
EMTE: An Enhanced Medical Terms Extractor
Using Pattern Matching Rules
Monah Bou Hatoum
1 a
, Jean-Claude Charr
1 b
, Christophe Guyeux
1 c
,
David Laiymani
1 d
and Alia Ghaddar
2 e
1
University of Bourgogne Franche-Comt
´
e, UBFC, CNRS, 90000 Belfort, France
2
Department of Computer Science, International University of Beirut, Beirut P.O. Box 146404, Lebanon
Keywords:
Deep Learning, Natural Language Processing (NLP), Computer-Aid Diagnosis, Chief Complaints, Text
Mining, Abbreviations, Negations, Phrases.
Abstract:
Downstream tasks like clinical textual data classification perform best when given good-quality datasets. Most
of the existing clinical textual data preparation techniques rely on two main approaches, removing irrelevant
data using cleansing techniques or extracting valuable data using feature extraction techniques. However, they
still have limitations, mainly when applied to real-world datasets. This paper proposes a cleansing approach
(called EMTE) which extracts phrases (medical terms, abbreviations, and negations) using pattern-matching
rules based on the linguistic processing of the clinical textual data. Without requiring training, EMTE extracts
valuable medical data from clinical textual records even if they have different writing styles. Furthermore,
since EMTE relies on dictionaries to store abbreviations and pattern-matching rules to detect phrases, it can
be easily maintained and extended for industrial use. To evaluate the performance of our approach, we com-
pared the performance of EMTE to three other techniques. All four cleansing techniques were applied to a
large industrial imbalanced dataset, consisting of 2.21M samples from different specialties with 1,050 ICD-10
codes. The experimental results on several Deep Neural Network (DNN) algorithms showed that our cleansing
approach significantly improves the trained models’ performance compared to the other tested techniques and
according to different metrics.
1 INTRODUCTION
The International Classification of Diseases, the 10
th
edition (ICD-10), is a standard tool to classify disease
diagnoses from a patient’s medical signs, symptoms,
and other health conditions. Governments, Health
Insurance Companies, and Healthcare providers also
use the ICD codes to report and communicate pa-
tients’ medical cases. ICD-10 codes are hierarchical
alphanumeric labels with a length between three to
seven characters depending on the depth of the hierar-
chy and the disease’s specificity level. The specificity
of the ICD-10 codes is crucial and controls which
treatment plan the physicians follow on the patients.
Physicians usually manually assign one or mul-
a
https://orcid.org/0000-0002-0773-8409
b
https://orcid.org/0000-0002-0807-4464
c
https://orcid.org/0000-0003-0195-4378
d
https://orcid.org/0000-0003-2580-6660
e
https://orcid.org/0000-0003-1363-6174
tiple ICD-10 codes to describe the patient’s illness
and symptoms during every patient visit. However,
this manual operation is time-consuming and error-
prone due to the large available number of ICD-10
codes. Consequently, hospitals and medical institutes
are motivated to turn to auto-diagnosis tools. In recent
years, researchers have provided several approaches
that tackle ICD-10 prediction from clinical textual
data, extracting medical terms using feature extrac-
tion techniques, and cleansing approaches to remove
the irrelevant data. Unfortunately, most of these ex-
isting approaches have limitations and show poor re-
sults when applied to industrial datasets (Dugas et al.,
2016). The significant limitations of these approaches
are: studying part of the ICD-10 codes without pre-
serving the specificity of the codes, applying data pre-
processing without studying its impact on the data,
and using small datasets to train models to extract fea-
tures from clinical textual data without studying other
essential features like the specialty , (Chraibi et al.,
2021)(Azam et al., 2020). We believe the main rea-
Hatoum, M., Charr, J., Guyeux, C., Laiymani, D. and Ghaddar, A.
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules.
DOI: 10.5220/0011717300003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 2, pages 301-311
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
301

31 year old female since one month ago has complained of epigastric
pain , increased after meals, associated with SOB , no nausea , no bowel
habits changes. no rectal bleeding,no weight loss,good appetite
no family history of CRC cancer.
Figure 1: An example of unprocessed chief complaint that
contains abbreviations like ”SOB”, ”CRC” and negations
like ”no weight loss”, ”no nausea” and terms like ”epigas-
tric pain”.
son behind this gap is the lack of knowledge about
the nature of the data. Clinical textual data are di-
verse, incomplete, and redundant. They includes ab-
breviations, periods, negations, and terms of one or
more words. Also, these data have different writing
styles. For example, negations could appear on the
left side like ”no rectal bleeding” or the right side
like ”smoker: no”. In addition, a medical term could
appear with its expanded form (”long-term”) ”Dia-
betes Mellitus” or using an abbreviated form (”short-
term”) ”DM”. For all these reasons, extracting valu-
able information from medical datasets is complex,
and it is essential to understand how physicians en-
code the medical phrases and the different writing
styles available.
This study aims to improve the ICD-10 predic-
tion performance by improving the data quality. In
particular, we provide a preprocessing data approach
for clinical textual data that enhances the quality of
data fed to state-of-art NLP models, while preserv-
ing the maximum information possible. Our approach
employs the power of Part of Speech (POS) tagging
(de Marneffe et al., 2021) and the pattern-matching
rules (PMRs) to extract valuable data and eliminate
irrelevant data. With the help of a medical team,
we built the pattern-matching rules to extract phrases
(negations with their different writing styles, abbre-
viations, and medical terms). In this work, the clini-
cal textual data consists of chief complaints (CC) and
History of Present Illness (HPI) written by physicians
as depicted in Figure 1. The main contributions of this
study can be summarized as follows:
• Provide a cleansing approach for clinical tex-
tual data using dictionary based pattern-matching
rules. Our approach extracts phrases (negations
and medical terms) from clinical textual data
and combines the detected words as one medical
phrase.
• Replace the short-term abbreviations with their
full-term forms, considering the specialties as an
additional feature to reduce abbreviation ambigu-
ity.
• Improve negation detection covering different
writing styles.
• Provide a comparative study that uses a large in-
dustrial dataset with 2.21M samples and 1,050
ICD-10 codes and shows that our cleansing ap-
proach has a better impact on the performance of
various NLP models for solving the multi-label
ICD-10 classification problem than three different
existing cleansing techniques.
The rest of this paper is organized as follows: Sec-
tion 3 presents some of the state-of-the-art text cleans-
ing techniques and feature extraction along with their
shortcomings. Our cleansing approach is detailed in
Section 4. The results of the comparative study be-
tween our approach and other cleansing methods are
exposed in Section 5. Section 6 details our findings
and recommendations to efficiently cleanse medical
text data. This article ends with a summary of the
contributions, and some future works are outlined.
2 BACKGROUND
This section presents a brief background about Nat-
ural Language Processing (NLP) and the significant
challenges to overcome when cleansing clinical tex-
tual data.
2.1 POS Tagging
Part of Speech tagging is a Natural Language Pro-
cessing (NLP) process in which every token in the
text is assigned a grammatical tag based on its def-
inition and context. A token in a text might be a
word, punctuation or space. ”POS” tags contain sev-
eral labels like ”ADJ” which stands for ”Adjective”
and ”PRON” stands for ”Pronoun” (Zeman, 2022).
In addition, Dependency Parsing (DEP) (de Marneffe
et al., 2021) is another NLP process that builds re-
lations between the words in the text, based on the
POS tags (Nguyen and Verspoor, 2019). Many avail-
able tools, like CoreNLP (Manning et al., 2014) and
Spacy (Honnibal et al., 2020), use Machine Learning
algorithms to offer many NLP functionalities like to-
kenization, POS tagging, sentence segmentation, de-
pendency parsing and entity recognition.
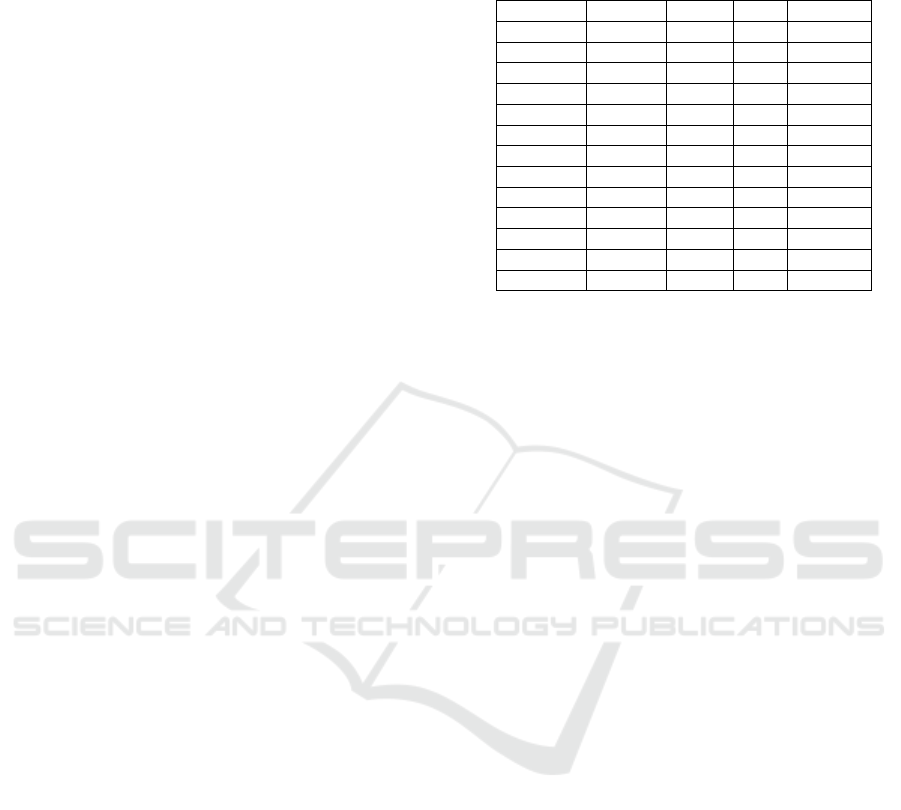
As an example, Table 1 shows a part of the lin-
guistic features output of the chief complaint show
in Figure 1, generated by the tool ”Spacy” (Honni-
bal et al., 2020). It shows the tokenization TEXT,
the lemmatization of the token LEMMA, POS, TAG,
and DEP for every token (word, punctuation, space)
in the chief complaint. This text tagging can be used
to build pattern-matching rules to discover negations,
multi-words phrases, etc.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
302
2.2 Abbreviations in Healthcare
Abbreviations are frequently used in healthcare to re-
duce time and typos. However, they create a signif-
icant challenge for the machine learning techniques
due to their ambiguity. Indeed, the same abbreviation
could have different meanings depending on the con-
text and the specialty. When analyzing clinical text,
three types of abbreviations can be found:
i) General abbreviations that have a common
meaning, such as ”dx” which stands for ”diag-
nosis” or ”c/o” which stands for ”complaining
of”.
ii) Specialty-specific abbreviations, where the
meaning differs from one specialty to another,
such as ”CLD” which stands for ”Chronic Liver
Disease” in the Gastroenterology department
and ”Chronic Lung Disease” in the Pulmonary
unit. Similarly, ”MS” is the abbreviation of
”Multiple Sclerosis” in the Neurology depart-
ment and ”Mitral Stenosis” in the Cardiology
and Radiology departments.
iii) An Ambiguous abbreviation that has a contex-
tual related meaning, such as ”LFT”, which ei-
ther stands for ”Lung Function Test” or ”Liver
Function Test”.
Many studies have tackled the abbreviation ambi-
guity using supervised (Koptient and Grabar, 2021)
and unsupervised (Marta Skreta, 2019) machine
learning approaches. However, most of these stud-
ies have limitations. They were only trained on clini-
cal textual data without taking other essential param-
eters like the specialty into consideration. Therefore,
these approaches perform poorly on multi-specialty
large datasets (Grossman Liu et al., 2021) because
they miss many abbreviations or suggest a wrong ex-
panded form of the abbreviated term.
2.3 Negation Detection
In clinical documents, the terms ”no”, ”nil”, ”ab-
sence of”, ”negative”, ”n’t”,”-ve” are often used
for negation. Hence, removing negations drastically
changes the semantics and the interpretation of clini-
cal notes. For instance, removing the word ”no” from
”patient has no cancer” will completely change its
meaning. Also, removing the punctuation ”−” or ”+”
from (”-ve” or ”+ve”) changes the meaning from
(”negative” or ”positive”) into ”ve” which refers to
”vaginal examination” in the ”Obstetrics and Gyne-
cology” department and ”ventricular extrasystoles”
in the Cardiology department. It is important to ex-
amine the negation indicators beside a word entity
Table 1: Tokenization, Lemmatization, POS tagging, and
dependency parsing result of the beginning of the chief
complaint in Figure 1 using Spacy.
TEXT LEMMA POS TAG DEP
31 31 NUM CD nummod
year year NOUN NN npadvmod
old old ADJ JJ amod
female female NOUN NN nsubj
since since SCONJ IN prep
one one NUM CD nummod
month month NOUN NN npadvmod
ago ago ADV RB pcomp
has have AUX VBZ aux
complained complain VERB VBN ROOT
of of ADP IN prep
epigastric epigastric ADJ JJ amod
pain pain NOUN NN pobj
and merge data with semantic spaces to appropriately
detect a negation. Moreover, physicians could write
the negations in different forms such as: Non smoker,
doesn’t smoke, smoker: no, or smoker: nil which
increases the complexity of detecting the negations.
Most of the existing solutions rely on the Dependency
Relation (DEP) like in (Mehrabi et al., 2015). Unfor-
tunately, most of the existing negation detectors fail to
detect all the potential negations since the DEP pro-
cess cannot handle all the negations writing styles in
clinical textual data (Wu et al., 2014).
3 RELATED WORK
3.1 Existing Cleansing Techniques
Many research studies analyzed clinical textual data
using machine learning techniques. In some of these
studies (Atutxa et al., 2019), (Shaalan et al., 2020)
and (Makohon and Li, 2021), researchers trained the
models directly on the raw data without any pre-
processing. They only relied on the power of the
machine learning techniques, like deep neural net-
works (DNN), to discover the relationship among the
data. The main limitations of such approaches are in-
creasing the training complexity and the dimensional
space, potentially leading to over-fitting problems and
low testing accuracy (Joachims, 1998).
On the other hand, other studies (Chen et al.,
2020), (Du et al., 2019), (Lucini et al., 2017) and
(Bai and Vucetic, 2019) have applied standard cleans-
ing (SC) techniques like stemming, lemmatization,
stop-words removal, and punctuation removal. Un-
fortunately, these preprocessing steps reduce the data
quality instead of improving it. For example, re-
moving stop-words like ”no”, ”has”, ”none”, ”not”
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules
303

change the meaning of the input data. As an exam-
ple, both complaints ”a patient has a colon cancer for
six months complaining from severe abdominal pain”
and ”a patient with severe abdominal pain, no colon
cancer in family history” would have the same mean-
ing if has and no were removed. Moreover, removing
punctuations from clinical textual data increases the
challenge of detecting the proper abbreviations and
distinguishing between dates and numbers.
3.2 Feature Extraction
Feature Extraction (FE) and Named Entity Recogni-
tion (NER) using machine learning are two tasks in
Natural Language Processing (NLP) that were widely
used in the last few years. The former is a process
of identifying and extracting important characteris-
tics from data, while the latter identifies and classi-
fies named entities in text. In healthcare, extracting
all essential features from data requires enormous re-
sources and is time-consuming due to the high dimen-
sionality of the data. FE and NER came into play
to help identify the relevant data such as diseases,
treatments, abbreviations, and symptoms; this helps
reduce the vocabulary size and hyperspace dimen-
sion of the data. However, these approaches require
massive labeled data for training using the super-
vised approaches (Adnan and Akbar, 2019). More-
over, the diversity of data makes these approaches
inefficient for both supervised and unsupervised ap-
proaches (Li et al., 2018), (Dugas et al., 2016). In
addition, the generated pre-trained models from these
approaches require large efforts to maintain and up-
date them to capture the new terms and to fix wrongly
predicted entities, which is time-consuming. Unfor-
tunately, with all these limitations, many of the exist-
ing FE and NER approaches are not ready for health-
care industrial use where the data are massive and
complex. On the other hand, other feature extrac-
tion approaches use pattern-matching rules (PMR).
PMRs are a set of rules manually written to iden-
tify patterns using lexico-syntactic patterns to iden-
tify the occurrence of similar entities in NLP. PMRs
are widely used for financial topics (Zheng et al.,
2021). Unfortunately, rare research topics investi-
gated these approaches in healthcare because since
2018 they mainly concentrated their work on machine
learning techniques (Bose et al., 2021). Unlike the
feature extraction tools using machine learning tech-
niques, PMRs are easy and faster to develop; they do
not require labeled datasets or downstream tasks.
3.3 Word Embeddings
Word Embedding is a technique used in natural lan-
guage processing (NLP) that represents words in a
vector form. Many techniques are available for word
embeddings such as Text Vectorization using Padding
sequences ”PS” (Abadi et al., 2015), Sentence2Vec
”S2V” (Pagliardini et al., 2017), BERT (Devlin et al.,
2019). Word Embedding is an essential step for con-
verting the textual data into numerical representation
for proceeding with the downstream tasks.
This paper aims to provide an approach for im-
proving the data quality from the data preparation
phase without losing vital information and consider-
ing crucial industrial requirements such as flexibility
and maintainability.
4 MATERIALS AND METHODS
EMTE (Enhanced Medical Terms Extractor) is an ap-
proach that extracts phrases and eliminates irrelevant
data from the clinical textual data. A phrase is a set
of one or more tokens that could be abbreviations,
negations, medical terms, other conditions, signs, and
symptoms. A token is a word, punctuation, or num-
ber. For example, ”no rectal bleeding” is a phrase
while ”no”, ”rectal”, and ”bleeding” are tokens.
EMTE depends on PMRs encoded using a combi-
nation of the linguistic features (POS, TAG, LEMMA,
and DEP) to detect phrases from clinical textual data.
In addition, it relies on JSON dictionaries to store
the phrases’ PMRs. Unlike the pre-trained models,
JSON dictionaries are simple, flexible, and maintain-
able, which are desirable solutions for industrial use.
EMTE has four main phases: (1) extraction of to-
kens from every chief complaint, (2) load dictionary
rules, (3) extract relevant phrases, (4) generate new
processed chief complaints. The dictionaries and al-
gorithms used by EMTE are presented in the next sub-
sections.
4.1 Dictionaries and Rules
The main objective of this paper is to detect the
medical terms, abbreviations and negations in chief
complaints during the cleansing phase using pattern
matching rules. It should improve the performance of
the machine learning models applied on the cleansed
data. For example, detecting the abbreviations and re-
placing them with their full-terms should reduce the
vocabulary size and the hyperspace dimension.
The PMRs were developed as follows: First, with
the help of a medical team and after analyzing many
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
304

chief complaints and discharge summaries, the dif-
ferent structures of clinical terms (length and syn-
tactical orders) were enumerated. Second, the dis-
covered structures were translated into linguistic key-
words (VERB, ADV, NOUN, PRONOUN, NEG, and
LEMMA). Finally, the patter-matching rules to detect
these structures were developed using the Spacy syn-
tax for the sake of experiments. The resulting PMRs
and the abbreviations were stored in two JSON dic-
tionaries denoted, R and A respectively.
4.1.1 Abbreviation Rules
As mentioned in Section 2.2, the existing solutions
have limitations and do not clarify the ambiguity of
abbreviations. To reduce the abbreviations ambiguity,
the physician’s specialty was considered while pro-
cessing the clinical textual data. Furthermore, to en-
sure flexibility and maintainability, the abbreviation
dictionary stores both general and specific abbrevia-
tions with their corresponding set of specialties.
Listing 1 shows a few entries in the JSON abbre-
viation dictionary. Every entry represents an abbre-
viation with three attributes: (”short”, ”specialties”,
and ”full”). The key ”short” stores the list of pos-
sible short-terms of the abbreviation, like ”dx” and
”pmh”. The key ”full” corresponds to the full expan-
sion of the abbreviated term. For example, ”hyper-
ventilation syndrome” is the full-term of ”hvs”.
Finally, the key ”specialties” stores the list of spe-
cialties where the abbreviated term can be used with-
out ambiguity. For example, the abbreviation ”hvs”
in Listing 1 has the same meaning in the ”Emergency
and Pulmonary” departments and can be used with
no ambiguity. On the other hand, it has a different
meaning when used in the ”Obstetrics and Gynecol-
ogy” department. For this reason, a second entry for
this abbreviation was added to the dictionary with the
”Obstetrics and Gynecology” specialties. It must be
noted that key ”specialties” could be empty if the ab-
breviation is a general non-ambiguous term. For in-
stance, it is empty for the abbreviation ”dx” because it
has the same meaning, ”diagnosis”, in all specialties.
4.1.2 Negations and Medical Terms Rules
Physicians use medical terms and negations in differ-
ent ways. For example, they may use the full-terms
(e.g., past medical history), or the short-terms (e.g.,
pmh). They may also put the negations on the left
side (e.g., no pmh), or the right side (e.g. pmh: no).
Fortunately, these different writing styles follow some
patterns, which can be captured using PMRs. For ex-
ample, the medical term, ”epigastric pain”, was de-
tected using a rule that catches the pattern: ”Adj”
[ { ” s h o r t ” : [ ” dx ” , ” d i a g ” ] ,
” f u l l ” : ” d i a g n o s i s ” ,
” s p e c i a l t i e s ” : [ ]
} , { ” s h o r t ” : [ ” c s ” , ” c / s ” , ” c / s e c ” , ” c . s . ” ] ,
” f u l l ” : ” c a e s a r e a n s e c t i o n ” ,
” s p e c i a l t i e s ” : [ ” O b s t e t r i c s and
Gyn ec olo gy ” ]
} , { ” s h o r t ” : [ ” h vs ” ] ,
” f u l l ” : ” h y p e r v e n t i l a t i o n sy nd ro me ” ,
” s p e c i a l t i e s ” : [ ” em ergen cy ” , ” p u lm o nar y ” ]
} , { ” s h o r t ” : [ ” h vs ” ] ] ,
” f u l l ” : ” h i g h v a g i n a l swab ” ,
” s p e c i a l t i e s ” : [ ” O b s t e t r i c s and
Gyn ec olo gy ” } ]
Listing 1: A sample of the abbreviations dictionary.
Figure 2: The detected phrases of chief complaint sample
in Figure 1 using our approach EMTE.
followed by a ”Noun”. If for the same phrase more
than one rule can be applied, the one with the most
tokens is applied. For example, the rule that detects
the negation, ”no epigastric pain” consisting of three
tokens, is applied instead of the one that just detects
the medical term, ”epigastric pain” consisting of just
two tokens. If two rules concern the same number of
tokens, the priority is given to the negation rule, oth-
erwise the first rule is selected. Figure 2 shows the
detected phrases from the raw chief complaint, pre-
sented in Figure 1, using EMTE.
Listing 2 shows a part of the rules dictionary.
These rules were built with the help of a medical team
after analyzing the different writing styles of physi-
cians working in a multinational Saudi private hospi-
tal. Every entry represents a rule definition that con-
tains two attributes (”type” and ”rule”). The first
key represents the type of the rule, which is either
a ”negation” or a ”medical term”. The second at-
tribute stores the pattern-matching rule. For example,
the second entry in Listing 2 detects a negation phrase
formed of a negation determiner followed by three
nouns, such as ”no bowel habits changes”. EMTE
can also detect the words that start with the ”non”
prefix, such as ”nonsmoker”, ”non-stick” and ”non-
fat”. These words are replaced by the following form:
”non smoker” ” non stick” and ”non fat”. Splitting
these words helps in unifying the terms and reducing
the vocabulary size.
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules
305

[ { ” t y p e ” : ” n e g a t i o n ” , ” r u l e ” : { ” l a b e l ” : ”
n e g a t i o n ” , ” p a t t e r n ” : [ [ { ’POS ’ : ’NOUN’ } , { ’
IS PUNCT ’ : T rue } , { ’DEP’ : ’ neg ’ } ] , [ { ’POS ’
: ’NOUN’ } , { ’ IS SPACE ’ : Tru e } , { ’DEP’ : ’ neg ’
} ] ] }
} , { ” t y p e ” : ” n e g a t i o n ” , ” r u l e ” : { ” l a b e l ” : ”
n e g a t i o n ” , ” p a t t e r n ” : [ [ { ’DEP’ : ’ neg ’ } , { ’
POS’ : ’NOUN’ } , { ’POS’ : ’NOUN’ } , { ’POS ’ : ’
NOUN’ } ] ] }
} , { ” t y p e ” : ” te r m ” , ” r u l e ” : { ” l a b e l ” : ” gen d e r
” , ” p a t t e r n ” : [ { ’LEMMA’ : { ’ IN ’ : [ ” g i r l ” , ”
boy ” , ”man” , ”woman” , ” l a d y ” , ” guy ” , ” f e m a l e ”
, ” male ” ] } } ] }} ]
Listing 2: A sample of the PMRs dictionary.
4.2 Definitions and Notations
Let Σ = {σ
j
}
z
j=1
be the set of the z available special-
ties. Let C = {S
i
}
n
i=1
be a raw data corpus consist-
ing of n samples, S
i
. Every S
i
∈ C is a tuple with
three attributes, S
i
= (τ
i
, σ
i
, λ
i
). S
i
contains the chief
complaint text τ
i
, the specialty σ
i
∈ Σ, and the set of
true labels λ
i
(i.e. ”ICD-10 codes”). Also, let Γ be
the annotation function that splits the chief complaint
τ
i
into tokens (word, punctuation, space) and applies
POS tagging on the resulting tokens. Finally, let Ψ
be the parser function that applies a set of rules on a
given set of tokens.
Token: A chief complaint τ
i
contains a set of to-
kens {t
i j
}
k
j=1
where k is the number of tokens. Ev-
ery token t
i j
contains five attributes (text
i j
, lemma
i j
,
pos
i j
, tag
i j
, dep
i j
) where text
i j
is the splitted token,
lemma
i j
is the lemmetization of t
i j
and pos
i j
, tag
i j
and dep
i j
) represent the linguistic features of t
i j
.
Abbreviation: Every abbreviation d
j
is a tuple
containing three attributes, d
j
= (α
j
, δ
j
, ε
j
), where α
j
contains the short-terms of the abbreviation, δ
j
cor-
responds to the full-term (i.e the expanded form) of
the abbreviation, and ε
j
stores the list of specialties
where the abbreviation α
j
can be used without am-
biguity. ε
j
is empty if α
j
is a general abbreviation.
Thus, ε
j
⊂ {φ}∪{σ
p
| σ
p
∈ Σ}
l
p=1
where l is the num-
ber of allowed specialties for the given abbreviation
α
j
.
Rule: A rule can be applied to detect negations or
medical terms. Thus, every rule r has two attributes,
i.e. r = (e, p), where e is the type of the rule and p is
the pattern-matching rule.
Dictionaries: The PMRs and abbreviations are
stored in two JSON dictionaries denoted, A and R
respectively. A = {d
j
}
s
j=1
is a dictionary of s abbre-
viations d
j
. While, R = {r
j
}
u
j=1
is the list of all nega-
tions and medical terms rules, r
j
. It is worth mention-
ing that the PMRs are independent from the special-
ties.
Phrase: Every phrase m
j
has two attributes, the
”label” from the available set of labels (”gender”,
”negation”, ”term”, ”period”, and ”abbreviation”)
and the ”phrase”, which is a set of detected tokens
{t
jp
}
c
p=1
of size c ≤ k. The labels are used for report-
ing and tracing purposes.
Let Ψ : (γ, ω) 7→ {m
j
}
b
j=1
be the parser function
that parses linguistically annotated set of tokens γ
based on the set of rules ω. The Ψ function gener-
ates b phrases.
For instance, ψ
i
= Ψ(γ
i
, ω
i
) = {m
i j
}
b
j=1
are the set
of detected phrases from sample S
i
.
4.3 Steps of EMTE Approach
Our aim is to apply the PMRs on the corpus in or-
der to extract the relevant phrases from every chief
complaint τ
i
. Algorithm 1 shows the pseudo-code of
EMTE which takes the abbreviations dictionary A ,
the negations and terms dictionary R , and the cor-
pus C as inputs. The algorithm returns the processed
corpus C
0
. EMTE loops on all S
i
∈ C and performs
several steps as follows:
- Extract all the tokens from every chief com-
plaint: First, EMTE splits every τ
i
into tokens. It uses
the linguistic annotator method Γ : τ 7→ γ, which is
available in many NLP tools (Honnibal et al., 2020)
and (Manning et al., 2014). The method returns a set
of tokens and generates their linguistic features. Thus,
for every chief complaint τ
i
, γ
i
= Γ(τ
i
) = {t
i j
}
w
i
j=1
is
the set of w
i
tokens in τ
i
.
- Generate and load the dictionary rules: Since
some abbreviations might depend of the specialties,
the abbreviation PMRs are built for every sample S
i
according to the specialty σ
i
. Therefore, EMTE first
finds all the abbreviations’ short-terms µ
i
that satisfy
the specialty σ
i
in sample S
i
. Then, it auto-generates
the PMRs ρ
i
that are specific to this specialty:
ρ
i
= {t
i j
| lemma
i j
∈ µ
i
}
k
j=1
where,
µ
i
=
[
{α
j
| ε
j
= φ ∨ σ
i
∈ ε
j
}
v
j=1
- Extract relevant phrases: The previously gener-
ated rules are used to extract phrases from each clin-
ical text. Let ω
i
= R ∪ {ρ
i
} be the set of PMRs to
be applied on the tokens γ
i
for each sample S
i
. The
parser Ψ(γ
i
, ω
i
) is called to generate ψ
i
, the list of all
detected phrases.
Figure 3 shows the result of all pattern matching
rules in action. For example, shortness of breath is
the result of the abbreviation pattern matching rule
that detected the abbreviation SOB, and replaced it
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
306

31_year female one_month epigastric_pain increased meals associated_with
shortness_of_breath no_nausea no_bowel_habits_changes no_rectal_bleeding no_weight_loss
good_appetite no_family_history colorectal_cancer cancer
Enhanced Medical Term Extractor (EMTE)
year female month epigastric_pain increased meals associated_with sob_no_nausea
no_bowel_habits changes rectal_bleeding weight_loss family_history crc_cancer
Default Sci-Spacy (DSS)
year old female since one month agao complained epigastric pain increased after meals
associated with SOB nausea bowel habits changes rectal bleeding weight loss good appetite
family history cancer
Standard Cleansing (SC)
31 year old female since one month ago has complained of epigastric pain, increased after
meals, associated with SOB , no nausea , no bowel habits changes. no rectal bleeding,no weight
loss,good appetite, no family history of CRC cancer.
Chief Complaint (CC)
Figure 3: A chief complaint sample before and after using
the cleansing methods SC, DSS, and EMTE.
with its full term while replacing the spaces with un-
derscores. Also, no nausea is a result of a negation
rule that detects the left side negations.
Algorithm 1: EMTE algorithm.
Input: A, R , C
Output: C
0
(the processed version of corpus C )
1: Initialize C
0
← φ
2: for each sample S
i
∈ C do
3: Annotate S
i
to build the POS tagging: γ
i
← Γ
i
(S
i
)
4: From A, load into ρ
i
all the abbreviation rules having
an empty specialties attribute or containing σ
i
5: Apply the abbreviation and medical terms detection
rules ω
i
= R ∪ {ρ
i
} on the annotated document γ
i
6: Replace the detected abbreviations with their full-
term
7: Convert the detected phrases to words by merging
their tokens with underscores
8: C
0
← C
0
∪ S
i
9: end for
10: return C
0
- Generate the new corpus: EMTE converts every
detected phrase m
i j
that has a label ”abbreviation”
from its short-term into its full-term representation
and obtains the updated sample S
0
i
. Then, it merges
the tokens of every detected phrase using underscores
to form one word as shown in Figure 3. Finally, it
reconstructs the sample S
0
i
using the detected phrases
and adds the processed sample S
0
i
to C
0
, the new pro-
cessed corpus.
5 EXPERIMENTS AND RESULTS
To evaluate the performance of our approach, a large
clinical textual dataset (Chief Complaints and History
of Present Illness) was cleansed using four cleans-
ing methods including our approach. The resulting
datasets were fed to different machine learning mod-
els to solve the ICD multi-label classification prob-
lem.
Besides ”EMTE”, the following cleansing meth-
ods were considered:
• RAW: no cleansing techniques were applied to the
original data.
• SC: the standard cleansing steps, such as lemmati-
zation, stemming, stop-words removal and punc-
tuation removal were applied on the RAW data.
• ”DSS”: it is based on the ”SciSpacy” NER pre-
trained model that extracts medical terms from the
RAW data.
Each one of the four cleansed datasets was fed to
the following word embedding techniques: Padding
Sequence (”PS” (Abadi et al., 2015)), Sentence2Vec
(”S2V” (Pagliardini et al., 2017)), and BERT-based
word embeddings (”Clinical
BERT” (Alsentzer
et al., 2019), ”BERT base” (Devlin et al., 2019)).
5.1 Industrial Medical Data
The experiments were applied to medical data re-
trieved from the outpatient departments of a private
Saudi hospital. The data covers three years and con-
sists of anonymous records. Each record corresponds
to a patient’s visit and contains the chief complaints
CCs (textual data), the list of diagnoses (represented
by ICD-10 codes), and the physician’s specialty. The
imbalanced dataset included samples from 24 special-
ties such as Pediatrics, Gastroenterology, etc. The
data consisted of over 2.21M records with 1,050 dif-
ferent ICD-10 codes.
5.2 Tools and Technical Challenges
To implement these experiments, the ”SciSpacy”
NER tool (Neumann et al., 2019) which is an exten-
sion from ”Spacy” (Honnibal et al., 2020), was used.
It contains a NER pre-trained model ”en core sci lg”,
consisting of around 785k vocabulary and trained on
biomedical data. This model generated the ”DSS”
corpus. Moreover, the ”EntityRuler” and ”Pattern-
Matcher” components of ”Spacy” were used in the
EMTE approach to implement our PMRs and exe-
cute them. In addition, the Deep Learning training
tasks were based on the Keras (Chollet et al., 2015)
and Tensorflow (Abadi et al., 2015) libraries. Table
2 shows the hyper parameters used in these experi-
ments.
Since the ”BERT Tokenizer” splits words into
chunks and subwords if they do not belong to the size
limited BERT vocabulary, it was modified to let the
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules
307

Table 2: The hyper parameters settings used with the Keras
deep learning training tasks.
Hyper parameters
Optimizer Adam
Loss function Binary Cross Entropy
Batch Size 64
Learning Rate 3e
−4
or 5e
−5
Threshold 0.5
Monitor val micro F1
Epsilon 1e
−8
Neurons 2/3 Input + Output
Patience 10
Minimum Delta 1e − 3
Maximum Epochs 200
Dropout 0.3
BERT embeddings work with the medical phrases de-
tected by EMTE.
5.3 Results
In this section, we show how EMTE outperforms both
cleansing methods, DSS and SC, in terms of features
extraction. In addition, we compare the impact of our
cleansing approach to the other considered cleansing
methods in improving the ICD-10 multi-label classi-
fication using four machine learning models.
Table 3: A qualitative comparison between the outputs of
EMTE and DSS when applied on a small dataset of 1,000
samples. EMTE detected 99.33% of the abbreviations and
98.87% of the negations.
Method
Abbreviations Negations Medical Terms
Total Samples Total Samples Total Samples
Gold
Truth
2,540 575 1,329 359 4,891 1,000
DSS
1,493
(58.79%)
411
(71.48%)
915
(68.85%)
312
(86.91%)
3,561
(72.81%)
904
(90.40%)
EMTE
2,523
(99.33%)
566
(98.43%)
1,314
(98.87%)
359
(100%)
4,843
(99.02%)
1,000
(100%)
5.3.1 Qualitative Analysis
Figure 3 shows the outputs of EMTE, DSS and SC
when applied to a given chief complaint. SC ap-
proach removed the determiner ”no”, which is a
negation, thus, it changed the meaning of the in-
put data. Moreover, this method fails to detect the
medical terms (phrases). DSS extracted many med-
ical terms, but some were inaccurate. For example,
DSS detected ”no bowel habits” instead of ”no bowel
habits changes”. It also combined the medical abbre-
viation ”SOB” that stands for ”shortness of breath”,
with the ”no nausea” term. Moreover, it inaccurately
identified ”weight loss” instead of detecting its nega-
tion.
On the other hand, with the help of the pattern-
matching rules, our approach could identify most
medical terms, negations, and abbreviations. For ex-
ample, the medical abbreviations ”SOB” and ”CRC”
were correctly replaced with their correct full-terms.
Furthermore, to qualitatively evaluate the perfor-
mance of ”EMTE” and compare it to ”DSS”, a
dataset of 1, 000 random samples was constructed.
First, the medical team manually counted the to-
tal number of abbreviations, negations, and medi-
cal terms found in the 1, 000 samples. They found
2, 540 abbreviations in 575 samples, 1, 329 negations
in 359 samples, and 4, 891 medical terms out of the
1, 000 randomly selected samples. This ”Gold Truth”
is compared to the results of ”EMTE” and ”DSS”
in Table 3. Our approach improved the abbrevia-
tions detection by 68.99%, the negations detection by
43.61%, and medical terms detection by 36% when
compared to DSS. Moreover, our approach detected
abbreviations, negations, and medical terms in more
samples than DSS by 37.71%, 15.06%, and 10.62%,
respectively. The major limitations in DSS is that it
does not detect abbreviations with punctuations such
as ”u/a”,”-ve”, ”+ve”. Moreover, DSS failed to de-
tect negations with ”nil” value and the negations that
were located on the right side like ”pmh: no”. In ad-
dition, DSS failed to detect medical terms like ”vagi-
nal discharge” and ”right sided breast pain” as one
phrase.
It is worth mentioning that even our approach
failed to detect some negations that contained typos.
For example, it did not detect the term ”noone” as
negation since it contained typos. In addition, our
approach did not catch ambiguous abbreviations like
”CLD” (Chronic Lung Disease or Chronic Liver Dis-
ease) since these abbreviations are ambiguous and
used in same specialty (Category iii 2.2).
5.3.2 ICD-10 Classification Results
In this section, the impact of EMTE on the ICD-
10 multi-label classification results are presented and
compared to the use of other cleansing techniques
(SC and DSS) and the RAW dataset. Table 4 presents
the results of the experiments with four different
DNN techniques. The columns present the results of
the evaluation metrics (Accuracy, Recall, Macro-F1,
Micro-F1, and Weighted-F1.
The experiments that applied EMTE outperformed
all the others for all the evaluated word embeddings
and for all the considered metrics. The percentage
of gain from applying EMTE instead of any other
method and according to any metric, was computed
as follows:
[%gain] = 100 ×
[EMT E%] − [otherMethod%]
[otherMethod%]
(1)
For instance, the Micro-F1, obtained with the test-
ing data and the BERT base model, was improved by
5.44% when using EMTE instead of DSS, 5.23% in-
stead of SC, and 4.46% instead of RAW. Moreover,
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
308

Table 4: The results of the classification experiments using different cleansing techniques and training models.
DNN
Data
Set
Training (%) Evaluation (%)
Accuracy Recall
F1-Score
Accuracy Recall
F1-Score
Macro Micro Weighted Macro Micro Weighted
PS
SC 70.07 54.93 67.03 68.31 67.20 59.55 46.85 57.11 58.58 57.27
DSS 70.35 55.00 67.06 68.38 67.27 61.41 48.38 58.41 60.17 58.83
RAW 71.89 55.13 67.88 69.23 68.51 62.37 48.98 59.42 60.78 59.07
EMTE 74.14 57.93 69.86 71.25 70.15 66.19 51.03 61.31 62.90 61.64
S2V
SC 71.13 54.81 70.76 70.66 69.99 59.78 49.98 60.13 61.20 59.71
DSS 72.12 55.47 70.29 70.77 69.93 61.35 50.19 62.10 62.85 61.75
RAW 72.51 55.84 70.76 71.05 70.20 61.48 50.80 62.81 63.27 62.13
EMTE 75.60 59.08 72.90 73.97 72.03 65.35 52.36 64.93 65.54 64.31
BERT
Base
SC 78.10 64.42 76.70 77.17 76.49 64.63 53.60 63.83 64.60 63.77
DSS 78.27 64.63 76.94 77.36 76.65 64.88 53.55 63.37 64.47 63.49
RAW 78.64 65.02 77.28 78.01 76.99 65.40 54.09 64.13 65.08 64.04
EMTE 79.76 65.72 77.93 78.46 77.74 67.46 55.93 66.05 67.98 66.19
Clinical
BERT
SC 78.18 60.19 73.99 76.29 74.17 66.83 55.63 63.20 64.69 64.79
DSS 78.42 60.57 74.51 77.56 75.51 66.69 56.12 64.62 64.76 65.84
RAW 79.83 61.89 74.10 78.00 75.68 66.99 56.59 65.18 65.36 66.18
EMTE 81.52 63.47 77.49 79.51 78.44 69.33 58.08 67.62 69.68 68.59
the same metric Micro-F1, obtained with the testing
data and the Clinical BERT model, showed a 7.61%
gain over DSS, 7.71% over SC, and 6.61% over RAW.
The gain in performance, when cleansing the
dataset with EMTE, was reflected on all the consid-
ered metrics in both training and evaluation datasets
and with the four considered word embeddings.
Moreover, the gain in performance was not lim-
ited to the evaluation metrics, the use of EMTE re-
duced the training time of the four models and the re-
quired number of epochs to converge. Table 5 shows
for each cleansed dataset, the average execution time
for an epoch and the number of epochs required for
each model to coverage.
The experiments that used EMTE, to cleanse the
dataset, converged faster than the others. For instance,
the BERT Base model required 19, 32, 24, and 26
epochs with the datasets cleansed by EMTE, RAW,
DSS, and SC respectively. Furthermore, the experi-
ments that used EMTE required 43.69% to 53.39%
less execution time per epoch than RAW, 8.63% to
12.09% less than DSS and 16.48% to 26.48% less
than SC for the different considered models.
Finally, the vocabulary size generated by EMTE
is smaller than the ones generated by the other
methods. For example, RAW vocabulary size
was 286,891 words while SC contained 229,102
words (−20.14%), DSS contained 183,403 words
(−36.07%), and EMTE contained 178,917 words
(−37.64%).
6 DISCUSSION
EMTE outperformed the other approaches for the fol-
lowing reasons: First, the experiments on RAW data
had the biggest vocabulary since the same medical
term might be represented by many data points. For
example, ”Blood Pressure” had two different data
points, ”BP” and ”Blood Pressure”. This large vo-
Table 5: The execution time per epoch and the number of
epochs per cleansing method and model.
DNN
Dataset
Time
(sec)
# Epochs
PS
SC 1167 23
DSS 976 22
RAW 1841 28
EMTE 858 20
Clinical
BERT
SC 1369 31
DSS 1164 26
RAW 2005 32
EMTE 1034 26
BERT
Base
SC 1347 26
DSS 1256 24
RAW 1998 32
EMTE 1125 19
S2V
SC 1297 51
DSS 1112 44
RAW 1912 58
EMTE 1016 33
cabulary required additional resources for training
and a longer execution time. Second, SC is also
inefficient since it leads to data loss and degrades
the data quality. For example, using SC, impor-
tant information such as the negations and impor-
tant punctuations like ”-” in the abbreviation ”-ve”
were lost. Third, DSS’s performance strictly de-
pends on the used feature extraction tool’s perfor-
mance. The ”SciSpacy” NER pre-trained model has
some limitations. For example, abbreviations could
be wrongly identified by DSS when studying datasets
including different specialties. Moreover, as shown
in Figure 3, DSS wrongly combined the abbreviations
”SOB” and ”CRC” with the medical terms ”no nau-
sea” and ”cancer” respectively. Furthermore, DSS
failed to detect ”no rectal bleeding” as a negation and
”good appetite” as a medical term.
In this paper, we studied the medical service spe-
cialty feature along with the textual data of a chief
complaint. More information can be added in the fu-
ture, like the body site which refers to the location of
the disease in the body like upper abdomen and lower
abdomen, gender and age, to reduce the abbreviation
ambiguity especially for those used in the same spe-
cialty.
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules
309

One of the main advantages of the EMTE ap-
proach is its flexibility and maintainability. The dic-
tionaries can be updated at any time without any need
to retrain the models on new medical terms. In ad-
dition, EMTE can be used as a document quality en-
hancer as it can unify the negations writing styles and
replace the abbreviations with their full-terms.
7 CONCLUSION AND FUTURE
WORK
This paper presented a cleansing approach that im-
proves the quality of medical terms extraction from
unstructured clinical data using pattern matching
rules based on dictionaries. The solution was con-
ceived with flexibility and maintainability in mind for
industrial use. The experiments showed that our ap-
proach helps solving the the ICD-10 prediction prob-
lem by improving the quality of the data fed to the
DNNs. As a result, the performance of the trained
models was improved according to various metrics.
The proposed approach also reduced the required re-
sources to train the models and decreased the training
time by accelerating the convergence of the models.
In future works and in order to improve further-
more the quality of the medical data, we aim to ex-
tend this work to improve data quality by tackling
several challenges like: medical term synonyms, im-
prove abbreviation detection by adding more features
(e.g. body site, gender, and age), and medical investi-
gation results (laboratory and radiology) in CCs.
ACKNOWLEDGEMENTS
All computations have been performed on the
M
´
esocentre of Franche-Comt
´
e, France and the med-
ical data was aquired from the Specialized Medical
Center Hospital in Riyadh, KSA.
REFERENCES
Abadi, M., Agarwal, A., et al. (2015). Tensorflow: Large-
scale machine learning on heterogeneous systems.
Adnan, K. and Akbar, R. (2019). Limitations of information
extraction methods and techniques for heterogeneous
unstructured big data. International Journal of Engi-
neering Business Management, 11.
Alsentzer, E., Murphy, J., et al. (2019). Publicly avail-
able clinical bert embeddings. In Proceedings of the
2nd Clinical Natural Language Processing Workshop,
pages 72–78, Minneapolis, Minnesota, USA.
Atutxa, A., de Ilarraza, A. D., et al. (2019). Inter-
pretable deep learning to map diagnostic texts to icd-
10 codes. International Journal of Medical Informat-
ics, 129:49–59.
Azam, S. S., Raju, M., et al. (2020). Cascadenet: An lstm
based deep learning model for automated icd-10 cod-
ing. In Advances in Information and Communication,
pages 55–74. Springer International Publishing.
Bai, T. and Vucetic, S. (2019). Improving medical code
prediction from clinical text via incorporating online
knowledge sources. In The World Wide Web Confer-
ence, pages 72–82, NY, USA.
Bose, P., Srinivasan, S., et al. (2021). A survey on re-
cent named entity recognition and relationship extrac-
tion techniques on clinical texts. Applied Sciences,
11(18):8319.
Chen, Q., Du, J., et al. (2020). Deep learning with sen-
tence embeddings pre-trained on biomedical corpora
improves the performance of finding similar sentences
in electronic medical records. BMC Medical Informat-
ics and Decision Making, 20.
Chollet, F. et al. (2015). Keras.
Chraibi, A., Delerue, D., et al. (2021). A deep learning
framework for automated icd-10 coding. Studies in
Health Technology and Informatics, 281.
de Marneffe, M.-C., Manning, C. D., et al. (2021). Uni-
versal dependencies. Computational Linguistics,
47(2):255–308.
Devlin, J., Chang, M.-W., et al. (2019). Bert: Pre-training
of deep bidirectional transformers for language under-
standing. In Proceedings of the 2019 Conference of
the North American Chapter of the Association for
Computational Linguistics, pages 4171–4186, Min-
neapolis, Minnesota.
Du, J., Chen, Q., et al. (2019). Ml-net: multi-label classifi-
cation of biomedical texts with deep neural networks.
Journal of the American Medical Informatics Associ-
ation, 26(11):1279–1285.
Dugas, M., Neuhaus, P., et al. (2016). Portal of medical
data models: information infrastructure for medical
research and healthcare. Database, 2016.
Grossman Liu, L., Grossman, R. H., et al. (2021). A deep
database of medical abbreviations and acronyms for
natural language processing. Scientific Data, 8(1).
Honnibal, M., Montani, I., et al. (2020). spacy: Industrial-
strength natural language processing in python.
Joachims, T. (1998). Text categorization with support vec-
tor machines: Learning with many relevant features.
Koptient, A. and Grabar, N. (2021). Disambiguation of
medical abbreviations in french with supervised meth-
ods.
Li, P., Wang, H., et al. (2018). Employing semantic context
for sparse information extraction assessment. ACM
Transactions on Knowledge Discovery from Data,
12(5).
Lucini, F. R., Fogliatto, F. S., et al. (2017). Text mining ap-
proach to predict hospital admissions using early med-
ical records from the emergency department. Interna-
tional Journal of Medical Informatics, 100:1–8.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
310

Makohon, I. and Li, Y. (2021). Multi-label classification of
icd-10 coding & clinical notes using mimic & codiesp.
Manning, C. D., Surdeanu, M., et al. (2014). The stanford
corenlp natural language processing toolkit.
Marta Skreta (2019). Training without training data: Im-
proving the generalizability of automated medical ab-
breviation disambiguation.
Mehrabi, S., Krishnan, A., et al. (2015). Deepen: A nega-
tion detection system for clinical text incorporating
dependency relation into negex. Journal of Biomed-
ical Informatics, 54:213–219.
Neumann, M., King, D., et al. (2019). Scispacy: Fast and
robust models for biomedical natural language pro-
cessing.
Nguyen, D. Q. and Verspoor, K. (2019). From pos tag-
ging to dependency parsing for biomedical event ex-
traction. BMC Bioinformatics, 20(1).
Pagliardini, M., Gupta, P., et al. (2017). Unsupervised learn-
ing of sentence embeddings using compositional n-
gram features.
Shaalan, Y., Dokumentov, A., et al. (2020). Ensemble
model for pre-discharge icd10 coding prediction.
Wen, Z., Lu, X. H., et al. (2020). Medal: Medical abbrevi-
ation disambiguation dataset for natural language un-
derstanding pretraining.
Wu, S., Miller, T., et al. (2014). Negation’s not solved:
Generalizability versus optimizability in clinical natu-
ral language processing. PLoS ONE, 9(11):e112774.
Zeman, D. (2022). Universal pos tags. Accessed: 2022-01-
20.
Zheng, Y., Si, Y.-W., et al. (2021). Feature extraction
for chart pattern classification in financial time series.
Knowledge and Information Systems, 63(7):1807–
1848.
EMTE: An Enhanced Medical Terms Extractor Using Pattern Matching Rules
311
