
Prediction of Antimicrobial Peptides Using Deep Neural Networks
Ümmü Gülsüm Söylemez
1a
, Malik Yousef
2b
and Burcu Bakir-Gungor
3c
1
Department of Software Engineering, Faculty of Engineering, Muş Alparslan University, Muş, Turkey
2
Department of Information Systems, Zefat Academic College, Zefat 13206, Israel
3
Department of Computer Engineering, Faculty of Engineering, Abdullah Gul University, Kayseri, Turkey
Keywords: Antimicrobial Peptides, Deep Neural Networks, Physicochemical Properties.
Abstract: Antimicrobial peptides (AMPs) are crucial elements of the innate immune system; and they are effective
against bacteria that cause several diseases. These peptides are investigated as potential alternatives of
antibiotics to treat infections. Since wet lab experiments are expensive and time-consuming, computational
methods become crucial in this field. In this study, we suggest a computational technique for AMP prediction
using deep neural networks (DNN). We trained a DNN classifier using physicochemical features that include
a sequential model; and evaluated the model with 10-fold cross-validation on a benchmark dataset. We
compared our method with other machine learning approaches and demonstrated that the method we
developed generates higher performance (accuracy: 92%, precision: 92%, recall: 93%, f1: 93%, AUC: 98%).
In our experiments, we have realized that there is a strong positive correlation between the ‘Normalized
Hydrophobic Moment’ feature and ‘Angle Subtended by the Hydrophobic Residues’ feature; and strong
negative correlations between ‘Normalized Hydrophobicity’ feature and ‘Disordered Conformation
Propensity’ feature, and between ‘Amphilicity Index’ - ‘Disordered Conformation Propensity’ features. We
believe that the approach we proposed could guide further experimental studies and could facilitate the
prediction of other types of AMPs having anticancer, antivirus, antiparasitic activities.
1 INTRODUCTION
Antimicrobial peptides (AMP), which are known as
crucial elements of the innate immune system, are
shown to be effective against a variety of pathogenic
microorganisms, including bacteria, viruses, fungi,
parasites, and others (Erdem Büyükkiraz & Kesmen,
2022). In recent years, AMPs have drawn attention as
an alternative to chemical antibiotics due to the
developing resistance of microbial infections
(Thomas et al., 2010). AMPs have crucial roles
including quick microbial elimination and later
immunological regulation (Wang, 2014). These
effects come into the scene since AMPs cause
multiple bacterial harm, such as disruption of
bacterial membranes, inhibition of protein, or
interaction with specific intracellular targets (Bahar
& Ren, 2013; Malmsten, 2014). As a result, AMPs
became popular as novel medications.
a
https://orcid.org/0000-0002-6602-772X
b
https://orcid.org/0000-0001-8780-6303
c
https://orcid.org/0000-0002-2272-6270
Numerous computational techniques have been
proposed recently to advance the identification and
synthesis of antimicrobial drugs, and to accelerate the
candidate selection (Hammami & Fliss, 2010).
Sequence-based models have been trained using
machine learning algorithms as the primary way to
distinguish AMPs from non-AMPs. For example,
Thomas et al. used supervised learning methods such
as Random Forest (RF), Support Vector Machines
(SVM) and Discriminant Analysis (DA) for
prediction of AMPs based on physico-chemical
features (Thomas et al., 2010). Their prediction
models have the accuracy values of 93.2% for RF,
91.5% for SVM, and 87.5% for DA. Lata et al.
utilized an SVM based model using amino acid
composition as features (Lata et al., 2010). Their
model achieved 92.14% accuracy for the antibacterial
peptide classification problem. Joseph et al.
developed an algorithm called ClassAMP, a
combination of RF and SVM to predict the ability of
188
Söylemez, Ü., Yousef, M. and Bakir-Gungor, B.
Prediction of Antimicrobial Peptides Using Deep Neural Networks.
DOI: 10.5220/0011690000003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 3: BIOINFORMATICS, pages 188-194
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

a sequence to have antibacterial, antifungal or
antiviral mode of action (Joseph et al., 2012). Several
other computational tools have been developed for
this purpose (Lata et al., 2007; Thakur et al., 2012;
Xiao et al., 2013).
During recent years, deep learning techniques
have been used for antimicrobial peptide prediction.
Bhadra et al. developed an approach for sequences
shorter than 30 amino acids (Bhadra et al., 2018).
They achieved 77% accuracy with their deepAMP
method, which combined a convolutional neural
network with an ideal feature set with reduced amino
acid composition. In addition, they evaluated their
outcomes using RF and SVM algorithms. The SVM
model reached 72% accuracy while the RF reached
75% accuracy. Schneider et al. presented a
feedforward neural network using self-organizing
maps as input layers on AMP data and achieved 92%
accuracy (Schneider et al., 2017). Lin et al. developed
a method called AI4AMP using 6 different
physicochemical properties for encoding and a deep
learning model (Lin et al., 2021). Witten et al.
proposed a convolutional neural network (CNN)
method for regression of AMP (Witten & Witten,
2019). Minimum Inhibition Concentration (MIC)
values are used for regression. k-Nearest Neighbour
(kNN) and Ridge Regression models are used for
comparison with CNN. They showed that CNN
model has better root mean square error performance
(0.501) than other models used. Veltri et al. utilized a
deep neural network model based on convolution and
lstm layers using sequence to vector model for input
layers (Veltri et al., 2018). They showed that their
model has better performance when compared with
state-of-the-art models.
In this study, we propose a deep neural network
prediction model based on physico-chemical
properties of sequences for identifying AMP. Various
classification models are applied on the dataset, and
the outcomes are compared using various
performance metrics.
2 METHODS
2.1 Dataset
The independent dataset used in this study was
obtained from (Xiao et al., 2013). The dataset consists
of 920 AMP sequences and 920 non-AMP sequences,
which forms a two-class data set.
2.2 Sequence Representation
The key to solving operational difficulties effectively
is understanding how to formulate peptides
mathematically into the fixed length features to create
a robust AMP classification model. Table 1 shows
example sequences included in the existing dataset.
Each sequence is represented with 11 features
obtained from DBAASP web server and labeled as
“pos” if the sequence is AMP and “neg” if the
sequence is Non-AMP.
2.2.1 Generation of Physicochemical
Features
Normalized Hydrophobic Moment, Normalized
Hydrophobicity, Isoelectric Point, Penetration Depth,
Tilt Angle, Disordered Conformation Propensity,
Linear Moment, Propensity to in vitro Aggregation,
Angle Subtended by the Hydrophobic Residues,
Amphiphilicity Index, Propensity to PPII coil are
used as physicochemical features. DBAASP
(Vishnepolsky et al., 2018) web server is used to
calculate these features.
2.3 Deep Neural Network
Deep Neural Network is a machine learning method
that allows us to train a model and to predict outputs
for a given dataset. The artificial intelligence model
can be trained using both supervised and
unsupervised learning techniques. Neurons make up
artificial neural networks, similar to the human brain.
All neurons are linked together and have an impact on
the result. There are three layers that make up
neurons:
1. Input Layer; 2. Hidden Layer(s) and 3. Output Layer
The first hidden layer receives the input data from
the input layer and transmits it. On our inputs, hidden
layers run mathematical computations. Choosing the
amount of hidden layers and neurons for each layer is
one of the challenges in creating neural networks. The
output layer returns the output data.
Dense layer allows neurons from one layer to be
connected to the next layer as an input.
Batch Normalization (BN) is an algorithmic
technique that speeds up and improves the stability of
Deep Neural Network’s training. BN is a
normalization technique used in multilayer deep
neural networks to reduce the covariance between
hidden layers. The input of each layer is used by
normalizing the output vector of the previous layer.
Activation function determines whether or not a
neuron should be activated by generating a weighted
Prediction of Antimicrobial Peptides Using Deep Neural Networks
189

Table 1: Representation of the physico-chemical characteristics of AMP and Non-AMP peptides that are included in the
dataset, obtained from DBAASP (Vishnepolsky et al., 2018).
Sequence
Norm.
Hyd.
Moment
Norm.
Hydrophobicity
Isoelectric
Point
Penet.
Depth
Tilt
Angle
Dis. Conf.
Propensity
Linear
Moment
Prop. to in
vitro
Aggregation
Angle Subt.
by the Hydr.
Residues
Amph.
Index
Prop. to
PPII coil
class
MPIAQIHILEG
RSDEQKETLIR
EVSEAISRSLD
APLTSVRVIIT
EMAKGHFGIG
GELASKVRR
0.42 -0.53 7.66 30 110 0.20 0.15 4.77 20.00 0.63 0.97 neg
GTLPCESCVWI
PCISSVVGCSC
KSKVCYKN
0.23 -0.70 7.83 30 44 0.33 0.39 12.84 40.00 0.81 1.14 pos
TPCGESCVYIP
CISGVIGCSCT
DKVCYLN
0.27 -0.99 3.94 30 136 0.49 0.32 7.67 60.00 0.52 1.11 pos
IWGIGCNP 0.93 -1.27 3.50 14 15 0.42 0.56 0.79 110.0 0.87 1.05 ne
g
… …
total and then including a bias with it. The activation
function's objective is to add non-linearity to a
neuron's output.
2.4 Machine Learning Models for
Comparison
In this study we have considered different Machine
Learning algorithms for comparisons. The Support
Vector Machines (SVM) find the optimal hyperplane
based on the super vectors from each class (Cortes &
Vapnik, 1995). k-Nearest Neighbour is defined as the
classification of the data that has not yet been
assigned to a class by setting it in the optimal class
based on the distance which is calculated by
comparing the data of the unknown class with the
other data in the training set (Fix & Hodges, 1989).
The bagging approach seeks to retrain the basic
learner by creating new training sets from an old
training set. By using estimators on the bootstrapped
samples collected from the original dataset, an
ensemble is created (Breiman, 1996). Gradient
Boosting algorithm is a machine learning technique
that creates prediction models similar to decision
trees for regression and classification problems
(Friedman, 2002). A decision tree is built using the
data set as it is, and a new decision tree is created
based on its errors. Thus, a large number of decision
trees are created. The gradient boosting technique
gives the final decision by finding the sum of the
decisions made by all these trees.
2.5 Model Construction
As the details are illustrated in Figure 1, a deep neural
network algorithm is utilized to classify a sequence as
AMP or non-AMP. In our experiments, we used 10-
fold cross validation. We use 90% of data as training
and 10% as testing. For the first two activation
functions ‘relu’ and for the last one ‘sigmoid’ are
applied for classification. ‘Binary Cross Entropy’ is
used as a loss function and ‘adam’ is applied as an
optimizer.
2.6 Performance Metrics
Several performance evaluation criteria, including
accuracy, precision, recall, AUC and F1 measure,
were computed to evaluate the performance of our
models. These measures are defined as follows:
(1)
(2)
(3)
(4)
where TP stands for “true positive”, TN for “true
negative”, FP for “false positive”, and FN for “false
negative”.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
190
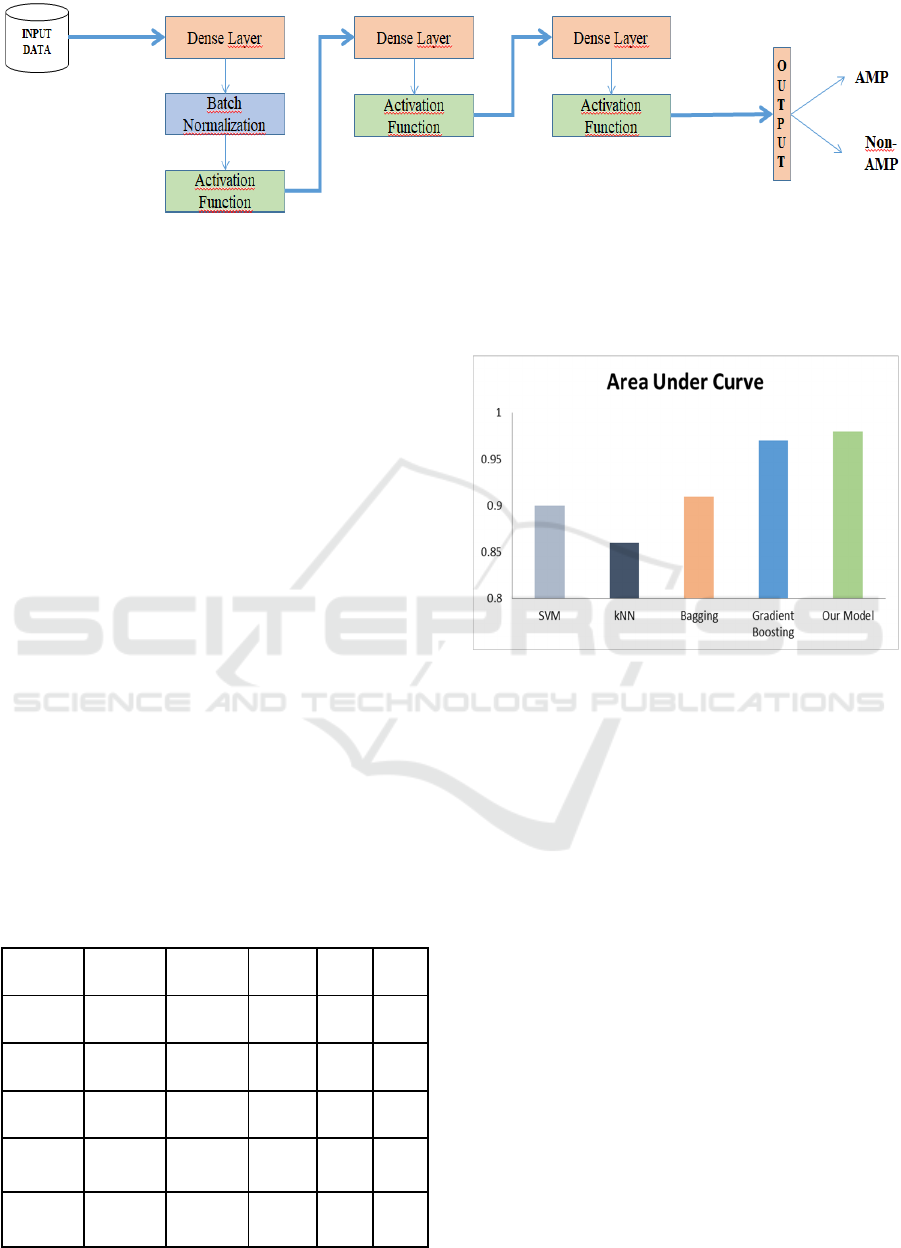
Figure 1: The architecture of the network consists of three blocks. First block consists of a dense layer, a batch normalization
layer, an activation layer and it is connected to the following one. The second block does not have a batch normalization layer
and it is connected to the previous and following blocks. The last block consists of a dense layer and an activation function
layer and it is connected to the previous block and output section.
3 RESULT
In our study, we have used 11 physicochemical
features that we mentioned above. We applied
different machine learning methods to classify each
peptide according to being AMP or not. We used
different performance metrics such as accuracy,
precision, recall, F1 measure, Area Under Curve
(AUC) for comparison. As shown in Table 2, our
methods achieve higher performances for all above-
mentioned metrics. Figure 2 represents the AUC
values for all tested classifiers. Our prediction method
yielded the best AUC result with 98%. In this study
we used a 10 fold cross validation technique;
calculated physicochemical properties and used them
as features. Other studies that use the same dataset do
not use these features and they use different peptide
properties (Meher et al., 2017; Veltri et al., 2017;
Xiao et al., 2013). They also apply different cross
validation techniques. For these reasons no
comparative evaluation could be made with these
above-mentioned studies.
Table 2: Comparison of our model with different machine
learning algorithms on the dataset in use.
Model Accuracy Precision Recall F1 AUC
SVM 0.77 0.71 0.93 0.80 0.90
kNN 0.80 0.87 0.79 0.80 0.86
Bagging 0.84 0.88 0.81 0.82 0.91
Gradient
Boostin
g
0.87 0.90 0.88 0.87 0.97
DNN
Model
0.92 0.92 0.93 0.93 0.98
Figure 2: Comparison of classifiers based on Area Under
Curve (AUC) metric.
The Pearson correlation values between all feature
pairs have been calculated using the Python Seaborn
Library in order to assess the pairwise correlations of
the features. These relations are depicted in Figure 3.
According to the correlation heatmap (shown in
Figure 3), there is a strong positive correlation
between ‘Normalized Hydrophobic Moment’ and
‘Angle Subtended by the Hydrophobic Residues’
features; and strong negative correlations between
‘Normalized Hydrophobicity’ and ‘Disordered
Conformation Propensity’ and between ‘Amphilicity
Index’ and ‘Disordered Conformation Propensity’
features. Except these, there are no other significant
correlations found between other feature pairs.
3.1 Feature Scoring and Feature
Ranking
Since deep neural networks do not allow us to
calculate importance scores for the features, we refer
to the Gradient Boosting model, which generated the
second highest performance in our experiments.
Prediction of Antimicrobial Peptides Using Deep Neural Networks
191
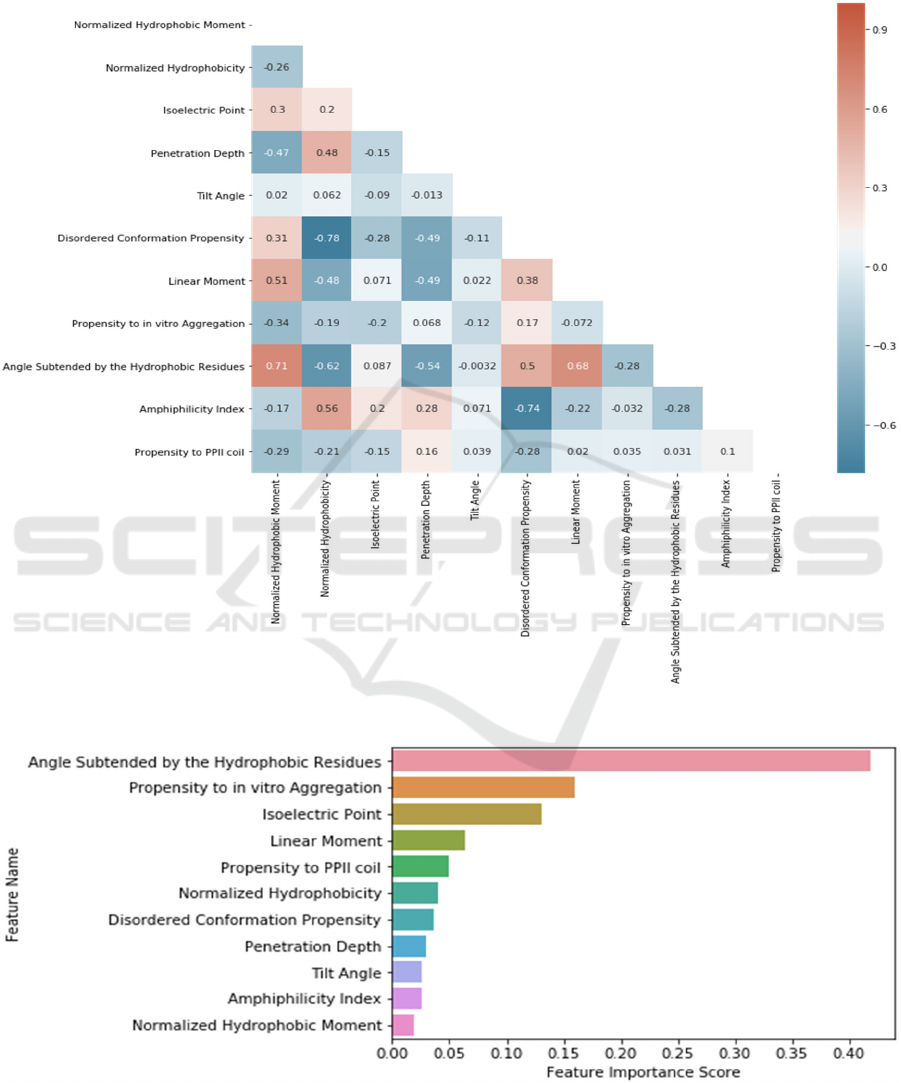
Hence, we scored each feature based on the Gradient
Boosting model. We have realised that Angle
Subtended by the Hydrophobic Residues, Propensity
to in vitro Aggregation and Isoelectric Point are more
important features than others when we examine the
score for each feature.
Figure 3: Correlation heatmap of physicochemical features, extracted from dataset in use.
Figure 4: Feature scoring according to their importances in classification using Gradient Boosting model.
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
192

4 DISCUSSION
Antimicrobial peptides (AMPs) are crucial elements
of the innate immune system and they are efficient
against bacteria that cause several diseases. These
peptides are investigated as potential alternatives to
antibiotics in order to treat infections. Since wet lab
experiments are expensive and time-consuming,
computational methods become crucial. In this study,
we suggest a precise computational technique for
AMP prediction using deep neural networks (DNN).
We evaluated the DNN classifier using
physicochemical features. Physicochemical
properties are one of the most frequently used
features for this problem(Lin et al., 2021; Moretta et
al., 2020; Vishnepolsky et al., 2019). In our previous
work, we have demonstrated that these features
perform better in predicting and describing the dataset
than other features and these properties should be
taken into account while developing novel models
(Söylemez et al., 2022). In this respect, we focused on
these features for this study and obtained satisfactory
results for different performance metrics (Table 2).
Additionally, it was found that Angle Subtended by
the Hydrophobic Residues is the greatest
distinguishing factor for antimicrobial peptide
prediction using the feature significance attribute of
the Gradient Boosting model.
5 CONCLUSION
AMPs are essential components of the innate immune
system and gaining importance in drug development.
Identification of AMPs has emerged as a critical
research area. The findings of this study suggest that
the model designed offers a reliable and practical
method.
We proposed a deep neural network based model
using different physicochemical features. We
demonstrated that our model outperformed its
competitors when we compared with regular machine
learning models such as SVM, kNN, Bagging and
Gradient Boosting. We believe that the approach we
proposed could guide further experimental studies
and could facilitate the prediction of other types of
AMPs having anticancer, antivirus, antiparasitic
activities.
REFERENCES
Bahar, A., & Ren, D. (2013). Antimicrobial Peptides.
Pharmaceuticals, 6(12), 1543–1575. https://doi.org/
10.3390/ph6121543
Bhadra, P., Yan, J., Li, J., Fong, S., & Siu, S. W. I. (2018).
AmPEP: Sequence-based prediction of antimicrobial
peptides using distribution patterns of amino acid
properties and random forest. Scientific Reports, 8(1),
1697. https://doi.org/10.1038/s41598-018-19752-w
Breiman, L. (1996). Bagging predictors. Machine
Learning, 24(2), 123–140. https://doi.org/10.1007/
BF00058655
Cortes, C., & Vapnik, V. (1995). Support-vector networks.
Machine Learning, 20(3), 273–297. https://doi.org/
10.1007/BF00994018
Erdem Büyükkiraz, M., & Kesmen, Z. (2022).
Antimicrobial peptides (AMPs): A promising class of
antimicrobial compounds. Journal of Applied
Microbiology, 132(3), 1573–1596. https://doi.org/10.
1111/jam.15314
Fix, E., & Hodges, J. L. (1989). Discriminatory Analysis.
Nonparametric Discrimination: Consistency Properties.
International Statistical Review / Revue Internationale
de Statistique, 57(3), 238. https://doi.org/10.23
07/1403797
Friedman, J. H. (2002). Stochastic gradient boosting.
Computational Statistics & Data Analysis, 38(4), 367–
378. https://doi.org/10.1016/S0167-9473(01)00065-2
Hammami, R., & Fliss, I. (2010). Current trends in
antimicrobial agent research: Chemo- and
bioinformatics approaches. Drug Discovery Today,
15(13–14), 540–546. https://doi.org/10.1016/j.drudis.
2010.05.002
Joseph, S., Karnik, S., Nilawe, P., Jayaraman, V. K., &
Idicula-Thomas, S. (2012). ClassAMP: A Prediction
Tool for Classification of Antimicrobial Peptides.
IEEE/ACM Transactions on Computational Biology
and Bioinformatics, 9(5), 1535–1538.
https://doi.org/10.1109/TCBB.2012.89
Lata, S., Mishra, N. K., & Raghava, G. P. (2010). AntiBP2:
Improved version of antibacterial peptide prediction.
BMC Bioinformatics, 11(S1), S19. https://doi.org/
10.1186/1471-2105-11-S1-S19
Lata, S., Sharma, B., & Raghava, G. (2007). Analysis and
prediction of antibacterial peptides. BMC
Bioinformatics, 8(1), 263. https://doi.org/10.1186/
1471-2105-8-263
Lin, T.-T., Yang, L.-Y., Lu, I.-H., Cheng, W.-C., Hsu, Z.-
R., Chen, S.-H., & Lin, C.-Y. (2021). AI4AMP: An
Antimicrobial Peptide Predictor Using
Physicochemical Property-Based Encoding Method
and Deep Learning. MSystems, 6(6), e00299-21.
https://doi.org/10.1128/mSystems.00299-21
Malmsten, M. (2014). Antimicrobial peptides. Upsala
Journal of Medical Sciences, 119(2), 199–204.
https://doi.org/10.3109/03009734.2014.899278
Meher, P. K., Sahu, T. K., Saini, V., & Rao, A. R. (2017).
Predicting antimicrobial peptides with improved
accuracy by incorporating the compositional, physico-
Prediction of Antimicrobial Peptides Using Deep Neural Networks
193

chemical and structural features into Chou’s general
PseAAC. Scientific Reports, 7(1), 42362.
https://doi.org/10.1038/srep42362
Moretta, A., Salvia, R., Scieuzo, C., Di Somma, A., Vogel,
H., Pucci, P., Sgambato, A., Wolff, M., & Falabella, P.
(2020). A bioinformatic study of antimicrobial peptides
identified in the Black Soldier Fly (BSF) Hermetia
illucens (Diptera: Stratiomyidae). Scientific Reports,
10(1), 16875. https://doi.org/10.1038/s41598-020-
74017-9
Schneider, P., Müller, A. T., Gabernet, G., Button, A. L.,
Posselt, G., Wessler, S., Hiss, J. A., & Schneider, G.
(2017). Hybrid Network Model for “Deep Learning” of
Chemical Data: Application to Antimicrobial Peptides.
Molecular Informatics, 36(1–2), 1600011.
https://doi.org/10.1002/minf.201600011
Söylemez, Ü. G., Yousef, M., Kesmen, Z., Büyükkiraz, M.
E., & Bakir-Gungor, B. (2022). Prediction of Linear
Cationic Antimicrobial Peptides Active against Gram-
Negative and Gram-Positive Bacteria Based on
Machine Learning Models. Applied Sciences, 12(7),
3631. https://doi.org/10.3390/app12073631
Thakur, N., Qureshi, A., & Kumar, M. (2012). AVPpred:
Collection and prediction of highly effective antiviral
peptides. Nucleic Acids Research, 40(W1), W199–
W204. https://doi.org/10.1093/nar/gks450
Thomas, S., Karnik, S., Barai, R. S., Jayaraman, V. K., &
Idicula-Thomas, S. (2010). CAMP: A useful resource
for research on antimicrobial peptides. Nucleic Acids
Research, 38(suppl_1), D774–D780. https://doi.org/
10.1093/nar/gkp1021
Veltri, D., Kamath, U., & Shehu, A. (2017). Improving
Recognition of Antimicrobial Peptides and Target
Selectivity through Machine Learning and Genetic
Programming. IEEE/ACM Transactions on
Computational Biology and Bioinformatics, 14(2),
300–313. https://doi.org/10.1109/TCBB.2015.2462364
Veltri, D., Kamath, U., & Shehu, A. (2018). Deep learning
improves antimicrobial peptide recognition.
Bioinformatics, 34(16), 2740–2747. https://doi.org/
10.1093/bioinformatics/bty179
Vishnepolsky, B., Grigolava, M., Zaalishvili, G.,
Karapetian, M., & Pirtskhalava, M. (2018). DBAASP
Special prediction as a tool for the prediction of
antimicrobial potency against particular target species.
Proceedings of 4th International Electronic Conference
on Medicinal Chemistry, 5608. https://doi.org/10.33
90/ecmc-4-05608
Vishnepolsky, B., Zaalishvili, G., Karapetian, M.,
Nasrashvili, T., Kuljanishvili, N., Gabrielian, A.,
Rosenthal, A., Hurt, D. E., Tartakovsky, M., Grigolava,
M., & Pirtskhalava, M. (2019). De Novo Design and In
Vitro Testing of Antimicrobial Peptides against Gram-
Negative Bacteria. Pharmaceuticals, 12(2), 82.
https://doi.org/10.3390/ph12020082
Wang, G. (2014). Human Antimicrobial Peptides and
Proteins. Pharmaceuticals, 7(5), 545–594.
https://doi.org/10.3390/ph7050545
Witten, J., & Witten, Z. (2019). Deep learning regression
model for antimicrobial peptide design [Preprint].
Bioinformatics. https://doi.org/10.1101/692681
Xiao, X., Wang, P., Lin, W.-Z., Jia, J.-H., & Chou, K.-C.
(2013). iAMP-2L: A two-level multi-label classifier for
identifying antimicrobial peptides and their functional
types. Analytical Biochemistry, 436(2), 168–177.
https://doi.org/10.1016/j.ab.2013.01.019
BIOINFORMATICS 2023 - 14th International Conference on Bioinformatics Models, Methods and Algorithms
194
