics-based due to the low
number of validated MedTech-NEs/eNEs.
6 DISCUSSION
We have discussed the modeling and implementation
of MedTech-eNER-IRS for the automatic recognition
of MedTech-NEs/eNEs. This constitutes the first
foundation for an IR system that is capable of
identifying entities that represent medical
technologies in unknown text documents. Since we
identified emerging technology to be a specific key
information need of medical experts, we designed
MedTech-eNER-IRS to distinguish between
MedTech-NEs and MedTech-eNEs, in order to
KEOD 2022 - 14th International Conference on Knowledge Engineering and Ontology Development
106

support the retrieval of the most recent MedTech
entities, before they are included in controlled
vocabularies. Within the restricted definition of
medical technology, we set in advance, the chosen
solution path – i.e., the combination of a learning-
based and a rule-based NER approach – and the
limited corpus size, we conclude the following: The
task is basically solvable using the approach of our
MedTech-eNER-IRS, but this needs to be improved
in terms of: (1) The size of the text corpus and the
number of MedTech-NE candidates for training
(learning-based NER): these restrictions led to an
impasse in terms of the use of metrics for MedTech-
eNER-IRS performance evaluation; (2) The
sophistication of the entity ruler (rule-based NER):
these restrictions led to the limitation of MedTech-
eNER-IRS in its recognition of simple MedTech
terms such as tubes, gloves, pipette tips, as well as in
its inability to recognize terms that are non-exact
wording and to avoid false-positives through
homonyms like e.g., chain (chain reaction); (3) The
consideration of naming patterns (rule-based NER):
the chosen approach led to meaningful hits, e.g.
Sysmex KX-21N, QIAmp DNA blood mini reagents,
Karius diagnostic test; restrictions led to false-
positives, mostly in cases where the name of a
MedTech product contains the manufacturer’s name.
7 CONCLUSION AND FUTURE
WORK
MedTech-eNER-IRS is being further developed
against the background of limitations discussed in
Section 6, as well as in terms of further observations
that go beyond the definition of medical technology
assumed here. We name three strategies for
improving the performance of MedTech-eNER-IRS:
(1) refining the machine-learning model, (2)
supporting annotation of training data by
automatically determining the descriptivity of NE
candidates, and (3) using procedural representations
of technological descriptions.
To improve our limited NER/eNER machine-
learning model, we propose to use fine-tuned, pre-
trained language models such as BioBERT (Lee et al.,
2019), and for this purpose also increase the volume
of training data. To alleviate work in manual labelling
in extensive training corpora and increase the
efficiency of an expert-based generation of gold
standards, the approach needs to be automated. To
prevent MedTech-NE candidates to be non-
descriptive and irrelevant to medical experts in
specific contexts, techniques such as TF-IDF (Term
Frequency - Inverse Document Frequency) (cf.
Sammut and Webb, 2011) and Word2Vec (Mikolov
et al., 2013) can be used.
For the sake of a first proof of concept and
simplicity of MedTech-eNER-IRS, we narrowed the
definition of medical technology down to technical
artifacts. The concept of medical technology – as is
the case with the concept of technology in general –
is actually more complex than representing tangibles
only. The more general definitions of health
technology show that intangible aspects are also
relevant, referring to the concepts of knowledge and
science. During corpus analysis and research of
scientific definitions of technology we found
evidence that: (1) Technological terms are embedded
in procedural descriptions within medical articles, in
particular in the common section “materials and
methods”, and (2) the systemic perspective on the
concept of technology supports this observation by
defining it based on the constituents, knowledge,
activities, artifacts (Bullinger 1994). Processing and
mining procedural knowledge from natural-language
data is an additional NLP task that can be used to
extract emerging medical technology. Procedural
knowledge can be described using a semantic
representation, “by specifying semantic elements of a
procedure and their interrelated information” (Zhang
et al., 2012, p. 522). This has been demonstrated by
task-based extraction of procedural knowledge from
text in case of cooking recipes (Schumacher et al.,
2012), with tasks being smaller units of activities, and
activities being indicated by verbs.
REFERENCES
Ali, Z., Waseem, S., Anis, R.A., Anees, M. (2020).
Assessment of cell free mitochondrial DNA as a
biomarker of disease severity in different viral
infections. In Pak J Med Sci, Vol. 36 No. 5, 860-866.
Bachmann, M. (2021). RapidFuzz 2.3.0 documentation.
https://maxbachmann.github.io/RapidFuzz/index.html.
Blauwkamp, T.A., Thair, S., Rosen, … Yang, S. (2018).
Analytical and clinical validation of a microbial cell-
free DNA sequencing test for infectious disease. In Nat
Microbiol, 4, 663-674.
Bullinger, H.-J. (1994). Einführung in das
Technologiemanagement. B.G. Teubner. Stuttgart.
Cheng et al., 2020: Cell-Free DNA in Blood Reveals
Significant Cell, Tissue and Organ Specific injury and
Predicts COVID-19 Severity. In medRxiv preprint.
EU (2012). Directive 98/79/EC of the European Parliament
and of the Council of 27 October 1998 on in vitro
diagnostic medical devices.
EU (2020). Regulation (EU) 2017/745 of the European
Parliament and of the Council of 5 April 2017 on
medical devices, amending Directive 2001/83/EC,
Regulation (EC) No 178/2002 and Regulation (EC) No
Named Entity Recognition for the Extraction of Emerging Technological Knowledge from Medical Literature
107

1223/2009 and repealing Council Directives
90/385/EEC and 93/42/EEC.
Explosion (2022). displaCy Named Entity Visualizer.
FDA (2017). Classification of Products as Drugs and
Devices and Additional Product Classification Issues:
Guidance for Industry and FDA Staff. U.S. Food and
Drug Administration.
FDA (2021a). Premarket Approval (PMA).
https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cf
PMA/pma.cfm.
FDA (2021b). Downloadable 510(k) Files.
https://www.fda.gov/medical-devices/510k-
clearances/downloadable-510k-files.
FDA (2021c). Device Registration and Listing.
https://www.fda.gov/medical-devices/how-study-and-
market-your-device/device-registration-and-listing.
Gan, Y.-J., Sullivan, J.L., Sixbey, J.W. (1993). Detection of
Cell-Free Epstein-Barr Virus DNA in Serum during
Acute Infectious Mononucleosis. In JID 170, 436-439.
Herschbach, D.R. (1995). Technology as Knowledge. In
Journal of Technology Education, Vol. 7 No. 1.
Honnibal, M., Montani, I. (2017). spaCy 2: Natural
language understanding with Bloom embeddings,
convolutional neural networks & incremental parsing.
International Network of Agencies for Health Technology
Assessment (INAHTA) (2022). Health technology.
http://htaglossary.net/health-technology.
Kanakry, A., Hegde, A.M., Durand, C.M., Massie, A.B.,
Greer, A.E., Ambinder, R.F., Valsamakis, A. (2016):
The clinical significance of EBV DNA in the plasma
and peripheral blood mononuclear cells of patients with
or without EBV diseases. In BLOOD, 21 APRIL 2016
x VOLUME 127, NUMBER 16.
Katsutoshi Shoda, K., Ichikawa, D, Fujita, Y., Masuda, K.;
Hiramoto, H., Hamada, J., Arita, T., Konishi, H.,
Kosuga, T., Komatsu, S., Shiozaki, A., Okamoto, K.,
Imoto, I., Otsuji, E. (2017). Clinical utility of
circulating cell-free Epstein–Barr virus DNA in
patients with gastric cancer. In Oncotarget, 2017, Vol.
8, (No. 17), 28796-28804.
Kimura, H., Nishikawa, K., Hoshino, Y., Sofue, A.,
Nishiyama, Y., Morishina, T. (1999). Monitoring of
cell-free viral DNA in primary Epstein-Barr virus
infection. In Med Microbiol Immunol 188, 197-202.
Kluyver, T., Ragan-Kelley, B., Fernando Perez, Granger,
B., Bussonnier, M., Frederic, J., … Willing, C. (2016).
Jupyter Notebooks. In F. Loizides. B. Schmidt (Eds.),
Positioning and Power in Academic Publishing:
Players, Agents and Agendas (87–90).
Laurent, D., Semple, F., Philip J., Lewis, S., Rose, E.,
Black, H.A., Coe, J., Forbes, S.J., Arends, M.J., Dear,
J.W., Aitman, T.J. (2020). Absolute measurement of
the tissue origins of cell-free DNA in the healthy state
and following paracetamol overdose. In BMC Medical
Genomics (2020), 13-60.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C.H.,
Kang, J. (2019). BioBERT: a pre-trained biomedical
language representation model for biomedical text
mining. In Bioinformatics, 2019, 1–7.
Mikolov, T., Chen, K., Corrado, G., Dean, F. (2013).
Efficient Estimation of Word Representations in Vector
Space. arXiv:1301.3781.
National Center for Health Statistics (2010). Health, United
States, 2009. Hyattsville, MD.
Nawroth, C., Engel, F., Eljasik-Swoboda, T., Hemmje, M.
(2018). Towards Enabling Emerging Named Entity
Recognition as a Clinical Information and
Argumentation Support. In DATA 2018, 47-55.
NCBI (2021). PubMed. https://pubmed.ncbi.nlm.nih.gov.
NLM (2021). Medical Subject Headings.
https://meshb.nlm.nih.gov.
Norman, D.A., Draper, S.W. (1986). User Centered System
Design, CRC Press. London.
Nunamaker, J., Chen, M., Purdin, T. (1991). Systems
development in information systems research. In J
Management Information Systems, 7, 89-106.
O’Grady, J. (2019). A powerful, non-invasive test to rule
out infection. In NATURE Microbiology, VOL 4,
APRIL 2019, 554-555.
Patel, N.V, Ghonheim, A. (2011). Managing emergent
knowledge through deferred action design principles:
The case of ecommerce virtual teams. In Journal of Ent
Info Management, 24(5), 424-439.
Perera, N., Dehmer, M., Emmert-Streib, F. (2020). Named
Entity Recognition and Relation Detection for
Biomedical Information Extraction. In Front. Cell Dev.
Biol., 28 August 2020.
Sammut, C., Webb, G.I. (eds.) (2011). Encyclopedia of
Machine Learning. Springer, Boston, MA.
Schumacher, P., Minor, M., Walter, K., Bergmann, R.
(2012). Extraction of Procedural Knowledge from the
Web. A comparison of two workflow extraction
approaches. In WWW 2012 Companion, April 16–20,
2012, Lyon, France. ACM 978-1-4503-1230-1/12/04.
SNOMED International (2021). SNOMED CT Release File
Specifications. https://www.snomed.org/rfs.
Szilágyi M, Pös O, Márton É, Buglyó G, Soltész B, Keserű
J, Penyige A, Szemes T, Nagy B (2020). Circulating
Cell-Free Nucleic Acids: Main Characteristics and
Clinical Application. In Int J Mol Sci. 2020 Sep
17;21(18):6827.
University of Bielefeld (2017). Rationalizing
Recommendations (RecomRatio). http://ratio.sc.cit-
ec.uni-bielefeld.de/projects/recomratio/.
WHO (2022). What is a health technology?
https://www.euro.who.int/en/health-topics/Health-
systems/health-technologies-and-medicines/policy-
areas/health-technology-assessment.
Yamamoto, M., Kimura, H., Hironaka, T., Hirai, K.,
Hasegawa, S., Kuzushima, K., Shibata, M., Morishima,
T. (1994). Detection and Quantification of Virus DNA
in Plasma of Patients with Epstein-Barr Virus-
Associated Diseases. In J Clin Microbiol, Vol. 33, No.
7, July 1994, 1765-1768.
Zhang, Z., Webster, P., Uren, V., Varga, A., Ciravegna, F.
(2012). Automatically Extracting Procedural
Knowledge from Instructional Texts using Natural
Language Processing. In LREC’12.
KEOD 2022 - 14th International Conference on Knowledge Engineering and Ontology Development
108
