
Towards Reducing the Need for Annotations in Digital Dermatology with
Self-supervised Learning
Fabian Gr
¨
oger
1 a
, Philippe Gottfrois
2
, Ludovic Amruthalingam
2 b
, Alvaro Gonzalez-Jimenez
2
,
Simone Lionetti
1 c
, Alexander A. Navarini
2,3 d
and Marc Pouly
1 e
1
Lucerne University of Applied Sciences and Arts, Rotkreuz, Switzerland
2
Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland
3
Department of Dermatology, University Hospital of Basel, Switzerland
Keywords:
Self-supervised Learning, Pre-training, Transfer Learning, Dermatology, Medical Imaging.
Abstract:
Training supervised models requires large amounts of labelled data, whose creation is often expensive and
time-consuming, especially in the medical domain. The standard practice to mitigate the lack of annotated
clinical images is to use transfer learning and fine-tune pre-trained ImageNet weights on a downstream task.
While this approach achieves satisfactory performance, it still requires a sufficiently large dataset to adjust
the global features for a specific task. We report on an ongoing investigation to determine whether self-
supervised learning methods applied to unlabelled domain-specific images can provide better representations
for digital dermatology compared to ImageNet. We consider ColorMe, SimCLR, BYOL, DINO, and iBOT,
and present preliminary results on the evaluation of pre-trained initialization for three different medical tasks
with mixed imaging modalities. Our intermediate findings indicate a benefit in using features learned by iBOT
on dermatology datasets compared to conventional transfer learning from ImageNet classification.
1 INTRODUCTION
Current artificial intelligence applications to medical
imaging often rely on Convolution Neural Networks
(CNNs) trained in a supervised way. These mod-
els have shown remarkable performance across many
tasks due to their ability to deal with raw image data
(Brinker et al., 2019). CNNs typically require vast
amounts of annotated data to achieve high levels of
performance and robustness. However, medical im-
ages can be hard to obtain: The acquisition process
needs to consider strict regulations, collection biases
should be appropriately mitigated (Groh et al., 2021),
and examples of rare conditions are challenging to
find.
Dermatology is a field of medicine where image
analysis can have a significant impact since many
pathologies are visible with a naked eye and can eas-
ily be photographed. Two main types of images are
a
https://orcid.org/0000-0002-9699-688X
b
https://orcid.org/0000-0001-5980-5469
c
https://orcid.org/0000-0001-7305-8957
d
https://orcid.org/0000-0001-7059-632X
e
https://orcid.org/0000-0002-9520-4799
usually considered: dermoscopy pictures taken with
a dedicated device called “dermatoscope”, which is
almost in direct contact with the skin and offers mag-
nification, and ordinary clinical images, which are in-
stead collected with cameras that are not specifically
designed for dermatology.
Although a diagnosis is usually reported in patient
files, one typically needs to recruit and train several
dermatologists to do a tedious labeling job if more de-
tailed information is required. Moreover, for several
medical conditions, it is hard to get experts to agree on
a common answer (Jacob et al., 2021). This explains
why the most popular general-purpose image dataset,
ImageNet (Deng et al., 2009), has 200 times more pic-
tures than the biggest public dermoscopy dataset and
2000 times more than the largest public clinical image
collection (ISIC, 2016).
Transfer learning can leverage knowledge from a
source task with plenty of data to perform a target
task where data is limited. The most common ap-
proach to transfer learning is pre-training large mod-
els such as residual neural network (ResNet) on huge
labeled image datasets. Models pre-trained on Ima-
geNet are also widely used in the medical domain,
Gröger, F., Gottfrois, P., Amruthalingam, L., Gonzalez-Jimenez, A., Lionetti, S., Navarini, A. and Pouly, M.
Towards Reducing the Need for Annotations in Digital Dermatology with Self-supervised Learning.
DOI: 10.5220/0011532100003523
In Proceedings of the 1st Workshop on Scarce Data in Artificial Intelligence for Healthcare (SDAIH 2022), pages 41-46
ISBN: 978-989-758-629-3
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
41

which is controversial since features extracted from
natural images, may not be ideal representations in
some medical contexts (Matsoukas et al., 2022).
Self-supervised learning (SSL) is a hybrid ap-
proach that strives to combine supervised and un-
supervised learning and is also often used through
pre-training followed by adaptation (Shurrab and
Duwairi, 2021). It aims at learning semantically use-
ful features by creating a supervised objective from a
pool of unlabelled data without the need for human
annotation. This objective is often called the pretext
task. Learnt features can then be used in downstream
tasks where annotated data is scarce. In recent years,
SSL became popular also in the medical domain as
large volumes of unlabelled data are easier to obtain
than their annotated counterparts. Several works have
demonstrated the effectiveness of this approach for
detection and classification (Li et al., 2021), for de-
tection and localization (Sowrirajan et al., 2021), and
for segmentation (Xie et al., 2020).
In this paper, we report intermediate results from
an ongoing investigation on using SSL to mitigate the
need for annotated samples in dermatology. We train
several prominent SSL algorithms on a mixed dataset
of 250’000 skin images, both from dermoscopy and
clinical pictures. We then research the impact of using
these representations as a starting point for different
medical imaging tasks in dermatology.
2 SELF-SUPERVISED LEARNING
TECHNIQUES
In this section, we briefly review the different SSL
techniques considered in this work.
2.1 ColorMe
ColorMe (Li et al., 2020) was specifically designed
for applications in the medical domain and embeds
inductive bias in its two pretext tasks. It uses an en-
coder to map the green channel of an image to a vec-
tor space. This is followed both by a decoder, which
learns to reconstruct the pixel values of the red and
blue channels and by a fully-connected layer, which
learns to predict the overall color distribution of the
red and blue channels.
2.2 SimCLR
A simple framework for contrastive learning of visual
representations (SimCLR) (Chen et al., 2020) pro-
poses to obtain different views of the same image with
heavy data augmentation, called positive samples, and
minimize the distance between their representations.
At the same time, the distance between views of dif-
ferent images in the same batch, the negative sam-
ples, is maximized. The encoder is typically a ResNet
architecture followed by a projection head to be dis-
carded after training.
2.3 BYOL
Bootstrap your own latent (BYOL) (Grill et al., 2020)
compares embeddings of image views obtained from
two networks, removing the need for negative sam-
ples. The first network is termed online and consists
of an encoder, a projection head, and a prediction
head. The second network is known as target, has the
same architecture as the online network except for the
prediction head, and its weights are an exponential
moving average of the online network weights. The
training objective is to match the online network’s
output with the target network’s output using mean
squared error.
2.4 DINO
Self-distillation with no labels (DINO) (Caron et al.,
2021) uses a similar principle to BYOL, but it passes
transformations of an image into two separate en-
coders, respectively the student and the teacher net-
works. Unlike previous SSL approaches the en-
coder is a vision transformer (ViT) (Dosovitskiy et al.,
2020). The loss compares the probability outputs of
both networks using cross-entropy. Only the weights
of the student are updated via backpropagation, while
the parameters of the teacher are an exponential mov-
ing average of the student.
2.5 iBOT
Image BERT pre-training with online tokenizer
(iBOT) (Zhou et al., 2022) exploits inherent proper-
ties of ViTs to learn representations capturing local
and global information. Similarly to DINO, iBOT
uses a student network that is trained and a teacher
network which is an exponential moving average. Its
loss function is the sum of two cross-entropies. The
first one compares the output of the two networks
when they are given different views of the same im-
age. And the second one, when the two networks
are both passed the same view, but some patches are
masked for the student, matches the outputs corre-
sponding to those patches.
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
42

Table 1: Hyperparameters of the different self-supervised learning techniques.
Algorithm Backbone # Params Optimizer Batch size Lr. Scheduler
ColorMe ResNet50 23 Mio. SGD 50 1 × 10
−3
-
SimCLR ResNet50 23 Mio. Adam 160 3 × 10
−5
cosine
BYOL ResNet50 23 Mio. Adam 150 3 × 10
−3
cosine
DINO ViT-tiny 5.4 Mio. AdamW 56 5 × 10
−4
cosine
iBOT ViT-tiny 5.4 Mio. AdamW 56 5 × 10
−4
cosine
3 EXPERIMENTAL SETUP
3.1 Pre-training Data
The training data includes both dermoscopy and clini-
cal images, with the hypothesis that there are common
patterns in skin pictures taken at different magnifica-
tions. In total, we use 242’039 images from both pub-
lic and private datasets as listed below.
• derm7pt (Kawahara et al., 2019), featuring 2’020
dermoscopy and clinical images of various skin
pigmentation lesions.
• ISIC (ISIC, 2016), which consists of 107’208 der-
moscopy images with a wide spectrum of pig-
mented skin lesions mostly on low-pigmentation
skin. We exclude pictures overlapping with
HAM10000 (Tschandl et al., 2018) as these are
used in a downstream task.
• MED-NODE (Giotis et al., ), which includes 170
clinical images of pigmented skin lesions.
• SD-260 (Sun et al., 2016), containing 12’583 clin-
ical images of 260 different skin conditions.
• Ph2 (Mendonc¸a et al., 2013), containing 200 der-
moscopy images of pigmented skin lesions.
• A private dataset of 119’858 clinical images, re-
flecting the data distribution encountered in a
Swiss hospital. Pictures were taken using diverse
reflex cameras by a trained photographer, were
anonymized, and used with approval (EKNZ-
2018-01074) from an ethical committee according
to Swiss regulations.
3.2 Downstream Tasks
To evaluate the performance of the pre-training meth-
ods, we use three different downstream tasks, which
were not present in the pre-training data.
• Fitzpatrick17k (Groh et al., 2021) is a public
benchmark dataset containing 16’577 clinical im-
ages with skin condition annotations and skin type
labels based on the Fitzpatrick scoring system.
The dataset contains labels with different granu-
larity. This study used the coarsest level, which
splits skin conditions into three main categories.
• PAD-UFES-20 (Pacheco et al., 2020) is a pub-
lic benchmark dataset composed of clinical im-
ages collected from smartphone devices and pa-
tient metadata. The dataset consists of 1’373 pa-
tients, 1’641 skin lesions, and 2’298 images for
six different diagnoses: three skin diseases and
three skin cancers.
• HAM10000 (Tschandl et al., 2018) is a public
benchmark dataset consisting of 10’015 dermo-
scopic images gathered from different cohorts.
The collected cases include a representative sam-
ple of seven diagnostic categories of pigmented
lesions.
All non-test data for downstream tasks were randomly
split into training and validation sets with size 85%
and 15% respectively. Each downstream task was
evaluated using the test set defined by the dataset au-
thors.
3.3 Architectures
The SSL algorithms considered can be split into two
groups based on the backbone architecture they work
with, which can be a CNN or a ViT. To ensure a fair
comparison, at least within the same group, we used
the very same model when possible. To further pro-
mote the correspondence between the two groups, we
selected architectures that perform similarly on Ima-
geNet. For the CNN-based models we chose ResNet-
50, and for the ViT-based ones a tiny vision trans-
former (Dosovitskiy et al., 2020) with patch size of
16 × 16. The number of trainable parameters is there-
fore roughly 23 Mio. for ResNet-50 and 5.4 Mio. for
ViT-tiny.
3.4 Hyperparameters
Table 1 gives some details about the hyperparameters
for pre-training. All images are resized to 224 × 224
pixels and normalized. Further, the models are trained
Towards Reducing the Need for Annotations in Digital Dermatology with Self-supervised Learning
43

Table 2: Macro-averaged F1 scores of various models and a baseline on the hold-out test set of the three open-source derma-
tology downstream tasks. After adding a linear layer, both freezing and fine-tuning the backbone are considered.
Evaluation Pre-training Fitzpatrick17k PAD-UFES-20 HAM10000
Linear Eval.
Stratified sampling 33.4 % 18.0 % 14.0 %
ImageNet 51.0 % 49.7 % 54.1 %
ColorMe 44.8 % 42.2 % 47.0 %
SimCLR 37.0 % 32.7 % 28.2 %
BYOL 48.1 % 34.4 % 44.4 %
DINO 46.7 % 44.2 % 57.2 %
iBOT 53.0 % 58.2 % 72.0 %
Fine-tuned
ImageNet 72.1 % 61.5 % 79.0 %
ColorMe 71.0 % 61.7 % 73.1 %
iBOT 73.9 % 62.3 % 82.0 %
until the validation loss does not improve consecu-
tively over five epochs. A full scan of hyperparameter
space is not performed as this exceeds the scope of
this paper and our available computational resources.
However, we ensure that all models converge to a
suitable solution by manually choosing an appropri-
ate optimizer and tuning the learning rate. We use
the same data augmentation policies described in the
original paper introducing each pretext task.
3.5 Evaluation
To compare the representations learned by different
pre-training strategies, we test them in two ways: us-
ing linear evaluation on frozen embeddings and fine-
tuning the whole model. Both experiments are per-
formed on all downstream tasks. For models using
a ResNet backbone, we add the linear layer after the
last average pooling layer. For ViT-based models, we
follow the (Caron et al., 2021) and add a linear layer
after the concatenation of the class token to the last
four blocks in the model. In the fine-tuning experi-
ment, the backbone and the linear classification head
are trained together.
Finally, to better understand the utility of self-
supervised pre-training in different low-data regimes,
we also train a simple k-nearest neighbor (kNN) clas-
sifier that acts as a few-shot learner for the down-
stream tasks. This classifier learns from a random
subset of the downstream task’s labeled data and is
evaluated on the same hold-out test set as the linear
evaluation and the fine-tuning.
4 RESULTS
Scores that probe the ability of self-supervised pre-
trained features to generalize to the three downstream
tasks are reported in table 2. The upper half shows
the performance of frozen pre-trained embeddings
upon linear evaluation. All models show better results
compared to the random stratified sampling base-
line, which randomly samples the prediction indepen-
dently of the input data with empirical class proba-
bilities determined from the training set. However,
we also observe that the results of ColorMe, SimCLR
and BYOL are worse than the performance achieved
by using ImageNet features. This indicates that mod-
els with a CNN backbone do not profit from domain-
specific pre-training in our setting and are better off
using general features such as the ones from Ima-
geNet. A possible explanation for this is that such
models have a strong inductive bias which bene-
fits more from initialization using general pre-trained
weights (Matsoukas et al., 2022). Features from a ViT
backbone yield less uniform patterns in comparison
with ImageNet. DINO is only able to outperform the
general features in one of three tasks and only by 3%.
On the other hand, the results clearly show that iBOT
performs well across all tasks and yields an improve-
ment over ImageNet initialization by 2%, 9.5%, and
17.9%, respectively.
The lower half of table 2 summarizes the per-
formance upon fine-tuning the pre-trained features.
Here we only report results for the reference Ima-
geNet model and the two models which obtained the
highest average scores in the CNN and ViT classes
of SSL approaches, i.e. ColorMe and iBOT. Simi-
lar to the results from the linear evaluation, we ob-
serve that using iBOT as initialization yields the best
performance for all downstream tasks. Furthermore,
we notice that the gap between ColorMe and iBOT
is smaller than using linear evaluation, indicating that
with enough training data and trainable parameters,
similar features can be learned. Comparing the per-
formance in the linear and fine-tuned evaluation, we
can also see that the improvement for iBOT on PAD-
UFES-20 and HAM10000 is noticeably smaller with
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
44
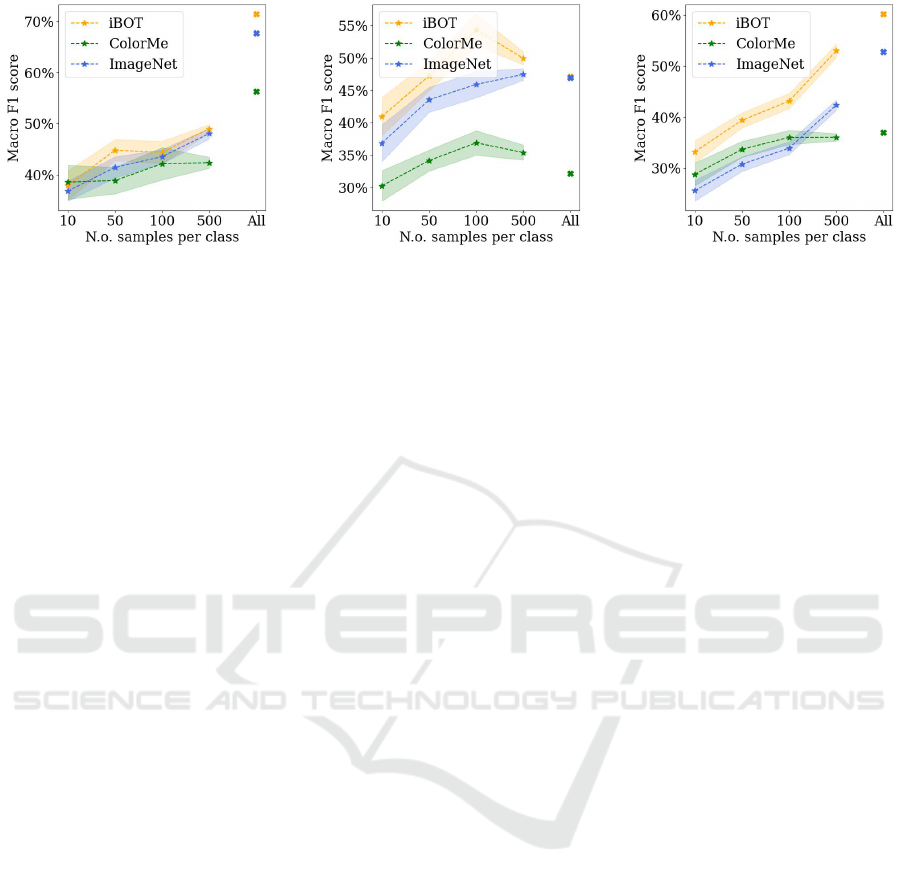
(a) Fitzpatrick17k (b) PAD-UFES-20 (c) HAM10000
Figure 1: Results of a kNN classifier on pre-trained representations when varying the number of samples per class for all three
downstream tasks.
respect to other methods. This suggests that the inher-
ent structure of the dataset labels was already learned
in the pre-training phase, and only minor adaptions to
the existing features could be done.
Finally, figure 1 shows the results of adding a kNN
classifier to the pre-trained ColorMe, iBOT and Im-
ageNet features upon changing the training dataset
size. The results achieved by iBOT outperform, on
average, the ones from ImageNet over all three down-
stream tasks, indicating that its features are very com-
petitive also in low data regimes.
5 CONCLUSION
In this paper, we set out to investigate whether
features from domain-specific self-supervised pre-
training yield a benefit over general-purpose ones
such as ImageNet weights, which are currently the
de facto standard in the medical domain. The re-
sults achieved so far indicate that there might be an
advantage in SSL initialization, especially when us-
ing iBOT. However, we currently cannot conclude
whether this benefit can be traced back to the pre-
training strategy or the difference in model architec-
ture. An indication in favor of the former is that
DINO, which is also based on ViTs, did not outper-
form ImageNet initialization. In the future, we plan
ablation experiments to determine if the performance
gain is really due to the pre-training task or influenced
by the different architecture.
REFERENCES
Brinker, T. J., Hekler, A., Enk, A. H., Berking, C., Hafer-
kamp, S., Hauschild, A., Weichenthal, M., Klode,
J., Schadendorf, D., Holland-Letz, T., von Kalle, C.,
Fr
¨
ohling, S., Schilling, B., and Utikal, J. S. (2019).
Deep neural networks are superior to dermatologists
in melanoma image classification. European Journal
of Cancer, 119:11–17.
Caron, M., Touvron, H., Misra, I., J
´
egou, H., Mairal, J., Bo-
janowski, P., and Joulin, A. (2021). Emerging Prop-
erties in Self-Supervised Vision Transformers. pages
9650–9660.
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020).
A Simple Framework for Contrastive Learning of Vi-
sual Representations. In Proceedings of the 37th In-
ternational Conference on Machine Learning, pages
1597–1607. PMLR.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). Imagenet: A large-scale hierarchical
image database. In 2009 IEEE Conference on Com-
puter Vision and Pattern Recognition, pages 248–255.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., Uszkoreit, J., and Houlsby,
N. (2020). An Image is Worth 16x16 Words: Trans-
formers for Image Recognition at Scale.
Giotis, I., Molders, N., Land, S., Biehl, M., Jonkman,
M., and Petkov, N. Med-node: A computer-
assisted melanoma diagnosis system using non-
dermoscopic images”. Expert Systems with Applica-
tions, 42:6578–6585.
Grill, J.-B., Strub, F., Altch
´
e, F., Tallec, C., Richemond, P.,
Buchatskaya, E., Doersch, C., Avila Pires, B., Guo,
Z., Gheshlaghi Azar, M., Piot, B., kavukcuoglu, k.,
Munos, R., and Valko, M. (2020). Bootstrap Your
Own Latent - A New Approach to Self-Supervised
Learning. In Advances in Neural Information Process-
ing Systems, volume 33, pages 21271–21284. Curran
Associates, Inc.
Groh, M., Harris, C., Soenksen, L., Lau, F., Han, R., Kim,
A., Koochek, A., and Badri, O. (2021). Evaluating
Deep Neural Networks Trained on Clinical Images in
Dermatology with the Fitzpatrick 17k Dataset. pages
1820–1828. IEEE Computer Society.
ISIC (2016). ISIC Archive. https://www.isic-archive.com/.
Accessed: 2022-05-20.
Jacob, J., Ciccarelli, O., Barkhof, F., and Alexander, D. C.
(2021). Disentangling human error from the ground
truth in segmentation of medical images. ACL.
Towards Reducing the Need for Annotations in Digital Dermatology with Self-supervised Learning
45

Kawahara, J., Daneshvar, S., Argenziano, G., and
Hamarneh, G. (2019). Seven-point checklist and skin
lesion classification using multitask multimodal neu-
ral nets. IEEE Journal of Biomedical and Health In-
formatics, 23(2):538–546.
Li, X., Hu, X., Qi, X., Yu, L., Zhao, W., Heng, P.-A.,
and Xing, L. (2021). Rotation-Oriented Collabora-
tive Self-Supervised Learning for Retinal Disease Di-
agnosis. IEEE Transactions on Medical Imaging,
40(9):2284–2294.
Li, Y., Chen, J., and Zheng, Y. (2020). A Multi-Task Self-
Supervised Learning Framework for Scopy Images. In
2020 IEEE 17th International Symposium on Biomed-
ical Imaging (ISBI), pages 2005–2009. ISSN: 1945-
8452.
Matsoukas, C., Haslum, J. F., Sorkhei, M., S
¨
oderberg, M.,
and Smith, K. (2022). What Makes Transfer Learning
Work For Medical Images: Feature Reuse & Other
Factors. Technical Report arXiv:2203.01825, arXiv.
Mendonc¸a, T., Ferreira, P. M., Marques, J. S., Marcal,
A. R. S., and Rozeira, J. (2013). PH2 - A dermo-
scopic image database for research and benchmark-
ing. In 2013 35th Annual International Conference of
the IEEE Engineering in Medicine and Biology Soci-
ety.
Pacheco, A. G. C., Lima, G. R., Salom
˜
ao, A. S., Krohling,
B., Biral, I. P., de Angelo, G. G., Alves Jr, F. C. R.,
Esgario, J. G. M., Simora, A. C., Castro, P. B. C.,
Rodrigues, F. B., Frasson, P. H. L., Krohling, R. A.,
Knidel, H., Santos, M. C. S., do Esp
´
ırito Santo, R. B.,
Macedo, T. L. S. G., Canuto, T. R. P., and de Barros,
L. F. S. (2020). PAD-UFES-20: A skin lesion dataset
composed of patient data and clinical images collected
from smartphones. Data in Brief, 32:106221.
Shurrab, S. and Duwairi, R. (2021). Self-supervised learn-
ing methods and applications in medical imaging
analysis: A survey. arXiv:2109.08685 [cs, eess].
arXiv: 2109.08685.
Sowrirajan, H., Yang, J., Ng, A. Y., and Rajpurkar, P.
(2021). MoCo-CXR: MoCo Pretraining Improves
Representation and Transferability of Chest X-ray
Models. arXiv:2010.05352 [cs].
Sun, X., Yang, J., Sun, M., and Wang, K. (2016). A
benchmark for automatic visual classification of clin-
ical skin disease images. In Leibe, B., Matas, J.,
Sebe, N., and Welling, M., editors, Computer Vision
– ECCV 2016, pages 206–222, Cham. Springer Inter-
national Publishing.
Tschandl, P., Rosendahl, C., and Kittler, H. (2018). The
HAM10000 dataset, a large collection of multi-source
dermatoscopic images of common pigmented skin le-
sions. Scientific Data, 5(1):180161.
Xie, Y., Zhang, J., Liao, Z., Xia, Y., and Shen, C.
(2020). PGL: Prior-Guided Local Self-supervised
Learning for 3D Medical Image Segmentation.
arXiv:2011.12640 [cs].
Zhou, J., Wei, C., Wang, H., Shen, W., Xie, C., Yuille, A.,
and Kong, T. (2022). ibot: Image bert pre-training
with online tokenizer. International Conference on
Learning Representations (ICLR).
SDAIH 2022 - Scarce Data in Artificial Intelligence for Healthcare
46
