
Altered Small-World Topological Properties of Functional Brain
Network in Patients with OSA
Ailin Hou
1,2,*
, Xueming Pang
2
and Quan Zhang
3,*
1
School of Precision Instrument & Opto-Electronics Engineering, Tianjin University, Tianjin 300072, China
2
School of Medical Imaging, Tianjin Medical University, Tianjin 300203, China
3
Department of Radiology, Characteristic Medical Center of Chinese People's Armed Police Force, Tianjin 300162, China
Keywords:
Obstructive Sleep Apnea, Resting-State, Functional Magnetic Resonance Imaging, Graph Theory, Small-
World Property.
Abstract: Obstructive sleep apnea (OSA) is a common disorder of sleep disorders in which patients often suffer from
cognitive impairment. Recent neurological imaging studies have shown that cognitive impairment in OSA
patients is closely related to the extensive brain regions with abnormal neural activity. However, it remains
unclear whether the topological properties of function networks in OSA patients have changed. Based on
resting-states functional magenetic resonance imaging (rs-fMRI) and graph-analysis methods, this study
explores the different orgnization in functional brain network between OSA patients and healthy volunteers.
The brain connectome of patients with OSA exhibited small-worldness, but existed significant statistical
difference compared with health controls. Besides, the betweenness centrality and degree centrality of right
dorsolateral superior frontal gyrus and right hippocampal gyrus were significantly different in OSA patient
group. The aberrant topological properties illustrated that the functional integration and segregation of brain
networks in patients with OSA were disrupted.
1 INTRODUCTION
Obstructive sleep apnea (OSA) is the most common
adult respiratory disease in which patients suffer from
frequent apnea or hypoventilation due to recurrent
complete or incomplete upper airway obstruction
during sleep (Zhang, 2012). The main manifestations
of OSA patients are snoring during sleep with apnea
and superficial breathing, recurrent hypoxemia,
hypercarbia and sleep architecture disorders at night.
This leads to daytime sleepiness, cardio-pulmonary
vascular complications and even multi-organ
damage, which seriously affects the quality of life and
life expectancy of patients (He, 2009). OSA is an
independent risk factor for a variety of systemic
diseases and may significantly affect cognitive
function in addition to increasing the incidence of
hypertension, diabetes, respiratory failure and
cardiovascular disease (He, 2009). Current studies
have shown that OSA can extensively impair
cognitive function, including attentional alertness,
executive ability, memory and motor coordination,
and severely affect patient outcomes and prognosis
(Verstraeten, 2007; Aloia, 2004; Decary, 2000).
Resting-state functional magnetic resonance
imaging (MRI) provides a non-invasive and effective
technique for studying brain function. Previous
studies based on fMRI data have found that some
functional measures of regions exhibited functional
abnormalities in patients of OSA, such as the regional
homogeneity (ReHo) in the right temporal, parietal
and frontal regions (Santarnecchi, 2013), the
decreased ALFF of regions in default mode network
(DMN) (Li, Ma, 2016; Li, Nie, 2016). And
researchers have found that all these abnormal
regional alterations may be correlated to cognitive
dysfunction.
However, brain is a complex information
processing system (Khazaie, 2017). Any neurological
activity can’t occur as a result of a single neuron or
brain region working alone, but rather a collection of
neurons or several brain regions acting in concert
with each other to transmit and integrate information.
The complex network-based graph theory analysis
has been widely applied to characterize such complex
systems. The so-called network is a mathematical
representation of a real complex system, defined as a
collection of nodes and edges. For a brain network,
Hou, A., Pang, X. and Zhang, Q.
Altered Small-World Topological Properties of Functional Brain Network in Patients with OSA.
DOI: 10.5220/0012012400003633
In Proceedings of the 4th International Conference on Biotechnology and Biomedicine (ICBB 2022), pages 27-30
ISBN: 978-989-758-637-8
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
27

the nodes can be brain regions or voxels, or even
neurons, while edges can be identified by anatomical
connectivity, functional connectivity depending on
the characteristics of the data set. Functional
connectivity indicates a correlation in time between
the neurophysiological activity of two points. Based
on graph theory, Chen found that the whole brain
network of OSA patients exhibited decreased
smallworld topological property (Chen, 2018).
However, Huang found that the smallworld property
of the OSA brain network was not significantly
different to HC’s, but OSA performed significantly
lower in Cp and higher in Lp (Huang, 2019). Hence,
it follows that the functional network organization of
OSA patients are not clear yet.
In this paper, we intend to explore the impact of
OSA disease on patients' functional brain networks
from a complex network perspective.
2 MATERIAL METHOD
2.1 Data Acquisition
The data of this experiment were divided into two
groups, the OSA patient group and the healthy
controls group (HC), which included 24 OSA patients
with typical indications of OSA disease and met the
diagnostic criteria for the disease in the relevant draft;
21 healthy volunteers were also recruited as the
control group.
In this study, MRI data were acquired using a GE
Signa HDx magnetic resonance scanner with a field
strength of 3.0 T. Scans and resting-state fMRI data
were acquired using Gradient Recalled Echo (GRE)
single excitation planar echo imaging (EPI) sequence
with the following parameters: TR=2000ms,
TE=30ms, FA=90°, FOV was 240*240mm², a 64*64
matrix was used, thickness =3mm, the layer spacing
(gap) was 1mm, a total of 38 layers were divided, and
180 time points were acquired in each volume.
2.2 Data Pre-Processing
The DPARSF (Data Processing Assistant for Resting-
State fMRI) software based on MATLAB was used
to preprocess the functional MRI data of both OSA
and HC related to the following steps.
The images of the first 10 time points were
excluded to avoid the potential noise and instability.
Slice timing was applied to correct this time
difference. Head movement correction was used to
avoid a slight head movement. The unified standard
spatial EPI template with a voxel size of 3×3×3mm3
was used for transformation in order to facilitate the
later study. The whole brain average signal,
cerebrospinal fluid and white matter signal and
motion signal were regressed out. The linear drift was
removed. Then a band-pass filtering (0.01-0.08Hz)
was used to remove the interference of low frequency
and high frequency signals.
2.3 Construction of Brain Networks
and Calculation of Topological
Properties
Brain networks were constructed and topological
properties were calculated. Based on the human
Brainnetome Atlas template (Fan, 2016), the
cerebrum of each subject was divided into 246 brain
regions (nodes). The Pearson correlation coefficient
of the average time series of each two brain regions
was calculated as the functional connectivity (edges),
then followed by the Fisher r-to-z transformation to
normalize it. The z-scored 246×246 correlation
coefficient matrix was obtained.
The correlation matrices were binarized by a pre-
selected value of sparsity K (0.05 ≤ K ≤ 0.5). For a
specific K, we got an undirected binarized network
for each subject, then we applied graph theory to
calculate the topological properties of each brain
network. The global network properties contain
clustering coefficient (C
p
), characteristic path length
(L
p
), normalized clustering coefficient (γ),
normalized characteristic path length (λ), and small-
world property (σ). The nodal properties contain the
betweenness centrality (BC) and degree centrality
(DC) of each brain regions.
2.4 Statistical Analysis
Two-samples t-test was performed for each parameter
corresponding to the two groups of subjects. p<0.05
is considered to be statistically different.
3 RESULTS AND DISCUSSIONS
3.1 Global Properties
The area under curve (AUC) of each global parameter
(C
p
, L
p
, γ,λ, and σ) in OSA group and HC group is
shown in Figure 1(a). The Cp, Lp,λ of OSA patients
were significantly lower than healthy controls (p <
0.05), while theγand σ of OSA patients were
significantly higher than healthy control (p < 0.05).
ICBB 2022 - International Conference on Biotechnology and Biomedicine
28
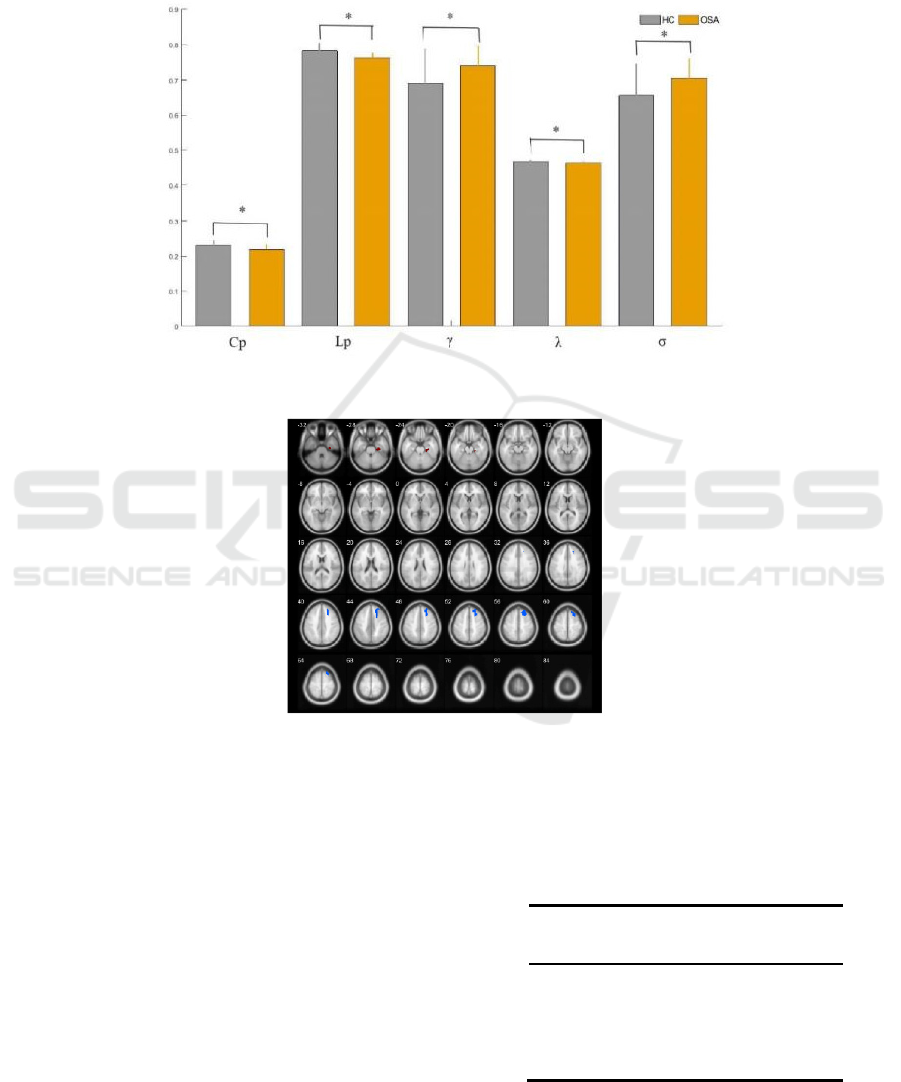
In each sparsity, the small-world property σ was
greater than 1, which indicated that the brain
functional network in both the HC and OSA groups
had the small-world property. But the small
worldness of OSA patients was significantly
increased compared with healthy controls. These
results suggest that the small-world properties of the
functional brain network of OSA patients are
significantly altered.
(a) The difference of global topological properties
(b) Regions whose DC and BC showed between OSA and HC significantly different
Note: * p<0.05, which was considered significantly difference.
Figure 1: The AUC of global properties and local properties comparisons with OSA and HC.
3.2 Local Properties
In this paper, we mainly analyzed the local properties:
BC and DC of every brain region, and the regions
significantly different between OSA patients and
healthy controls are shown in Fig. 1(b) and Table 1.
The BC and DC of right dorsolateral superior frontal
gyrus (Frontal_Sup_R) was significantly lower in the
OSA patient group. The BC and DC of right
hippocampal gyrus (ParaHippocampal_R) was
significantly increased in OSA group.
Table 1: The P-value of brain regions whose BC and DC
were significantly different between OSA and HC..
Brain Regions BC DC
Frontal_Sup_R 0.0074 0.0048
ParaHippocampal_R 0.0010 0.0015
Altered Small-World Topological Properties of Functional Brain Network in Patients with OSA
29

4 CONCLUSION
Based on the results and discussions presented above,
the conclusions are obtained as below:
(1) The resting-state brain functional networks of
both OSA patients and normal controls exhibit small-
world property. But the small-world property of OSA
patients are significantly altered, which suggest that
the functional network organization of OSA patients
is altered.
(2) The local property of OSA patients was
significantly decreased in superior frontal gyrus,
which indicated that the impairment of the superior
frontal gyrus is associated with behavioral cognitive
dysfunction in OSA patients.
(3) The local property of OSA patients was
significantly increased in parahippocampal gyrus,
which indicated that the significantly higher value of
BC and DC may be related to compensatory
mechanisms of memory function impairment.
REFERENCES
Aloia MS, Amedt JT, Davis JD, et al. Neuropsychological
sequelae of obstructive sleep apnea-hypopnea
syndrome: a critical review [J]. J Int Neuropsychol Soc.
2004, 10(5): 772-785.
Chen L, Fan X, Li H, et al. Topological Reorganization of
the Default Mode Network in Severe Male Obstructive
Sleep Apnea[J]. Frontiers in neurology 2018; 9: 363.
Decary A, Rouleau 1, Montplaisir J. Cognitive deficits
associated with sleep apnea syndrome: a proposed
neuropsychological test battery [J]. Sleep. 2000, 23(3):
369-381.
Fan L, Li H, Zhuo J, et al. The Human Brainnetome Atlas:
A New Brain Atlas Based on Connectional Architecture
[J]. Cerebral Cortex, 2016, 26(8):3508-3526.
He Q Y, Chen B Y. Sleep and respiratory disease [M].
People's Medical Publishing House.2009; 89-421.
Huang Y, Liu Y, Zhao D, et al. Small-world properties of
the whole-brain functional networks in patients with
obstructive sleep apnea-hypopnea syndrome[J]. Sleep
medicine 2019; 62: 53-8.
Khazaie H, Veronese M , Noori K, et al. Functional
Reorganization in Obstructive Sleep Apnoea and
Insomnia: A Systematic Review of the Resting-State
fMRI[J]. Neuroscience & Biobehavioral Reviews,
2017, 77:219-231.
Li, C., Ma, X., Dong, M., Yin, Y., Hua, K., Li, M., Li, C.,
Zhan, W., Li, C., Jiang, G., 2016a. Abnormal
spontaneous regional brain activity in primary
insomnia: a resting-state functional magnetic resonance
imaging study. Neuropsychiatr. Dis. Treat. 12, 1371–
1378.
Li, H.J., Nie, X., Gong, H.H., Zhang, W., Nie, S., Peng,
D.C., 2016b. Abnormal resting-state functional
connectivity within the default mode network
subregions in male patients with obstructive sleep
apnea. Neuropsychiatr. Dis.Treat. 12, 203–212.
Santarnecchi, E., Sicilia, I., Richiardi, J., Vatti, G.,
Polizzotto, N.R., Marino, D., Rocchi, R., Van De Ville,
D., Rossi, A., 2013. Altered cortical and subcortical
local coherence in obstructive sleep apnea: a functional
magnetic resonance imaging study. J. Sleep Res. 22,
337–347.
Verstraeten E. Neurocognitive effects of obstructive sleep
apnea syndrome [J]. Curr Neurol Neurosci Rep. 2007,
7(2): 161-166.
Zhang Q. Resting-sate function magnetic resonance
imaging of obstructive sleep apnea-hypopnea syndrome
[D]. Tianjin Medical University, 2012.
ICBB 2022 - International Conference on Biotechnology and Biomedicine
30
