
Assessing the Impact of Deep End-to-End Architectures in Ensemble
Learning for Histopathological Breast Cancer Classification
Hasnae Zerouaoui
1a
, Ali Idri
1,2 b
and Omar El Alaoui
2c
1
Modeling, Simulation and Data Analysis, Mohammed VI Polytechnic University, Benguerir, Morocco
2
Software Project Management Research Team, ENSIAS, Mohammed V University in Rabat, Morocco
Keywords: Deep Learning, Machine Learning, Ensemble Learning, Computer Vision, Breast Cancer, Whole Slide
Images.
Abstract: One of the most significant public health issues in the world and a major factor in women's mortality is breast
cancer (BC). Early diagnosis and detection can significantly improve the likelihood of survival. Therefore,
this study suggests a deep end-to-end heterogeneous ensemble approach by using deep learning (DL) models
for breast histological images classification tested on the BreakHis dataset. The proposed approach showed a
significant increase of performances compared to their base learners. Thus, seven DL architectures (VGG16,
VGG19, ResNet50, Inception_V3, Inception_ResNet_V2, Xception, and MobileNet) were trained using 5-
fold cross-validation. Thereafter, deep end-to-end heterogeneous ensembles of two up to seven base learners
were constructed based on accuracy using majority and weighted voting. Results showed the effectiveness of
deep end-to-end ensemble learning techniques for breast cancer images classification into malignant or
benign. The ensembles designed with weighted voting method exceeded the others with an accuracy value
reaching 93.8%, 93.4%, 93.3%, and 91.8% through the BreakHis dataset's four magnification factors: 40X,
100X, 200X, and 400X respectively.
1 INTRODUCTION
Cancer is considered among the most serious health
issues in the world. In 2020, more than 19.3 million
new cancer cases are diagnosed and nearly 10 million
deaths are declared (Sung et al., 2021). By far the
most eminent and leading cause of death in women
worldwide is breast cancer (BC), with 2.3 million
women affected by it in 2020 (Sung et al., 2021).
Early detection and diagnosis of this disease are
essential to minimise morbidity in women. Even
though X-ray, MRI (Magnetic Resonance Imaging),
ultrasound, and other imaging techniques have been
used for more than 40 years to detect breast cancer
(Stenkvist et al., 1978), biopsy techniques have
always been the most commonly used method for
correctly diagnosing breast cancer. The procedure
entails collecting tissue samples, mounting them on
microscopic glass slides, and staining them for
visualization purposes (Mitko Veta, 2014).
Pathologists then examine and diagnose the
a
https://orcid.org/0000-0001-7268-8404
b
https://orcid.org/0000-0002-4586-4158
c
https://orcid.org/0000-0002-4395-9989
histopathological images to affirm the diagnosis of
breast cancer. (Mitko Veta, 2014). Manual
examination of large-scale histological images,
however, is a difficult process due to changes in
appearance, structure, and textures(Li et al., 2019), it
is time-consuming, and usually depends on human
subjective interpretation since the level of experience
of the pathologists involved may have an impact on
the results of the analysis. Therefore, computer-aided
(Aswathy and Jagannath, 2017) analysis of
histological images are crucial in the diagnosis of
breast cancer.
Deep learning (DL) has recently outperformed a
variety of machine learning (ML) models for the
medical image analysis tasks, such as classification
(Mardanisamani et al., 2019), detection (Herent et al.,
2019), and segmentation (Lateef and Ruichek, 2019).
When compared to other types of ML classifiers, DL
has the advantage of being able to achieve results that
are similar or better than human performance. DL
techniques have been used in computer vision (Xie et
Zerouaoui, H., Idri, A. and El Alaoui, O.
Assessing the Impact of Deep End-to-End Architectures in Ensemble Learning for Histopathological Breast Cancer Classification.
DOI: 10.5220/0011574400003335
In Proceedings of the 14th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2022) - Volume 1: KDIR, pages 109-118
ISBN: 978-989-758-614-9; ISSN: 2184-3228
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
109

al., 2018), biological science (Gulshan et al., 2016),
and many other domains to solve problems of
traditional feature extraction. More specifically the
Deep convolutional neural networks (DCNNs) have
been widely recognized as one of the most efficient
tools for image classification since they provide
numerous advantages over traditional solutions,
including an end-to-end architecture that relieves
users from hand-crafted feature extraction tasks. (Jia
et al., 2020).
Despite its popularity, a single DCNNs model can
only extract a limited amount of discriminative
features, resulting in suboptimal classification
performance. To improve classification accuracy,
ensembles of DCNN architectures have been
designed to learn the representation of
histopathological images from various perspectives.
Since then, many researchers started investigating
deep ensemble learning techniques to ameliorate the
performances. For instance, the study (El Ouassif et
al., 2021), proposed multiple heterogeneous
ensembles for breast cancer classification on three
datasets (WDBC, Wisconsin, WPBC). From seven
classifiers (KNN, Decision Three, MLP, SVM, SVM-
PUK, SVM-RBF, S-LK, SVM-NP), the authors,
selected and constructed ensembles based on two
selection strategies: (1) selection by accuracy and
diversity, and (2) selection by only accuracy. Then,
the constructed ensembles were combined using the
majority voting method. According to the findings of
this study, investigating both accuracy and diversity
to select ensemble members often resulted in better
performance than designing and building ensembles
without member selection. In (Vo et al., 2019),
authors proposed an ensemble constructed with three
DL models for breast cancer classification tested on
the BreakHis and Bioimaging-205 datasets.
Inception_ResNet_V2 was used to extract features
and gradient boosting trees for classification. the
classifiers were combined using the majority voting
strategy to improve the performance. The accuracy
values reached: 95.1%, 96.3%, 96.9%, 93.8% and
86.75% for the magnification factors (MF) MFs of the
BreakHis dataset and bioimaging dataset respectively.
Some limitations have been revealed in the
studies(Idri et al., 2020),(Vo et al., 2019): (1) the
design of heterogenous ensemble using only one
combination method, (2) except of the study (El
Ouassif et al., 2021), a lack of statistical analysis to
select the outperforming proposed model is
noticeable.
To elevate the burden of those limitations, this
study proposes a deep end-to-end heterogenous
ensemble technique (DEHtE) using seven end-to-end
DL models as base learners for breast
histopathological images classification over the
BreakHis dataset. The proposed approach consists of
combing seven DL techniques of two up to seven DL
models as base learners, based on accuracy using two
voting methods: majority voting by taking the mode
of the distribution of predicted labels, and weighted
voting by taking the average of predicted
probabilities. The seven DL techniques were based on
fine-tunned VGG16, VGG19, ResNet50,
Inception_V3, Inception_ResNet_V2, Xception, and
MobileNet, using a 5-fold cross-validation evaluation
technique.
The performance of the proposed approach was
evaluated using four classification performance
measures (Hosni et al., 2019; Zerouaoui and Idri,
2021a) (accuracy, precision, recall, and F1-score),
Scott Knott (SK) statistical test to group the proposed
ensembles and identify the best cluster, and the Borda
Count voting method to sort and identify the best
performing ensemble. So far as we are
knowledgeable, this study is the first to construct
DEHtE of DL models as base learners based on
accuracy and combined with two voting methods for
histopathological BC classification.
The current study focuses on two research
questions. (RQs):
- (RQ1): Does the deep end-to-end heterogenous
ensembles using voting methods outperform
their base learners?
- (RQ2): What are the suitable number of base
learners to design the deep end-to-end
heterogenous ensembles and the suitable voting
combination method used?
The following are the study's main contributions:
1. Assessing and comparing the performance of the
seven fine-tunned DL end-to-end architectures
over the BreakHis dataset.
2. Constructing DEHtE using one selection criteria:
selection by accuracy.
3. Combining the constructed ensembles using
majority and weighted voting methods.
4. Assessing and comparing the performance of the
designed DEHtE with their base learners over the
BreakHis dataset.
The remainder of this paper is structured as
follows: Section 2 provides an overview of the deep
learning models and ensemble learning techniques
used to develop the proposed approach. Section 3
presents data preparation process. Section 4 provides
the details of the experiment configuration, the
empirical methodology and the abbreviations
followed in this empirical study. Section 5 reports and
discusses the empirical results. Section 6 covers the
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
110

threats of validity of this study. Lastly, Section 7
Outlines the conclusion and ongoing work.
2 BACKGROUND
This section delves into some of the key principles
used in this empirical study, starting with the concept
underlying the experiment's various DCNN
architectures, then discussing ensemble learning
techniques.
2.1 DCNNS Architectures
This subsection introduces and defines the different
DCNNs used in this proposal.
VGGNet (2014): VGGNet finished first in the
2014 ImageNet Challenge (Simonyan and Zisserman,
2015). There is a total of six VGGNet architectures.
The most common are VGG-16 and VGG-19. The
VGG architectures are made up of convolutional
layers with the ReLU, one max pooling layer, and
multiple fully connected layers.
ResNet (2015): Resnet was designed to avoid the
vanishing gradient problem that previous deep
learning models had. (He and Sun, 2016). ResNet is
built with a variety of layer counts: 34, 50, 101, 152,
and 1202. ResNet50. The most popular ResNet have
49 convolution layers and one fully connected layer
at the end.
Inception_V3: Inception V3 is a member of the
Inception deep architectures, has the same
architecture as InceptionV1 and InceptionV2 with a
few changes. Inception V3 has 42 layers anwithd a
fixed input size of 299x299 by default (Szegedy et al.,
2014). It has a parallel convolutional layer block with
three different filter sizes (1x1, 3x3, 5x5).
Inception_ResNet_V2: Inception ResNet V2 is a
convolutional neural network with 164 layers and an
input size of 299x299. It is based on a combination of
the Inception architecture and the Residual
connection. Multiple convolutional filters are
combined with residual connections in this
architecture(Szegedy et al., n.d.).
MobileNet_V2: As a source of non-linearity,
MobileNet V2 filters features using lightweight
depthwise convolution layers. It begins with a fully
convolutional layer with 32 filters, then proceeds to
19 residual bottleneck layers(Sandler et al., 2018).
Xception: Xception is an architecture with 36
convolutional layers that serve as the network's
feature extraction foundation. It consists of a linear
stack of separable depth-wise convolution layers with
residual connections. This makes it very simple to
define and modify the architecture. Xception has a
total of 22.8 million trainable parameters. (Chollet,
2017).
2.2 Ensemble Learning
In 1965, Ensemble Learning was proposed for
classification tasks(Nilsson, 1965). It is based on the
concept of training several base learners as ensemble
members and combining their predictions into a
single output that should outperform any other
ensemble member with uncorrelated error on the
target dataset on average(Zhou, 2012). An ensemble
is composed of several base learners. A base learning
algorithm, which can be a decision tree, a neural
network, or another type of learning algorithm, is
typically used to generate base learners from training
data. Learners of the same type, leads to
homogeneous ensembles, and learner of different
algorithms are leading to heterogenous ensembles.
An ensemble's generalization ability is commonly
much higher than that of base learners.
The current study uses heterogenous ensembles with
majority and weighted voting methods to combine
predictions of the DL base learners. Every classifier
vote for one class label in majority voting, and the
final output class label is the one that receives more
than half of the votes. Weighted voting, on the other
hand, takes into account the probabilities thrown by
each classifier; these probabilities are weighted and
averaged, and the winning class is the one with the
highest weighted and averaged probability. (Zhou,
2012)
3 DATA PREPARATION
In this section, we will present the process to prepare
the histological BreakHis dataset consisting of five
steps: data acquisition, data pre-processing using
Contrast Limited Adaptive Histogram Equalization
(CLAHE), intensity normalization and data
augmentation. Since the same process was followed
in the studies (Zerouaoui et al., 2021; Zerouaoui and
Idri, 2022), we therefore summarize it as described
below.
The BreakHis dataset contains haematoxylin-eosin-
stained breast histological slides which refer to
microscopic examination of a biopsy to study the
appearances of the cancer. It is constituted of 7,909
breast histopathological images collected from 82
patients at different magnification factors (MF) such
as 40X, 100X, 200X, and 400X with effective pixel
sizes of 0.49 m, 0.20 m, 0.10 m, and 0.05 m All
images are stored in TrueColor (24-bit colour depth,
8 bits per colour channel) three channel format
Assessing the Impact of Deep End-to-End Architectures in Ensemble Learning for Histopathological Breast Cancer Classification
111

(RGB). A pathologist examined the images and
identified the most relevant region of interest,
excluding out-of-focus images and images with
undesirable areas such as black borders or text
annotations. Each image's final size is 700x460 pixels
in PNG format (Gandomkar et al., 2018)(Spanhol et
al., 2016). The dataset is divided into benign tumor
(adenosis, fibroadenoma, phyllodes tumor, and
tubular adenoma) and malignant tumor (ductal
carcinoma, lobular carcinoma, mucinous carcinoma,
and papillary carcinoma) (Zhu et al., 2019). One of
the benefits of the BreakHis dataset is the use of four
magnification factors, which allows for the detection
of various cancer types and subtypes (Alom et al.,
2019).
We used intensity normalization and CLAHE
(Zerouaoui and Idri, 2021b, 2022) to improve the
image quality. The data augmentation was used to
deal with the imbalanced data by incorporating
geometric transformations (Jiang et al., 2019)(Hosni
et al., 2019) since the number of images in each
category non-cancerous (2480 images) and cancerous
(5429 images) are imbalanced with 70% of the
images represent the malignant class.
4 EMPIRICAL DESIGN
In this section, the empirical design proposed to build
the DEHtE is provided, starting with the process
tailed to construct and evaluate the designed proposed
approach including the experiment configuration and
the statistical tests such as Scott Knott (SK) and borda
count voting method (Bhering et al., 2008; Black,
1976). Next, the abbreviations employed to refer to
the designed ensembles and their base learners are
given. At last, a framework of the experiment and the
empirical design will be described.
4.1 Experiment Configuration
After preprocessing, the data is divided into two sets
(train and test sets) with partitions of (80%, 20%),
respectively. Then, using transfer learning for fine-
tunning, seven DL techniques (VGG16, VGG19,
ResNet50, Inception V3, Inception ResNet V2,
Xception, and MobileNet) were trained for each MF
of the BreakHis dataset (Nguyen et al., 2020). The
models were trained on the train set and tested on the
test set using stratified K-fold cross validation with
k=5.
The seven DL techniques were trained using the
following configurations:
1. The histological images were resized to 224x224
pixels for all DL architectures except for both
Inception_V3 and Inception-ResNet_V2 that
was resized to 299x299 pixels.
2. The transfer learning technique for fine tuning
was used for the training of the seven DL models.
The pre-trained architecture's top convolutional
layers were frozen (ImageNet weights were used)
and the extracted features were fed to an
Artificial neural network (ANN) classifier.
3. The ANN classifier used is built with a fully
connected layer of 256 neurons with RELU
activation function. To avoid overfitting, we
added a dropout layer with rate set to 50%, a
dense layer of two neurons with SoftMax
activation function, and a dense layer of two
neurons with SoftMax activation function. The
batch size was set to 32 and the number of epochs
to 200. As for the optimization, we used the
Adam optimizer with a learning rate of 0.0001
that decreases during training. Finally, we added
L2 regularization to penalize large weight values
and reducing model overfitting.
The end-to-end architectures of this experiment are
trained and tested in Python using the Keras and
Tensorflow deep learning frameworks and run on a
TPU processing unit with 8 cores, 35 GB of RAM,
and a Linux-based OS provided by Google in Colab
Notebook. The training process of the seven DL
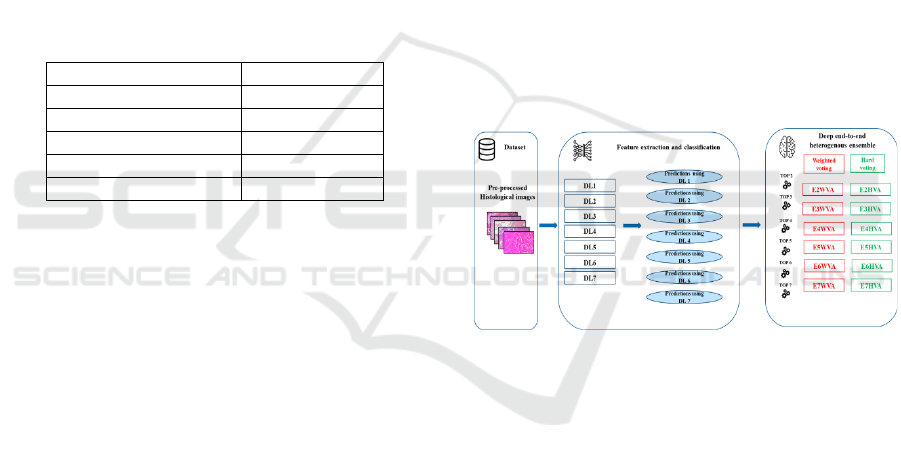
models is depicted in Figure 1.
Figure 1: Training process of the seven DL techniques.
4.2 Statistical Tests
Scott Knott (SK) is a hierarchical clustering algorithm
proposed by Scott and Knott in 1974 (Idri et al.,
2018). It is a quick and efficient way to perform
multiple comparisons with no ambiguity (Bhering et
al., 2008). Because of its simplicity and robustness,
the SK test is the most commonly used hierarchical
clustering algorithm when compared to other
statistical tests (Spanhol et al., 2016)(Hamza and
Larocque, 2005). The SK test was used in this study
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
112
to cluster the base learners and DEHtE techniques
based on accuracy to see if there was a significant
difference between them.
Borda count is a single-winner election method in
which candidates are assigned points in descending
order based on their ranking. The point values for all
ranks and votes are added up, and the candidate with
the most points is declared the winner (Emerson,
2013). The Borda count method was used in this
study to find the best performing DEHtE based on
four classification performance measures with equal
weights (accuracy, precision, recall, and f1-score).
4.3 Abbreviation
To help the reader and abbreviate the names of the
various techniques used in this research, we shorten
the names of each DL technique as described in Table
1:
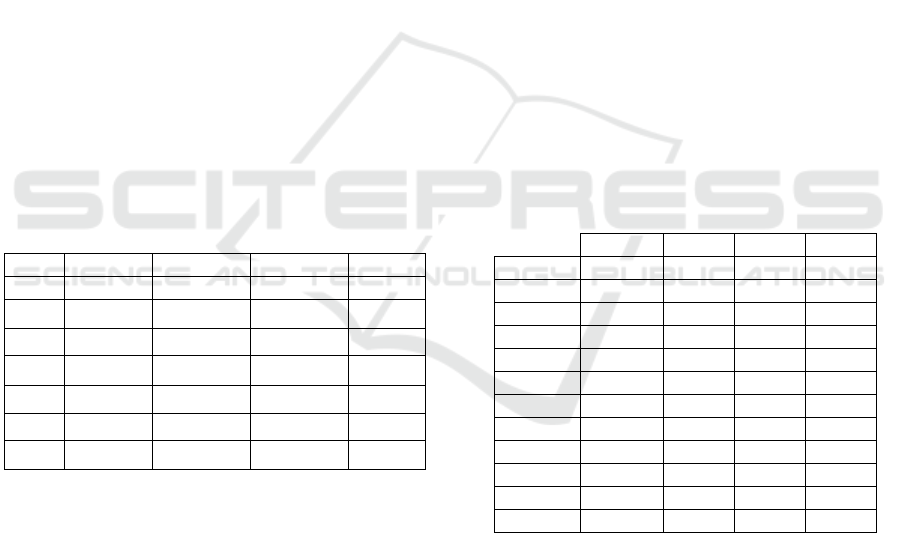
Table 1: The abbreviation of the DL architectures
DL architecture Abbreviations
Xception XCEP
ResNet50 RES50
MobileNet MOBN
Inception_ResNet_V2 INRESV2
Inception_V3 INV3
As for the DEHtE names the following abbreviations
were chosen:
EnHVA: Ensemble of size n combined with hard
voting method and constructed with selection by
accuracy strategy.
EnWVA: Ensemble of size n combined with
weighted voting method and constructed with
selection by accuracy strategy.
Exemple: Ensemble of 6 base learners using weighted
voting with the selection by accuracy strategy is
E6WVA
4.4 Empirical Design
This subsection describes the methodology followed
for the DEHtE method. This experiment consists of
the following seven steps:
• Step 1: evaluating the performance of the seven
deep learning techniques based on accuracy.
• Step 2: Constructing for each MF, DEHtE of 2
up to 7 DL models used as base learners
(combinations of 2, 3, 4, 5, 6, and 7) following
the selection by accuracy strategy which consists
of ranking the seven DL techniques with Borda
count in terms of accuracy, precision, recall and
f1-score, then from top to down constructing
combinations of two, four, five, six and seven. At
the end of this step, we obtain 6 DEHtE for each
MF.
• Step 3: For each MF, apply the majority and
weighted voting methods on all combinations
obtained in step 2, to obtain then, 12 ensembles
for each MF (6 DL end-to-end architecture x 2
voting methods). Then, evaluate their
performance in term of accuracy.
• Step 4: For each MF, apply the SK test on each
12-ensemble obtained in step 3 with the seven
DL end-to-end architectures based on accuracy.
• Step 5: This step entails using Borda count to
rank the variants of the best clusters (obtained in
step 4) based on accuracy, precision, recall, and
F1-score.
Figure 2 describes the process to design the
DEHtE, it consists of three main steps: 1) data
preprocessing, 2) DL models training for both feature
extraction and classification and 3) combining the
DEHtE using the selection by accuracy strategy and
using the two com bination rules: majority (Idri et al.,
2020) and weighted voting .
Figure 2: The main steps to design the deep ent-to-end
heterogenous ensemble.
5 RESULTS AND DISCUSSION
This section describes and compares the performance
of the DEHtE designed using the selection by
accuracy strategy and their base learners over the four
MFs of the BreakHis dataset (40X, 100X, 200X and
400X) and defines the suitable number of base
learners and voting combination methods. To do so,
(1) the performances were analyzed and compared
based on accuracy of all DEHtE, then (2) the
difference of performances was observed using SK
statistical test, (3) the best clusters obtained using SK
test were ranked using Borda count voting method.
Assessing the Impact of Deep End-to-End Architectures in Ensemble Learning for Histopathological Breast Cancer Classification
113
5.1 Performance of Deep End-to-End
Heterogenous Ensembles
In a previous study (Alaoui et al., 2022), the seven
DL architectures including VGG16, VGG19,
ResNet50, Inception_V3, Inception_ResNet_V2,
Xception, and MobileNet were evaluated and
compared using the BreakHis dataset. The main
results have showed that the DL end-to-end
architecture XCEP achieved the highest accuracy
using the 40X MF with a value of 90.91 %, the DL
end-to-end architecture VGG16 achieved 90.43 % on
100X MF, and the DL end-to-end architecture
MOBN performed best on 200X and 400X with
accuracy values of 90.90 % and 89.91 %,
respectively. Furthermore, it is noticeable that the DL
model RES50 underperformed compared to the other
architectures regardless the MF used.
To assess and compare the performances of DEHtE
constructed based on the selection by accuracy
strategy and combined using two voting techniques
(majority and weighted voting), we first rank the
seven DL architectures based on accuracy and then
combine them using majority and weighted voting
following the ranking presented in Table 2. As results
we obtained DEHtE of 2, 3, 4, 5, 6 and 7 DL models
as base learners.
Table 2: The ranking of the seven DL models to design the
DEHtE based on the selection by accuracy strategy.
Rank 40X 100X 200X 400X
1 XCEP VGG16 MOBN MOBN
2 MOBN MOBN XCEP INRES
3 INRES XCEP VGG16 VGG16
4 VGG16 INV3 VGG19 XCEP
5 INV3 INRES INRES INV3
6 VGG19 VGG19 INV3 VGG19
7 RES50 RES50 RES50 RES50
Table 3 indicates the accuracy values of the DEHtE
selected by accuracy using majority and weighted
voting methods over the BreakHis dataset four MFs.
It is revealed that:
- For 40X MF, the best accuracy value obtained
using majority voting was 93.5% reached by
ensemble of size six, and the best accuracy
obtained using weighted voting was 93.8%
reached by ensembles of size six and seven.
Moreover, ensemble of size two shows the worst
accuracy value in both voting methods: majority
and weighted voting with values: 90.9% and 92%,
respectively.
- For 100X MF, the best accuracy value obtained
using majority voting was 92.8% reached by
ensemble of size six, and the best accuracy value
obtained using weighted voting was 93.4%
reached by ensemble of size four. Moreover, the
worst accuracy value obtained using majority
voting was 90.4% reached by ensemble of size
two, and the worst accuracy value obtained using
weighted voting was 92.5% reached by ensemble
of size three.
- For 200X MF, ensemble of size seven shows the
best accuracy value in both voting methods:
majority and weighted voting with values: 93.1%
and 93.3%, respectively. Moreover, ensemble of
size two shows the worst accuracy value in both
voting methods: majority and weighted voting
with values: 90.9% and 92.3%, respectively.
- For 400X MF, the best accuracy value obtained
using majority voting was 91.5% reached by
ensemble of size six, and the best accuracy
obtained using weighted voting was 91.8%
reached by ensembles of size four and seven.
Moreover, ensemble of size two shows the worst
accuracy value in both voting methods: majority
and weighted voting with values: 89.9% and
90.9%, respectively.
Table 3: Results based on accuracy of DEHtE selected by
accuracy over the BreakHis dataset.
40X 100X 200X 400X
E2HVA 90.90 % 90.40% 90.90% 89.90%
E3HVA 93.20% 92.50% 92.80% 91.30%
E4HVA 93.30% 92.50% 92.70% 91.40%
E5HVA 93.40% 91.80% 93.00% 91.30%
E6HVA 93.50% 92.80% 93.00% 91.50%
E7HVA 93.00% 92.10% 93.10% 91.40%
E2WVA 92.00% 93.00% 92.30% 90.90%
E3WVA 93.20% 92.70% 92.90% 91.50%
E4WVA 93.50% 93.40% 93.00% 91.80%
E5WVA 93.70% 92.90% 93.20% 91.40%
E6WVA 93.80% 93.00% 93.20% 91.60%
E7WVA 93.80% 93.10% 93.30% 91.80%
In order to determine whether the DEHtE outperform
their singles, we clustered the constructed ensembles
following the selection by accuracy strategy and
using two voting methods (majority and weighted
voting) with their seven DL models used as base
learners. Figure 3 illustrates the outcomes of the SK
statistical test on the BreakHis dataset. It is observed
that:
- For 40X MF, 6 SK clusters were obtained. The
best cluster includes 10 ensembles out of 12 (all
ensembles except E2WVA which belongs to the
second cluster, and E2HVA which belongs to the
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
114
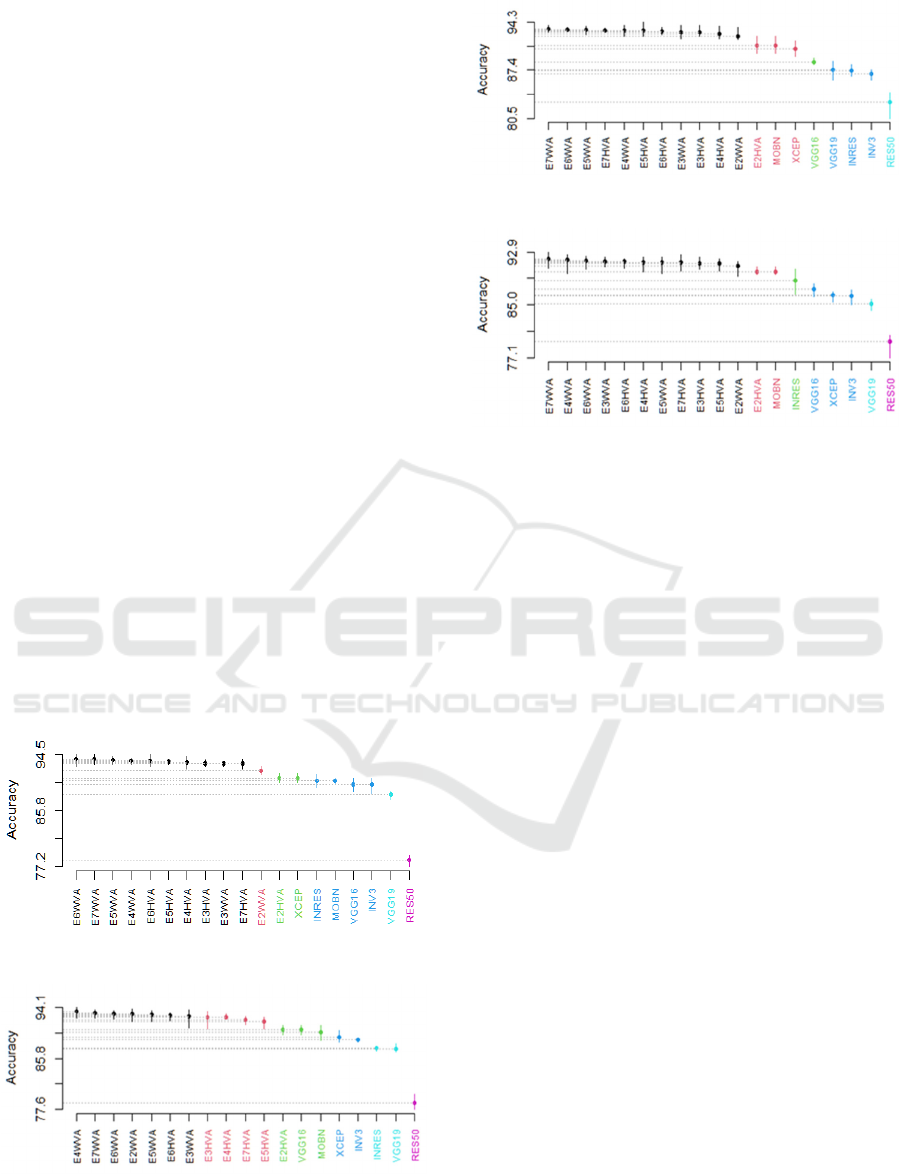
third cluster with the DL model XCEP). The
fourth cluster contains VGG16, INRES, MOBN
and INV3. Moreover, the fifth and the last
clusters contain VGG19 and ResNe50,
respectively.
- For 100X MF, 6 SK clusters were obtained. The
best cluster involves 7 ensembles out of 12
(E7WVA, E6WVA, E5WVA, E4WVA,
E3WVA, E2WVA, E6HVA). The second cluster
contains 4 ensembles (E7HVA, E5HVA,
E4HVA, and E3HVA). E2HVA belongs to the
third cluster with VGG16 and MOBN, the fourth
cluster contains XCEP and INV3. Moreover,
INRES and VGG19 belong to the fifth cluster,
and the last cluster contains RES50.
- For 200X MF, 5 SK clusters were obtained. The
best SK cluster groups 11 ensembles out of 12
(all ensembles except E2HVA which was shown
in the second cluster with MOBN and XCEP).
The third cluster contains VGG16. The fourth
cluster contains VGG19, INRES and INV3.
Lately, RES50 was presented in the last cluster.
- For 400X MF, 6 SK clusters were obtained. The
best SK cluster comprises 11 ensembles out of 12
(all ensembles except E2HVA which was shown
in the second cluster with MOBN). The third
cluster contains INRES. The fourth cluster
contains VGG16, XCEP and INV3. The fifth and
the last clusters contain VGG19 and ResNe50,
respectively.
A) 40X
B) 100X
C) 200X
D) 400X
Figure 3: The SK test of ensembles selected by accuracy
over BreakHis dataset.
To sum up the obtained results, the SK test showed
that:
- For 40X, 10 out of 12 ensembles gave significant
results compared to their base learners (all
ensembles except E2HVA and E2WVA).
- For 100X, 7 out 12 ensembles gave significant
results compared to their base learners (E7WVA,
E6WVA, E5WVA, E6HVA, E4WVA, E3WVA,
E2WVA).
- For 200X and 400X MFs, all ensembles except
E2HVA gave significant results compared to
singles.
The analysis above proves that the designed DEHtE
significantly outperformed their base learners since
they almost always belong to the best or second-best
SK cluster. As results it is recommended to use the
proposed approach to ameliorate the performances.
5.2 Number of Base Learners and
Combination Rule to Use
This subsection is to determine the suitable number
of base learners and combination rule to use in the
design of the DEHtE. To do so, Borda Count voting
method was applied on the basis of the four
performance measures to rank the ensembles
belonging to the best SK cluster. Table 4 displays the
ranking results for the BreakHis dataset.
Assessing the Impact of Deep End-to-End Architectures in Ensemble Learning for Histopathological Breast Cancer Classification
115

Table 4: Borda Count Ranking of Ensembles Selected by
Accuracy on Breakhis.
Ensembles 40X 100X 200X 400X
E7WVA 1 2 1 1
E6WVA 2 3 2 3
E5WVA 3 5 3 6
E6HVA 4 6 4 5
E4WVA 4 1 5 2
E4HVA 5 --- 8 7
E5HVA 6 --- 5 10
E7HVA 7 --- 4 8
E3WVA 7 7 6 4
E3HVA 8 --- 7 9
E2WVA --- 4 9 11
As a summary, the Borda count voting method
showed that E7WVA outperformed all other DEHtE
over the BreakHis dataset MFs, since it was ranked
top one on the three dataset MF (40X, 200X, and
400X) and ranked second for the 100X MF. As results
this study has shown the importance of designing
DEHtE to ameliorate the performances of the
histopathological classification for BC diagnosis
compared to the 7 DL models used as base learners.
Furthermore, it showed that the number of base
learners to design DEHtE plays a significant role
since the ensemble of 7 base learners outperformed
the others, and the ensembles of two base learners
underperformed compared to the others.
Finally, the best combination rule to design the
DEHtE is the weighted voting since the ensemble
E7WVA outperformed the others and achieved an
accuracy of 93.8%, 93.3%, 93.1%, and 91.8% over
the MFS values 40X, 100X, 200X and 400X,
respectively of the BreakHis dataset.
6 THREATS OF VALIDITY
Internal Validity: This study applied a 5-fold cross
validation evaluation technique, which is commonly
used in machine learning to assess a model's ability to
predict new data points(Hosni et al., 2019). Another
internal threat is the use of transfer learning for fine
tuning, which involves freezing all convolutional
base layers with ImageNet weights. Freezing or
tuning some convolutional layers may affect the
performance of classifiers.
External Validity: The external threat's aim is to see
if the study's findings are applicable to other
contexts(Idri et al., 2016). Since this study used only
one dataset of histological images with four
magnification factors, we cannot generalize the
results to all datasets of the same image type. As a
consequence, it is essential to test this study on other
public or private datasets in order to confirm or refute
the study's findings.
Construct Validity: The construct validity seeks to
provide an answer to the measurement validity
question(Hosni et al., 2018), or, more precisely, the
reliability of the measurements chosen to assess the
performance of the proposed techniques. As a result,
this study employs four performance measures
(accuracy, precision, recall, and F1-score), the SK test
to cluster statistically indifferent models and the
Borda count voting technique, which takes into
account the four-evaluation metrics, ensures that no
performance metric is favored over another.
7 CONCLUSION
This paper addresses the problem of breast
histopathological images binary classification over
the BreakHis dataset. It designed and proposed a deep
end-to-end heterogenous ensemble learning approach
based on seven DL models using fine-tuned VGG16,
VGG19, RES50, INV3, INRESV2, XCEP, and
MOBN. The proposed approach consists of using two
voting methods (majority and weighted voting) and
constructing DEHtE of two up to seven models based
on the selection by accuracy strategy. The following
evaluation techniques were used to assess and rank
the proposed ensembles over the BreakHis dataset:
four classification performance criteria (accuracy,
precision, recall, and F1-score), SK statistical test,
and Borda Count. The following are the study's main
findings:
(RQ1): Does the heterogenous ensembles using
voting methods outperform the singles?
The deep end-to-end heterogeneous ensembles
outperformed the DL base learners in all dataset MFs,
with the accuracy value increasing from 90.91 % (the
best accuracy value achieved on 40X by XCEP) to
93.8 % when using weighted voting. For 100X MF,
the accuracy value increased by 3.07% (from 90.43%
to 93.5% achieved by ensemble of six when using
weighted voting). For 200X and 400X MFs, the
accuracy value increased from 90.9%, 89.91% to
93.3%, 91.8%, respectively. As a result, the DEHtE
outperformed their base learners significantly.
(RQ2): What are the suitable number of base
learners to design the deep end-to-end heterogenous
ensembles and the suitable voting combination
method used?
The results have proved that the deep end-to-end
heterogenous ensemble designed using the weighted
voting combination rule outperformed the ones with
majority voting. In addition to that, the increase of
number of base learners to design the DEHtE plays an
important role in the amelioration of the
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
116

performances since the best performing designed
DEHtE is the E7WVA across the BreakHis Mf
values.
Ongoing works will focus on proposing DEHtE
techniques using different selection strategies such as
selection by diversity and selection by both accuracy
and diversity, in order to determine the best criteria to
select the base learners in order to propose the most
performing DEHtE.
ACKNOWLEDGEMENTS
This work was conducted under the research project
“Machine Learning based Breast Cancer Diagnosis
and Treatment”, 2020-2023. The authors would like
to thank the Moroccan Ministry of Higher Education
and Scientific Research, Digital Development
Agency (ADD), CNRST, and UM6P for their
support.
This study was funded by Mohammed VI polytechnic
university at Ben Guerir Morocco.
Compliance with ethical standards.
Conflicts of interest/competing interests Not
applicable.
Code availability not applicable.
REFERENCES
Alaoui, O. El, Zerouaoui, H. and Idri, A. (n.d.). “Deep
stacked ensemble for breast cancer diagnosis”, pp. 1–
10.
Alom, M.Z., Yakopcic, C., Nasrin, M.S., Taha, T.M. and
Asari, V.K. (2019), “Breast Cancer Classification from
Histopathological Images with Inception Recurrent
Residual Convolutional Neural Network”, Journal of
Digital Imaging, Journal of Digital Imaging, available
at:https://doi.org/10.1007/s10278-019-00182-7.
Aswathy, M.A. and Jagannath, M. (2017), “Detection of
breast cancer on digital histopathology images: Present
status and future possibilities”, Informatics in Medicine
Unlocked, Elsevier Ltd, Vol. 8 No. October 2016, pp.
74–79.
Bhering, L.L., Cruz, C.D., De Vasconcelos, E.S., Ferreira,
A. and De Resende, M.F.R. (2008), “Alternative
methodology for Scott-Knott test”, Crop Breeding and
Applied Biotechnology, Vol. 8 No. 1, available
at:https://doi.org/10.12702/1984-7033.v08n01a02.
Black, D. (1976), “Partial justification of the Borda count”,
Public Choice, Vol. 28 No. 1, pp. 1–15.
Chollet, F. (2017), “Xception: Deep learning with
depthwise separable convolutions”, Proceedings - 30th
IEEE Conference on Computer Vision and Pattern
Recognition, CVPR 2017, Vol. 2017-Janua, available
at:https://doi.org/10.1109/CVPR.2017.195.
Emerson, P. (2013), “The original Borda count and partial
voting”, Social Choice and Welfare, Vol. 40 No. 2, pp.
353–358.
Gandomkar, Z., Brennan, P.C. and Mello-Thoms, C.
(2018), “MuDeRN: Multi-category classification of
breast histopathological image using deep residual
networks”, Artificial Intelligence in Medicine, Elsevier
B.V., Vol. 88, pp. 14–24.
Gulshan, V., Peng, L., Coram, M., Stumpe, M.C., Wu, D.,
Narayanaswamy, A., Venugopalan, S., et al. (2016),
“Development and validation of a deep learning
algorithm for detection of diabetic retinopathy in retinal
fundus photographs”, JAMA - Journal of the American
Medical Association, Vol. 316 No. 22, available
at:https://doi.org/10.1001/jama.2016.17216.
Hamza, M. and Larocque, D. (2005), “An empirical
comparison of ensemble methods based on
classification trees”, Journal of Statistical Computation
and Simulation, Vol. 75 No. 8, pp. 629–643.
He, K. and Sun, J. (2016), “Deep Residual Learning for
Image Recognition”, available
at:https://doi.org/10.1109/CVPR.2016.90.
Herent, P., Schmauch, B., Jehanno, P., Dehaene, O.,
Saillard, C., Balleyguier, C., Arfi-Rouche, J., et al.
(2019), “Detection and characterization of MRI breast
lesions using deep learning”, Diagnostic and
Interventional Imaging, Société française de radiologie,
Vol. 100 No. 4, pp. 219–225.
Hosni, M., Abnane, I., Idri, A., Carrillo de Gea, J.M. and
Fernández Alemán, J.L. (2019), “Reviewing ensemble
classification methods in breast cancer”, Computer
Methods and Programs in Biomedicine, Vol. 177, pp.
89–112.
Hosni, M., Idri, A., Abran, A. and Nassif, A.B. (2018), “On
the value of parameter tuning in heterogeneous
ensembles effort estimation”, Soft Computing, Vol. 22
No. 18, available at:https://doi.org/10.1007/s00500-
017-2945-4.
Idri, A., Abnane, I. and Abran, A. (2018), “Evaluating
Pred(p) and standardized accuracy criteria in software
development effort estimation”, Journal of Software:
Evolution and Process, Vol. 30 No. 4, pp. 1–15.
Idri, A., Bouchra, E.O., Hosni, M. and Abnane, I. (2020),
“Assessing the impact of parameters tuning in ensemble
based breast Cancer classification”, Health and
Technology, Health and Technology, Vol. 10 No. 5, pp.
1239–1255.
Idri, A., Hosni, M. and Abran, A. (2016), “Improved
estimation of software development effort using
Classical and Fuzzy Analogy ensembles”, Applied Soft
Computing Journal, Elsevier B.V., Vol. 49, pp. 990–
1019.
Jia, H., Xia, Y., Song, Y., Zhang, D., Huang, H., Zhang, Y.
and Cai, W. (2020), “3D APA-Net: 3D Adversarial
Pyramid Anisotropic Convolutional Network for
Prostate Segmentation in MR Images”, IEEE
Transactions on Medical Imaging, Vol. 39 No. 2,
available
at:https://doi.org/10.1109/TMI.2019.2928056.
Assessing the Impact of Deep End-to-End Architectures in Ensemble Learning for Histopathological Breast Cancer Classification
117

Jiang, Y., Chen, L., Zhang, H. and Xiao, X. (2019), “Breast
cancer histopathological image classification using
convolutional neural networks with small SE-ResNet
module”, PLoS ONE, Vol. 14 No. 3, pp. 1–21.
Lateef, F. and Ruichek, Y. (2019), “Survey on semantic
segmentation using deep learning techniques”,
Neurocomputing, Vol. 338, available
at:https://doi.org/10.1016/j.neucom.2019.02.003.
Li, C., Wang, X., Liu, W., Latecki, L.J., Wang, B. and
Huang, J. (2019), “Weakly supervised mitosis detection
in breast histopathology images using concentric loss”,
Medical Image Analysis, Elsevier B.V., Vol. 53, pp.
165–178.
Mardanisamani, S., Maleki, F., Kassani, S.H., Rajapaksa,
S., Duddu, H., Wang, M., Shirtliffe, S., et al. (2019),
“Crop lodging prediction from UAV-acquired images
of wheat and canola using a DCNN augmented with
handcrafted texture features”, IEEE Computer Society
Conference on Computer Vision and Pattern
Recognition Workshops, Vol. 2019-June, available
at:https://doi.org/10.1109/CVPRW.2019.00322.
Mendel, K., Li, H., Sheth, D. and Giger, M. (2019),
“Transfer Learning From Convolutional Neural
Networks for Computer-Aided Diagnosis: A
Comparison of Digital Breast Tomosynthesis and Full-
Field Digital Mammography”, Academic Radiology,
Elsevier Inc., Vol. 26 No. 6, pp. 735–743.
Mitko Veta. (2014), Breast Cancer Histopathology Image
Analysis, Vol. 6.
Nguyen, M.-T., Le, D.T., Son, N.H., Minh, B.C., Duong,
D.H.T. and Linh, L.T. (2020), “Understanding
Transformers for Information Extraction with Limited
Data”, Proceedings of the 34th Pacific Asia Conference
on Language, Information and Computation, No.
October, pp. 478–487.
Nilsson, N.J. (1965), “Learning machines; foundations of
trainable pattern-classifying systems”, McGraw-Hill
Series in Systems Science.
El Ouassif, B., Idri, A. and Hosni, M. (2021), “Investigating
Accuracy and Diversity in Heterogeneous Ensembles
for Breast Cancer Classification”, pp. 263–281.
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. and
Chen, L.C. (2018), “MobileNetV2: Inverted Residuals
and Linear Bottlenecks”, Proceedings of the IEEE
Computer Society Conference on Computer Vision and
Pattern Recognition, IEEE, pp. 4510–4520.
Simonyan, K. and Zisserman, A. (2015), “Very deep
convolutional networks for large-scale image
recognition”, 3rd International Conference on
Learning Representations, ICLR 2015 - Conference
Track Proceedings, pp. 1–14.
Spanhol, F.A., Oliveira, L.S., Petitjean, C. and Heutte, L.
(2016), “A Dataset for Breast Cancer Histopathological
Image Classification”, IEEE Transactions on
Biomedical Engineering, Vol. 63 No. 7, pp. 1455–1462.
Stenkvist, B., Westman-Naeser, S., Holmquist, J., Nordin,
B., Bengtsson, E., Veaelius, J., Eriksson, O., et al.
(1978), “Computerized Nuclear Morphometry as an
Objective Method for Characterizing Human Cancer
Cell Populations”, Cancer Research, Vol. 38 No. 12,
pp. 4688–4697.
Sung, H., Ferlay, J., Siegel, R.L., Laversanne, M.,
Soerjomataram, I., Jemal, A. and Bray, F. (2021),
“Global Cancer Statistics 2020: GLOBOCAN
Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries”, CA: A Cancer Journal for
Clinicians, Vol. 71 No. 3, pp. 209–249.
Szegedy, C., Ioffe, S., Vanhoucke, V. and Alemi, A.A.
(n.d.). “the Impact of Residual Connections on
Learning”, pp. 4278–4284.
Szegedy, C., Vanhoucke, V., Shlens, J. and Wojna, Z.
(2014), “Rethinking the Inception Architecture for
Computer Vision”.
Vo, D.M., Nguyen, N.Q. and Lee, S.W. (2019),
“Classification of breast cancer histology images using
incremental boosting convolution networks”,
Information Sciences, Vol. 482, available
at:https://doi.org/10.1016/j.ins.2018.12.089.
Xie, J., Hou, Q., Shi, Y., Lü, P., Jing, L., Zhuang, F., Zhang,
J., et al. (2018), “The Automatic Identification of
Butterfly Species”, Jisuanji Yanjiu Yu
Fazhan/Computer Research and Development, Vol. 55
No. 8, available at:https://doi.org/10.7544/issn1000-
1239.2018.20180181.
Zerouaoui, H. and Idri, A. (2021a), “Reviewing Machine
Learning and Image Processing Based Decision-
Making Systems for Breast Cancer Imaging”, Journal
of Medical Systems.
Zerouaoui, H. and Idri, A. (2021b), “Reviewing Machine
Learning and Image Processing Based Decision-
Making Systems for Breast Cancer Imaging”, Journal
of Medical Systems, Vol. 45 No. 1, p. 8.
Zerouaoui, H. and Idri, A. (2022), “Biomedical Signal
Processing and Control Deep hybrid architectures for
binary classification of medical breast cancer images”,
Biomedical Signal Processing and Control, Elsevier
Ltd, Vol. 71 No. PB, p. 103226.
Zerouaoui, H., Idri, A., Nakach, F.Z. and Hadri, R. El.
(2021), “Breast Fine Needle Cytological Classification
Using Deep Hybrid Architectures BT - Computational
Science and Its Applications – ICCSA 2021”, in
Gervasi, O., Murgante, B., Misra, S., Garau, C., Blečić,
I., Taniar, D., Apduhan, B.O., et al. (Eds.), , Springer
International Publishing, Cham, pp. 186–202.
Zhou, Z.H. (2012), Ensemble Methods: Foundations and
Algorithms, Ensemble Methods: Foundations and
Algorithms, available
at:https://doi.org/10.1201/b12207.
Zhu, C., Song, F., Wang, Y., Dong, H., Guo, Y. and Liu, J.
(2019), “Breast cancer histopathology image
classification through assembling multiple compact
CNNs”, BMC Medical Informatics and Decision
Making, Vol. 19 No. 1, pp. 1–17.
KDIR 2022 - 14th International Conference on Knowledge Discovery and Information Retrieval
118
