
MedCC: Interpreting Medical Images Using Clinically Significant
Concepts and Descriptions
Xuwen Wang
a
, Zhen Guo
b
, Ziyang Wang
c
and Jiao Li
d
Institute of Medical Information and Library, Chinese Academy of Medical Sciences and Peking Union Medical College,
Beijing, China
Keywords: Concept Detection, Fine-Grained Multi-Label Classification, Pattern-Based Caption Prediction.
Abstract: This paper aims to identify valuable semantic concepts and predict descriptions automatically for medical
images to assist doctors in image reading. A simple framework called MedCC is proposed for medical image
concept detection and caption prediction. MedCC employed multiple fine-grained multi-label classification
(MLC) models trained on manually annotated datasets, which contain image-concept pairs of different
semantic types, such as Imaging Type, Anatomic Structure, and Findings. We validate the performance of
MedCC based on the open sourced concept detection dataset and achieved the best F1 score of 0.419, which
is comparable with the SOTA models. Combining the detected concepts into sentences according to the
manually defined sentence patterns resulted in a BLEU score of 0.257, which still has room for improvement.
1 INTRODUCTION
Diversified medical imaging technologies have
produced massive medical images of multiple modes,
providing rich evidence and perspectives for clinical
diagnosis. The automatic processing and analysis of
multimodal medical images can help relieve the
doctors’ pressure of image reading and effectively
improve the efficiency and accuracy of Clinical
Decision Support (CDS).
Due to the highly heterogeneous nature of medical
images, such as various anatomic structures,
abnormalities, and diagnostic procedures, it is crucial
and challenging to identify comprehensive and
interpretable biomedical semantic concepts as well as
fluent descriptions for providing clear understanding
of medical images. Considering these problems,
concept detection and caption prediction have gained
increasing attention in recent years. The former task
aims to identify various biomedical entities from
medical images (Miranda, Thenkanidiyoor, and
Dinesh 2022), and the latter further predicts brief
expressive textual descriptions.
a
https://orcid.org/0000-0003-3022-6513
b
https://orcid.org/0000-0002-7454-0750
c
https://orcid.org/0000-0002-0368-9590
d
https://orcid.org/0000-0001-6391-8343
This paper aims to interpret medical images using
clinically significant concepts and descriptions. Our
contributions are summarized as follows: (1) We
proposed MedCC, a simple and useful framework for
identifying medical image concepts and predicting
concise captions. The transfer learning-based multi-
label classification (MLC) model (Szegedy et al.
2016) was employed as our baseline concept
detection model. (2) To retrain multiple MLC models
separately with fine-grained semantic concepts, we
divide concepts into three categories based on their
semantic types, namely Imaging Types, Anatomical
Structure, and Findings. Then we manually re-
annotated the open sourced medical images with
different types of concepts via a self-developed
platform. (3) We review the expression of 3256 image
captions and conclude two major sentence patterns
for combining detected concepts to readable
sentences.
In section 2, we summarize the recent works on
concept detection and caption prediction for medical
images. Section 3 provides an overview of proposed
MedCC framework, and introduces the main
functional modules in detail. In section 4, we
518
Wang, X., Guo, Z., Wang, Z. and Li, J.
MedCC: Interpreting Medical Images Using Clinically Significant Concepts and Descriptions.
DOI: 10.5220/0011954800003612
In Proceedings of the 3rd International Symposium on Automation, Information and Computing (ISAIC 2022), pages 518-525
ISBN: 978-989-758-622-4; ISSN: 2975-9463
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
specifically describe our experimental data,
experimental settings and the evaluation criteria.
Section 5 shows the experimental results on the open
sourced ImageCLEFmedical 2021 dataset, and a
preliminary case analysis was conducted. Section 6 is
a brief summary and outlook of this work.
2 RELATED WORK
ImageCLEF hosts annual challenges for medical
image concept detection, medical caption prediction,
Tuberculosis type detection and multi-drug resistance
detection. As one of the representative tracks, the
ImageCLEFmedical Caption 2021 challenge consists
of two tasks: Concept Detection and Caption
Prediction, with the goal of mapping visual
information of radiology images to textual
descriptions of different granularity (Pelka et al.
2021). The concept detection task aims to identify
semantically relevant UMLS (Bodenreider 2004)
Concept Unique Identifiers (CUIs) from radiology
images, whereas the caption prediction task requires
describing the entirety of a medical image and
generating coherent reasonable captions.
The methods of concept detection mainly include
multi-label classification, sequence-to-sequence
learning, entity recognition from captions, and
similarity-based image-text searching approaches
(Miranda, Thenkanidiyoor, and Dinesh 2022). It is
worth noting that due to the heterogeneity and
similarity of medical images, the concept detection
technology used in natural images cannot be applied
directly for medical images. Heterogeneity refers to
the fact that one concept may have completely
different image characteristics, which are constructed
by different imaging techniques. The similarity
means that similar appearances may be associated
with difference concepts. Therefore the concept
detection model needs to identify the inter concept
similarities and intra concept heterogeneity.
Supervised learning such as multi-label classification
(MLC) (Rio-Torto et al. 2022), convolutional neural
network (CNN) (Beddiar, Oussalah, and Seppänen
2021), and concept retrieval were commonly used for
detecting medical concepts. (Rio-Torto et al. 2022;
Serra et al. 2022).
Concept detection is also the premise of image
caption prediction. Using natural language processing
(NLP) technology to combine a group of concepts is
the most concise method to produce textual
descriptions of images. Further, these captions can be
used as components for generating imaging reports.
Transformer-based models are generally selected as
image decoders to generate semantically coherent
captions. (Dalla Serra et al. 2022)
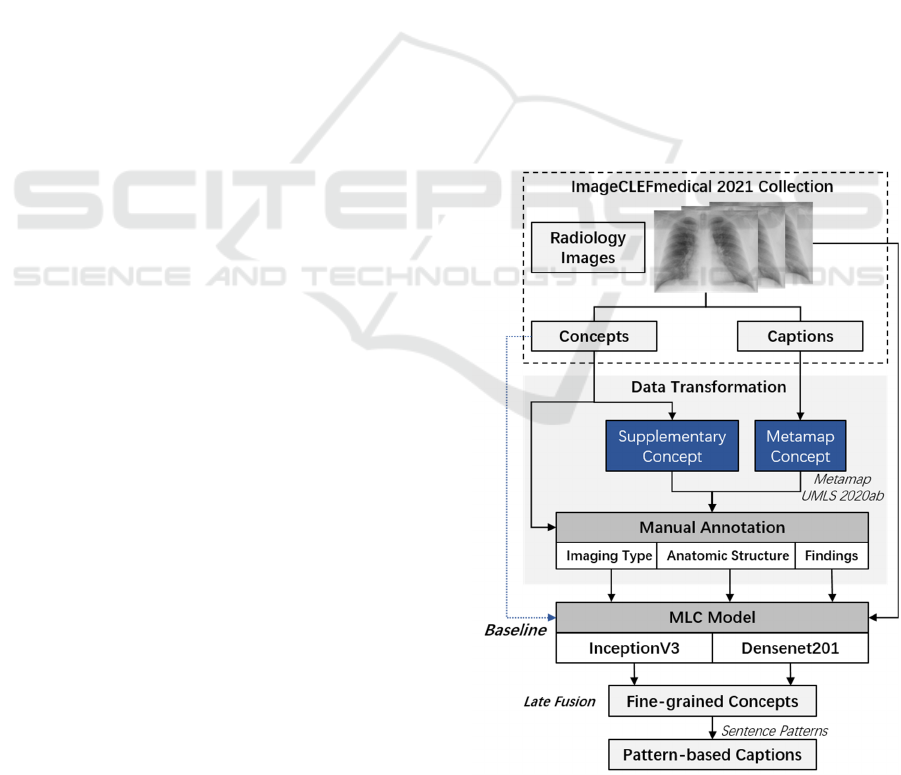
3 METHODOLOGY
The motivation of this work is to build a simple
architecture that provides comprehensible semantic
concepts and descriptions for interpreting multimodal
radiology images. Figure1 shows our workflow of
Medical Image Concepts Detection and Captions
Prediction (Abbreviated as MedCC); including the
analysis and transformation of the ImageCLEF
dataset consisting of elaborately collected medical
images, concepts, and descriptions from PMC
literatures, as well as methods for medical concept
detection and caption prediction.
A transfer learning-based MLC method is utilized
as baseline for modelling overall concepts. In
addition, considering the distinction of concepts with
different semantic types, we further divided the
original concept detection dataset into three subsets
according to their semantic types, which supported us
to train fine-grained MLC models and reveal clinical
insights of radiology images.
Figure 1: Workflow of proposed MedCC framework.
MedCC: Interpreting Medical Images Using Clinically Significant Concepts and Descriptions
519

3.1 Data Analysis and Transformation
3.1.1 Details of Dataset
The ImageCLEFmedical 2021 track (Ionescu et al.
2021) released a collection of 3,256 radiology images
with 3,256 captions and 1,586 no duplicate UMLS
CUIs (Concept Unique Identifiers). The training set
contains 2756 radiology images with captions and
concepts extracted from PMC literatures. Figure 2
shows a training sample of medical image with
associated caption and concept CUIs. Specifically,
concepts in the training set are automatically labelled
from image captions, and then filtered and mapped to
UMLS CUIs. While the development set consists of
500 radiology images annotated by professional
radiologists.
Figure 2: A sample of medical images from the training set
of ImageCLEFmedical Caption track, each with the
associated caption and CUIs.
According to the officially provided CUIs, we
backtracked biomedical terms from the
UMLS2020ab thesaurus, and collected TUIs (Unique
Identifier of Semantic Type) together with semantic
type strings for each term. We observed that most
images were accompanied with concepts on behalf of
imaging modality, such as the Diagnostic Procedure
or Medical Device, and some concepts representing
the anatomic structures or clinical findings.
Table 1 shows the distribution of high-frequency
concepts, and it is evident that the semantic types of
these concepts are relatively concentrated on a few
specific TUIs. For example, terms like ‘Tomography,
Emission-Computed’, ‘Plain x-ray’, ‘Magnetic
Resonance Imaging’ and ‘Ultrasonography’ have the
same semantic type, i.e. T060 that refers to the
Diagnostic Procedure. Similarly, terms like ‘Lesion’,
‘Thickened’ and ‘Mass of body structure’ belongs to
the T033 that refers to Finding; while terms like
‘Appendix’, ‘Right Kidney’ and ‘Spinal epidural
space’ that associated with body parts or organ
components can be classified as Anatomic Structure.
Obviously, medical concepts of different semantic
types can reveal the clinical significance of medical
images from different perspectives.
Table 1: Part of high-frequency UMLS concepts in the
ImageCLEFmedical caption 2021 collection. The
abbreviations are as follows: CUI refers to Concept Unique
Identifiers, TF refers to Term Frequency, TUI refers to
Unique Identifier of Semantic Type, and SEMTYPE refers
to Semantic Type.
CUI TF Term
String
TUI SEMTYPE
C00403
98
1400 Tomography
, Emission-
Compute
d
T060 Diagnostic
Procedure
C00244
85
796 Magnetic
Resonance
Imaging
T060 Diagnostic
Procedure
C13066
45
627 Plain x-ray T060 Diagnostic
Procedure
C00416
18
373 Ultrasonogr
aphy
T060 Diagnostic
Procedure
C00099
24
283 Contrast
Media
T130 Indicator,
Reagent, or
Diagnostic Ai
d
C05775
59
274 Mass of
body
structure
T033 Finding
C00029
78
119 angiogram T060 Diagnostic
Procedure
C02211
98
108 Lesion T033 Finding
C13226
87
107 Endoscopes,
Gastrointesti
nal Tract,
Upper Tract
T074 Medical
Device
C02054
00
92 Thickened T033 Finding
.. .. .. .. ..
C00036
17
52 Appendix T023 Body Part,
Organ, or
Organ
Component
C02281
34
50 Spinal
epidural
space
T030 Body Space or
Junction
C00166
58
47 Fracture T037 Injury or
Poisoning
C00058
89
47 Body Fluids T031 Body
Substance
C02276
13
47 Right
kidney
T023 Body Part,
Organ, or
Organ
Component
3.1.2 Data Transformation
As previous experiences show that too many labels
may reduce the accuracy of the classifier, an
alternative strategy is to divide the label set into
multiple subcategories for training fine-grained
multi-label classification models. In this work, we
manually divided the original concepts into three
categories according to the UMLS semantic types,
namely Imaging Type (IT), Anatomic Structure (AS),
and Finding (FD). Concepts that do not belong to the
above categories are classified as ‘others’.
A secondary data annotation was performed based
on the official training set as well as development set
ISAIC 2022 - International Symposium on Automation, Information and Computing
520

via a self-developed medical data annotation platform.
As shown in Figure 3, there are three sources of
relevant concepts for a given radiology image. The
first category contains the original ImageCLEF
concepts annotated by official tools and radiologists.
These concepts are semantically related but are often
incomplete because many images have only one label.
We take such concepts as preferred labels. As long as
there are preferred concepts assigned to the three
major categories, i.e. Imaging Type (IT), Anatomic
Structure (AS) and Finding (FD), we no longer
expand them to ensure the accuracy. The second
source of concepts are automatically annotated from
the given image captions using the MetaMap tool
(Aronson 2001) and the UMLS 2020ab vocabulary.
Therefore, we call them candidate META tags, which
are more comprehensive but also introduce noise
words. If the preferred concepts are insufficient for a
given image, we seek for appropriate concepts from
META tags for supplementing corresponding
categories. The third source provides alternative
supplementary concepts summarized manually from
the high-frequency ImageCLEF concepts. The
purpose of collecting such concepts is to facilitate
dragging and supplementing high-frequency words
that are not included in the caption and concept
annotations during manual annotation.
Graduate students majoring in medical imaging
were invited to annotate images by consulting visual
information, textual descriptions and the three kinds
of concepts described above. The labelling protocol
is that each radiology image should be assigned at
least one IT label, zero or more AS labels, and zero or
more FD labels. In addition, ImageCLEF concepts
that are indefinite to be classified in the above
categories can be assigned to the ‘Others’.
By collecting the annotated image-concept pairs,
three subsets were constructed for training
subsequent fine-grained MLC models. These re-
annotated subsets consist of same images from the
original training set and development set, but differ in
related concepts. Table 2 shows the amount of
concepts in different subsets. It can be seen that the
smallest subset is the Imaging Type, which contains
99 no duplicate concepts related to imaging
diagnostic procedure and devices. The other two
subsets include 786 and 854 concepts respectively,
about half of the original concept scale. Empirically,
with the same number of medical images, the more
concentrated the semantic concepts to be predicted,
the more effective the multi-label classification
model will be trained. Our subsequent experiments
also verified this issue.
Figure 3: Data flow in the process of secondary data
annotation; there are three sources of related concepts for a
given medical image, i.e. the official ImageCLEF concepts,
META tags and supplementary concepts.
Table 2: Count of concepts in different subsets, in which the
CNT refers to the counted number of no duplicate concepts
in the corresponding subset.
Subset CNT Concept Sample
Imaging
T
yp
e
99
C0040398 Tomography
Emission-Com
p
ute
d
Anatomic
Structure
786
C0228134 Spinal epidural
s
p
ace
Finding 854
C0577559 Mass of body
structure
3.2 Concept Detection
3.2.1 Transfer Learning-Based Multi-Label
Classification
Multi-label Classification (MLC) is a common
method for concept detection of medical images.
However, limited scale of annotated medical images
prevent us from training effective deep models from
scratch. Therefore, the MLC method based on
transfer learning (Szegedy et al. 2016) was used to
assign multiple concept CUIs to medical images.
Consider a dataset including 𝑛 unique concepts
C={𝑐
,𝑐
,…𝑐
} that appear in the context of
medical images, a MLC model predicts a set of 𝑙
labels Y=
{
𝑦
,𝑦
,…𝑦
}
,Y ⊂C associated with a
given image X (Miranda, Thenkanidiyoor, and
Dinesh 2022). Previous studies generally
implemented MLC using CNN networks that pre-
trained on the ImageNet dataset (Russakovsky et al.
2015). Specifically, the output sigmoid layer has n
MedCC: Interpreting Medical Images Using Clinically Significant Concepts and Descriptions
521

nodes representing the concepts to be predicted, and
produces a set of n probabilitiesP={𝑝
,𝑝
,…𝑝
},
inwhich 𝑝
is the probability of the input image
associated with the concept 𝑐
.
To compare the classification effects of different
CNN networks, two classic models, i.e. the Inception-
V3 (Szegedy et al. 2016) and DenseNet 201 (Huang
et al. 2017) were separately used as the backbone
network of our MLC framework. The parameters of
the pre-trained CNN model were transferred as the
initial parameters of the MLC model.
We reuse the pre-trained CNN architecture,
replace the layers used for classification, and retrain
the network to predict multiple relevant concepts of
medical images. The convolutional layers realizes
image feature extraction. The last learnable layer and
the classification layer are used for classification,
which combine the image features into class
probabilities and predict highly correlated concepts.
To obtain the distribution of the relevant probability
of medical concepts, the network should be retrained
as a regression task. Specifically, the final fully
connected layer, the softmax layer, and the
classification output layer were transformed into a
fully connected layer and a regression layer. Then we
fine tune the weights based on medical images, and
assign concepts with probabilities above a certain
threshold to the test images.
3.2.2 Fine-Grained Multi-Label
Classification
Inspired by the idea of multimodal data fusion, we go
further to train multiple fine-grained MLC models
based on the secondary annotation subsets, which
label the same images with a smaller number but
more focusing concepts. The transformation of CNN
networks are same as Section 3.2.1. Therefore, three
types of semantic concepts can be obtained for each
medical image. Further, the late fusion strategy is
employed together with predefined threshold and
concept selecting rules to fuse the predicted results.
The concept selection strategy would be introduced in
detail in the experiments section.
3.3 Pattern-Based Caption Prediction
To obtain readable image captions, a simple and
direct way is to combine the semantic concepts
identified in the previous stage, simulating that
human beings compose sentences by keywords.
According to the expression characteristic of captions
in the ImageCLEF dataset, a few sentence patterns are
concluded for combining identified concepts to
descriptions, see Table 3. Obviously, the accuracy
and comprehensiveness of concept detection will
directly affect the quality of synthesized sentences.
Table 3: Pre-defined sentence patterns for combining
concepts as captions.
Pattern Caption Sample
<Imaging Type> of
<Anatomic Structure>
demonstrate/show/suggest
<Finding>
FigID_synpic31919: Longitudinal
sonographic image of the left kidney
shows hyperechoic renal pyramids
with faint shadowing.
<Imaging Type>
demonstrate/show/suggest
<Finding> in/of/within
<Anatomic Structure >
FigID
_
synpic41602: Axial CT
images demonstrate a rounded mass
in the right upper quadrant.
4 EXPERIMENTS
4.1 Dataset
Both of the original ImageCLEF dataset and the
secondary re-annotated dataset are utilized as our
experimental data for the subsequent comparative
experiment.
For the original da
Dallataset, 3,256 radiology
images were separately resized to 299*299 pixels for
training Inception V3, and 224*224 pixels for the
DenseNet model. Concept CUIs associated with
overall images are collected as the label set. We used
the official 2756 training images for the training
process. The development set containing 500 human-
labelled images is randomly divided equally into
validation set and test set, each collection contains
250 images and related text descriptions.
For the secondary re-annotated dataset, each
subset contains the same 3,256 radiology images and
associated concepts of different semantic types. We
did the same resize processing for the images as
mentioned above. The division of training set,
validation set and test set is also the same as above.
4.2 Experimental Settings
All experiments were implemented on a Windows
Sever 2012 R2, with detailed configurations of
Intel(R) Xeon(R) Gold 6130 64 CPU, 512GB
memory, and NVIDIA Tesla P100 16GB * 4 GPUs.
The transfer learning-based MLC model trained
on the original ImageCLEF dataset was taken as the
baseline model. The label set contains more concepts
to be predicted, resulting in a larger scale of the
corresponding concept probability matrix. During the
training process, pre-trained models including
DenseNet201 and Inception v3 were re-trained on the
ISAIC 2022 - International Symposium on Automation, Information and Computing
522

current dataset. The parameters were set as follows:
both models used the SGDM as Gradient descent
algorithm, the epoch is set as 30, the initial learning
rate is 0.005 with a drop period of 20. We further fine-
tuned the models based on the validation set. Then,
predicted concepts with high probabilities above the
predefined threshold were selected as the preferred
labels for a given test image. The concept selection
rules according to the output score matrix includes the
Term Frequency, the Threshold of probabilities and
the Top Rank of probabilities. Based on the validation
set, we gradually adjusted the threshold from zero to
0.5, and the lowest term frequency is set to 5, while
the top rank of probabilities ranges from 1 to 5,
increasing by 1 each iteration.
As comparative experiments, both of the
DenseNet and Inception v3-based MLC models were
separately trained and verified on the three secondary
annotated subsets. The parameters were set as
follows: the Gradient Descent algorithm include
SGDM, ADAM and RMS; the epoch is set as 20,
initial learning rate is 0.001 with the drop period of
20. The threshold gradually increased from zero to
0.5 with an interval of 0.1, the term frequency is set
to 10 while the top rank ranges from 1 to 5, with an
interval of 1. Then with refer to the late fusion
strategy, the best results of the above methods are
combined as predicted concepts for test images.
Finally, the preferred concepts are filled into the
sentence template to form a comprehensible
description. A classical Dual path CNN model
(Zheng et al. 2020) was taken as a comparison
method.
4.3 Evaluation Criteria
In this study, the evaluation criteria follows the
ImageCLEFmedical 2021 track (Pelka et al. 2021).
For the concept detection task, balanced precision and
recall trade-off were measured in terms of F1 scores
between predicted and ground truth concepts, which
were calculated by the Python's scikit-learn library.
The caption evaluation is based on BLEU score
(Papineni et al. 2002), an automatic evaluation
method for machine learning that implemented by the
Python’s NLTK (v3.2.2) BLEU scoring method.
5 RESULTS
Based on re-annotated subsets, we validated the
performance of fine-grained MLC models separately.
Preliminary results show that the Inception V3 model
outperforms DenseNet in predicting Imaging Type
labels, with an F1 score of 0.9273. However, the
identification of other types of concepts, such as
Findings, is far from satisfactory. One possible reason
is that hundreds of candidate labels in a training
subset are still too many to adequately train an
effective MLC model compared to a limited number
of thousands of images.
Intuitively, it is understandable that images of
similar cases may have similar anatomical structures
or findings labels. However, since the images in the
original ImageCLEF dataset come from PMC
literatures, and the diversity and heterogeneity of
image content as well as context determine that it is
not suitable for specific disease detection tasks, which
makes it difficult to predict accurate body parts,
organs, or findings.
Table 4 shows the experimental results of our
MLC models on the concept detection task. Among
them, MLC_baseline represents the MLC model
trained on the overall concept set based on the
Inception-V3 backbone network. The MedCC_FD
represents the fine-grained MLC model trained on the
subset including the Findings (FD) concepts, similar
to this, MedCC_* represents the combination of the
concepts predicted by different MLC models. We
also combined the fine-grained predicted concepts
with the baseline.
Unexpectedly, the fine-grained MLC model
trained based on the subset of Imaging Types, i.e.
MedCC_IT obtained the best F1 score of 0.419,
indicating that concepts of this type have a high
coverage in radiology images, and are relatively
concentrated and suitable for training an effective
classification model. Whereas MedCC_FD and
MedCC_AS that predicted body-related concepts or
clinical findings introduced more unmentioned words
and reduced the overall score. However, previous
experience gained through manual annotation
suggests that some unmentioned terms are also worth
referring to interpret given medical images. Figure 4
shows an example in the validation set. For a given
medical image, MedCC produced a few medial
concepts as well as a concise caption, in which red
concepts are consistent with the Ground Truth (GT),
and unmatched concepts such as ‘Appendix’, ‘Mass
of body structure’ are also meaningful and related to
the given image.
Table 5 shows the performance of MedCC on the
caption prediction task. A Dual path CNN model was
taken as baseline, and achieved a BLEU score of
0.137. Due to the limited predefined sentence patterns
and the influence of concept detection results, our
pattern-based caption prediction model received a
BLEU score of only 0.257. Case analysis shows that
MedCC: Interpreting Medical Images Using Clinically Significant Concepts and Descriptions
523

the sentences generated by MedCC are coherent and
in line with the logic of medical image description.
However, it still does not meet doctors’ need for
quick reading and reasonable interpretation of
images.
Table 4: Experimental results of MedCC on the concept
detection task.
Method F1
MedCC
_
FD 0.019
MedCC
_
AS 0.037
MedCC
_
IT
_
AS
_
FD 0.327
MedCC
_
IT
_
FD 0.355
MedCC
_
IT_AS 0.370
MLC
_
baseline 0.380
MedCC
_
baseline 0.396
MedCC
_
IT
_
baseline 0.400
MedCC
_
IT 0.419
Table 5: Experimental results of MedCC on the caption
prediction task.
Method BLEU
Dual Path CNN 0.137
MedCC
_
Pattern1 0.203
MedCC_Pattern2 0.257
Figure 4: An example in the validation set, comparing the
concepts and captions predicted by MedCC with official
Ground Truth.
6 CONCLUSIONS
This article introduces MedCC, a simple architecture
that provides understandable semantic concepts and
descriptions for interpreting multimodal radiology
images. The MLC method based on transfer learning
is mainly used to detect UMLS concepts for medical
images. We manually annotated three subsets
according to different semantic types of concepts,
namely Imaging Type, Anatomic Structure and
Finding. Then we trained multiple fine-grained MLC
models based on different subset separately for
identifying semantic concepts of specific types.
Further, the detected concepts were combined into
sentences according to predefined sentence patterns.
Through this study, we acquired a more intuitive
and in-depth understanding of biomedical concepts
related to the clinical interpretation of radiology
images. In order to obtain more relevant concepts for
medical images, the set of semantic concepts should
be more focused and specific, which is crucial for
training effective models. In addition, it is still worth
exploring how to generate more readable and
reasonable descriptions on the basis of clear and
clinically significant concepts.
ACKNOWLEDGEMENTS
This work was supported by the National Natural
Science Foundation of China (Grant No.61906214),
Chinese Academy of Medical Sciences (Grant
No.2021-I2M-1-56), the Beijing Natural Science
Foundation (Grant No. Z200016).
REFERENCES
Aronson, A. R. 2001. “Effective Mapping of Biomedical
Text to the UMLS Metathesaurus: The MetaMap
Program.” Proceedings. AMIA Symposium: 17–21.
Beddiar, Djamila-Romaissa, Mourad Oussalah, and Tapio
Seppänen. 2021. “Attention-Based CNN-GRU Model
For Automatic Medical Images Captioning:
ImageCLEF 2021.” In Proceedings of the Working
Notes of CLEF 2021 - Conference and Labs of the
Evaluation Forum, CEUR Workshop Proceedings, eds.
Guglielmo Faggioli et al. Bucharest, Romania: CEUR,
1160–73.
Bodenreider, Olivier. 2004. “The Unified Medical
Language System (UMLS): Integrating Biomedical
Terminology.” Nucleic Acids Research 32(Database
issue): D267-270.
Dalla Serra, F., Deligianni, F., Dalton, J., and O’Neil, A. Q.
(2022).Cmre-uog team at imageclefmedical caption
2022:Concept detection and image captioning. page
1381–90
Djamila-Romaissa, B., Mourad, O., and Tapio, S. (2021).
Attention-based cnn-gru model for automatic medical
images captioning: Imageclef 2021. In Proceedings of
the Working Notes of CLEF 2021 - Conference and
Labs of the Evaluation Forum, pages 1160––73.
Huang, Gao, Zhuang Liu, Laurens Van Der Maaten, and
Kilian Q. Weinberger. 2017. “Densely Connected
Convolutional Networks.” In 2017 IEEE Conference on
Computer Vision and Pattern Recognition (CVPR), ,
2261–69.
Ionescu, Bogdan et al. 2021. “The 2021 ImageCLEF
Benchmark: Multimedia Retrieval in Medical, Nature,
Internet and Social Media Applications.” In Advances
in Information Retrieval: 43rd European Conference
on IR Research, ECIR 2021, Virtual Event, March 28 –
April 1, 2021, Proceedings, Part II, Berlin, Heidelberg:
Springer-Verlag, 616–23.
ISAIC 2022 - International Symposium on Automation, Information and Computing
524

Miranda, Diana, Veena Thenkanidiyoor, and Dileep Aroor
Dinesh. 2022. “Review on Approaches to Concept
Detection in Medical Images.” Biocybernetics and
Biomedical Engineering 42(2): 453–62.
Papineni, Kishore, Salim Roukos, Todd Ward, and Wei-
Jing Zhu. 2002. “BLEU: A Method for Automatic
Evaluation of Machine Translation.” In Proceedings of
the 40th Annual Meeting on Association for
Computational Linguistics, ACL ’02, USA:
Association for Computational Linguistics, 311–18.
Pelka,O., Abacha,A.B.,Obioma et al. 2021. “Overview of
the ImageCLEFmed 2021 Concept & Caption
Prediction Task.” In CLEF(Working Notes),pages
1101-1112.
Rio-Torto, Isabel, Cristiano Patrício, Helena Montenegro,
and Tiago Gonçalves. 2022. “Detecting Concepts and
Generating Captions from Medical Images:
Contributions of the VCMI Team to
ImageCLEFmedical 2022 Caption.” In Proceedings of
the Working Notes of CLEF 2022 - Conference and
Labs of the Evaluation Forum, CEUR Workshop
Proceedings, eds. Guglielmo Faggioli, Nicola Ferro,
Allan Hanbury, and Martin Potthast. Bologna, Italy:
CEUR, 1535–53.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh,
S.,Ma, S., Huang, Z., Karpathy, A., Khosla, A.,
Bernstein.,M., et al.2015. “ImageNet Large Scale
Visual Recognition Challenge.” International Journal
of Computer Vision 115(3): 211–52.
Serra, Francesco Dalla, Fani Deligianni, Jeffrey Dalton, and
Alison Q. O’Neil. 2022. “CMRE-UoG Team at
ImageCLEFmedical Caption 2022: Concept Detection
and Image Captioning.” In Proceedings of the Working
Notes of CLEF 2022 - Conference and Labs of the
Evaluation Forum, CEUR Workshop Proceedings, eds.
Guglielmo Faggioli, Nicola Ferro, Allan Hanbury, and
Martin Potthast. Bologna, Italy: CEUR, 1381–90.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and
Wojna, Z.2016. “Rethinking the Inception Architecture
for Computer Vision.” In 2016 IEEE Conference on
Computer Vision and Pattern Recognition (CVPR), ,
2818–26.
Zheng, Z., Zheng, L., Garrett, M., Yang, Y., Xu, M., and
Shen, Y.-D. (2020). Dual-path convolutional imagetext
embeddings with instance loss. ACM Transactions on
Multimedia Computing, Communications, and
Applications (TOMM), 16(2):1–23.
MedCC: Interpreting Medical Images Using Clinically Significant Concepts and Descriptions
525
