
Explainability of MLP Based Species Distribution Models: A Case
Study
Imane El Assari
1a
, Hajar Hakkoum
1
and Ali Idri
1,2 b
1
ENSIAS, Mohammed V University in Rabat, Morocco
2
AlKhwarizmi, Mohammed VI Polytechnic University, Benguerir, Morocco
Keywords: Interpretability, xAI, Biodiversity, Species Distribution Modelling, PDP, SHAP, Machine Learning.
Abstract: Species Distribution models (SDMs) are widely used to study species occurrence in conservation science and
ecology evolution. However the huge amount of data and its complexity makes it difficult for professionals
to forecast the evolutionary trends of distributions across the concerned landscapes. As a solution, machine
learning (ML) algorithms were used to construct and evaluate SDMs in order to predict the studied species
occurrences and their habitat suitability. Nevertheless, it is critical to ensure that ML based SDMs reflect
reality by studying their trustworthiness. This paper aims to investigate two techniques: SHapley Additive
exPlanations (SHAP) and the Partial Dependence Plot (PDP) techniques to interpret a Multilayer perceptron
(MLP) trained on the Loxodonta Africana dataset. Results demonstrate the prediction process and how in-
terpretability techniques could be used to explain misclassified instances and thus increase trust between ML
results and domain experts.
1 INTRODUCTION
Environment scientists assert that the magnitude, pace
and severity of the current environmental crisis are
unprecedented (B.Daley and R.Kent, 2005). Several
regions are losing their biodiversity to make way for
human dwelling and industry. Therefore, in order to
safeguard the environment, it’s crucial to implement
well planned policies that take into consideration the
environmental characteristics and biological
outcomes of each region.
As part of the conservation science, having a well-
thought-out strategy for managing the environment
requires a thorough understanding of its components,
namely its climatic conditions and species distribu-
tion. However, it’s hard to pinpoint the exact loca-
tion of each individual of each species at any moment.
Therefore, species distribution models (SDMs) are
used to find whether a species is likely to be present or
absent in a geographic location based on its environ-
mental conditions. Their objective is to understand a
particular ecosystem, its number of species, the com-
position of its population, and to predict the spatial
and temporal pattern of species occurrence.
a
http://ensias.um5.ac.ma
b
https://msda.um6p.ma/home
The new technologies and the data they gener- ate
hold great potential for large-scale environmental
monitoring, however traditional statistical approaches
limits its usage which inefficiently distill data into rel-
evant information (D.Tuia et al., 2022). Conversely,
data science community works to apply information
technologies to gather, organize, and analyze biolog-
ical data (American Museum of Natural History, ).
Basically, they try to use machine learning (ML) to
discover new insights and patterns from all the avail-
able expeditions and remote sensing data. ML tech-
niques are useful to perform predictive analytics since
it gives a variety of tools to support complex data
structures, and thus provides a powerful approach for
assessing SDMs challenges. However, according to a
review made by Beery et al. despite the considerable
use of ML techniques in ecology, SDMs has received
relatively little attention from the computer science
community (S.Beery et al., 2021).
In fact, ML contributed to this field in a cou-ple
of areas, namely climate models that repre- sent our
understanding of Earth and climate physics (Rolnick
et al., 2019), forest management based on satellite
imagery and 3D Deep Learning tech- niques(Liu et
690
El Assari, I., Hakkoum, H. and Idri, A.
Explainability of MLP Based Species Distribution Models: A Case Study.
DOI: 10.5220/0011745300003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 3, pages 690-697
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

al., 2021), mapping wetlands distribu- tion in highly
modified coastal catchments (Wen and Hughes,
2020),urban vegetation(Abdollahi and Prad- han,
2021)and many more.
There are two categories of ML algorithms: legi-
ble white-box algorithms that provide readable rules
(SEON, 2022), , and black-boxes which are opaque
yet more powerful since they can identify nonlinear
relationships in data. To remedy this limitation, in-
terpretability attempts to explain model predictions to
understand the reasoning behind the prediction pro-
cess (AI, 2020), avoid bias and gain trust whether
globally or locally. Global interpretability, exam- ines
a model’s general behavior, whereas local inter-
pretability concentrates on a particular scenario that
was supplied as input to the model.
Interpretability techniques have been used to im-
prove the explainability of ML based species distri-
bution models. For instance, Ryo et al. investi- gated
explainable AI (xAI) techniques in the context of
SDMs based on the African elephant dataset (Ryo et
al., 2021). They used random forest (RF) as a black
box algorithm to predict the presence/absence val-
ues of the studied species. In terms of interpretabil-
ity, they used: (1) feature permutation importance
(FI) and the (2) Partial Dependence Plot (PDP) as
global interpretability techniques, FI classifies the
predictor variables according to their importance,
while PDP demonstrate the marginal effect a feature
may have on the ML model predicted outcome
(Molnar, 2019), along with (3) Local Interpretable
Model-Agnostic Explanation (LIME) (Ribeiro et al.,
2016) as a local interpretability technique. According
to this study, the most relevant feature based on FI
was the precipi- tation of the wettest quarter.
This article aims to evaluate and interpret a basic
Multilayer Perceptron (MLP) model trained to fore-
cast the African elephant distribution (GBIF, 2021)
using: (1) the SHapley Additive exPlanations (SHAP)
(Lundberg and Lee, 2017), a local interpretability
method that explains individual predictions based on
the game theoretically optimal Shapley values; and
(2) the Partial Dependence Plot (PDP): a global inter-
pretability technique that visualizes the marginal ef-
fect of an individual feature to the predictive value of
the studied model (Molnar, 2019). This paper an-
swers and discusses the following research questions:
• RQ1: What is the overall performance of the
con- structed MLP models?
• RQ2: What is the local interpretability of the
best performing model?
• RQ3: How global interpretability enhances
local explanations?
The main contributions of this study are the fol-
lowing:
• Assessing and comparing the performance of
12 MLP based classifiers that were
generated by combining 3 different
Hyperparameters using GridSearch.
• Interpreting 3 randomly chosen instances
locally using SHAP and comparing them with
their true labels.
• Explanation of misclassified instances using
PDP.
The remainder of this paper is organized as fol-
lows: Section2 introduces explainable AI and the
difference between Global and Local Interpretability.
Section 3 describes the used dataset and performance
metrics to select the best performing model. Section4
presents the experimental design followed during this
study. A discussion about the obtained results and
findings is presented in Section 5. Section 6 covers
the threats to validity and the conclusion.
2
BACKGROUND
This section presents the feed-forward neural net-
work used in this research, along with the used in-
terpretability techniques namely SHAP and PDP.
2.1
Artificial Neural Networks: MLP
ANNs are a collection of simple computational units
interlinked by a system of connections (Cheng and
Titterington, 1994) that were inspired from the
brain’s neuron architecture. They are frequently used
in data modelling, as they are perceived as better
substitute to standard nonlinear regression or cluster
analysis prob- lems (Gurney, 1997).
In general, ANNs are organized in layers and this
is where the MLP comes in, it is a typical example of
the feed-forward ANN where information travels in
one direction from input to output (Hakkoum et al.,
2021). A MLP is constituted of 3 types of layers: the
input layer, which receives the data to be processed,
one or more hidden layers, which together constitute
the network’s true engine, and the output layer.
As to optimize the MLP Hyperparameters tuning
phase, GridSearch a technique that specifies a search
space as a grid of Hyperparameters and evaluates ev-
ery position in the grid (Brownlee, 2020)was used.
Explainability of MLP Based Species Distribution Models: A Case Study
691

Table 1: Bioclimatic variables.
Variable Description Abbreviations Range
Elev Elevation Elev [-61 ; 3508]
Bio1 Annual Mean Temperature AMT [7,810688 ; 29,427000]
Bio2 Mean Diurnal Range MDR [6,640182 ; 18,048876]
Bio3 Isothermality (BIO2/BIO7) (×100) Isothermality [26,918064 ; 92,384308]
Bio4 Temperature Seasonality TempSeasonality [15,199914;1039,296265]
Bio5 Max Temperature of Warmes
t
Month MaxTempWM [17,118000;41,708752]
Bio6 Min Temperature of Coldes
t
Month MinTempCM [-9,583250 ; 22,415842]
Bio8 Mean Temperature of Wettes
t
Quarte
r
MeanTempWQ [6,682958 ; 31,448957]
Bio12 Annual Precipitation AnnualPrecip [3,000000 ; 3369,000000]
Bio13 Precipitation of Wettes
t
Month PrecipwettestM [1,000000 ; 535,000000]
Bio14 Precipitation of Dries
t
Month PrecipDriestM [0,000000 ; 105,000000]
Bio15 Precipitation Seasonality PrecipSeasonality [12,457452 ; 153,643448]
Bio18 Precipitation of Warmes
t
Quarte
r
PrecipWQ [0,000000 ; 728,000000]
Bio19 Precipitation of Coldes
t
Quarte
r
PrecipCQ [0,000000 ; 948,000000]
2.2
Interpretability
Interpretability is determined by whether the model
has a transparent process that allows the users to un-
derstand how inputs are mathematically mapped to
outputs (Doran et al., 2017). It represents the degree
to which a human can consistently predict the model’s
result, and evaluate the forecasting process by giving
the relative importance of each variable.
Interpretability methods can be categorized ac-
cording to various criteria, depending on how they are
used: Intrinsic/ post-hoc; model-specific/ model-
agnostic; global/ local (Molnar, 2019). Global inter-
pretability describes how the entire model behaves,
meanwhile local interpretability focuses on the pre-
diction of a particular instance; it is similar to a zoom
in on a single instance and then examining the reasons
behind the model’s prediction for this input.
This paper uses SHAP to examine the local inter-
pretability of the best performing MLP model. It ex-
plains the model’s individual predictions using Shap-
ley values, a cooperative game theory concept that
calculates the contribution of each feature to the dif-
ference between the predicted value and the aver- age
of all predictions. Shapley values compute the
marginal contribution of each feature to the end out-
come by perturbing the input features and observ- ing
how these changes correspond to the final model
prediction. The Shapley value is then calculated by
taking the average of all marginal contributions
(Gopinath and Kurokawa, 2021). SHAP is a model-
agnostic method, meaning that SHAP’s process re-
main the same regardless of the used ML algorithm.
In addition to SHAP, PDP was used to study the
marginal effect a feature may have on the target vari-
able. It shows whether the relationship between the
target and a feature is linear, monotonic or more com-
plex(Molnar, 2019).
3
DATA DESCRIPTION AND
PERFORMANCE CRITERIA
The following section describes the used datasets, and
introduces the performance measures.
3.1
Data Description
To run a SDM, two types of data are needed:
occurrence data, which presents the coordinates of the
locations where the studied species occurs, and
environ- mental data, that describes the bioclimatic
conditions of those locations (EcoCommons, 2022).
For occurrence data GBIF LoxodontaAfricana
tabular data were used. It contains 10494 rows and
257 columns, its relevant features are decimal Lon-
gitude, decimal Latitude and the occurrenceStatus.
The decimal longitude and latitude define the species
geographical locations, while the occurrenceStatus
presents the occupancy / absence values at those lo-
cations.
The original record contains 10466 row of pres-
ence data and 28 row of absence data. To resolve this
imbalanced data problem a sample of 8019 back-
ground points were randomly generated using dismo
package in R to sum up with 16511 occurrences of
which 8979 are presence data and 8019 are pseudo-
absences.
Despite the existence of different methods
to generate background points, the randomly
selected pseudo-absences yielded the most reliable.
distribution models (Barbet-Massin et al., 2012).
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
692

Figure 1: Experimental Design.
It is critical to consider the credibility of the generate
background points, a location with no occurrences of
the species does not imply the absence of the species
in this loca- tion, it could be an area with no data or a
location out of land. To avoid such problems the
dismo package provides different methods dedicated
to SDMs challenges, such as sampling the random
points from a study area to form the pseudo-absences
afterwards or using a mask to exclude the areas not on
land (Spatial Data Science, 2021).
For the environmental data, the WorldClim stan-
dard bioclimatic variables were extracted from 19
GeoTiff (.tif) file using ‘biovars’ function in the dismo
R package. Table1 presents the used bioclimatic vari-
ables and how they were encoded in this work.
3.2
Performance Criteria
The MLP based models performance is reported and
compared in terms of 4 different metrics: Accuracy,
Precision, Recall, and AUC. After calculating these
metrics, the Borda Count, a ranked voting technique
that ranks the models in order of preference, was used
to select the best MLP classifier using the accuracy,
precision, recall, and AUC metrics as voters (Lipp-
man, ).
4
EXPERIMENTAL DESIGN
This section describes in details the steps followed in
this case study. It starts with model configuration and
selection, then the local interpretation using SHAP’s
Waterfall plots, and finally the PDP plots generation.
The steps are showed in detail in Figure1.
4.1
Data Retrieval and Cleaning
As mentioned in section 3, this study used two types
of data: Loxodonta Africana occurrence data from
GBIF and environmental data from WorldClim, which
were concatenated using R based on their com- mon
geographic points. Missing values, outliers, data
balancing, and normalization were all resolved during
the Data Cleaning phase.
4.2
Models Construction
The MLP classifiers were built using one hidden
layer. Three different hyper-parameters were used to
control the model’s learning process: the batch size,
which represents the number of samples processed
before the model is updated, the solver for weight op-
timization where SGD refers to Stochastic Gradient
Descent, known as the most basic form of gradient
descent while ‘Adam’ is an extension to SGD that
provides faster results (Brownlee, 2017) , however
different studies argue that although Adam converges
faster, SGD generalizes better than Adam and thus
results in improved final performance (Park, 2021).
Finally, we have the hidden layer size that represents
the number of hidden nodes on the first hidden layer,
its selected range was chosen as to provide good
performance but without requiring a huge amount of
time in the training phase since the aim of this
empirical evaluation was interpretability and not
performance.
Explainability of MLP Based Species Distribution Models: A Case Study
693

GridSearch was employed to optimize the Hyper-
parameter tuning phase with 10 cross validation
train
ing. Table 2 presents the used configuration. Note
that
the hidden layer sizes were selected based on
previous experimentations where the highest
performance scores were found close to the mentioned
range in table 2. The batch size and the solver values
were chosen as a standard configuration commonly
found in literature.
Table 2: Hyperparameters configuration.
H
y
perparameters Selected Ran
g
e
Hidden La
y
e
r
Size [(20,),(25,),(30,)]
Batch size (32,64)
Solve
r
[’s
g
d’, ’adam’]
4.3
Interpretability
This step tries to interpret the MLP classifier results
locally using SHAP’s waterfall plot. It explains a set
of 3 different instances that were randomly selected
in a way to have one true positive where the model
agrees with reality and predicts a high habitat suit-
ability, one true negative where the model accurately
predicts a low habitat suitability and one false nega-
tive, where the classifier predicts low habitat suitabil-
ity, while it’s not the case in reality.
After the interpretability of the MLP classifier
PDP was used to determine if there is an agreement
between the classifier predictions, the instances true
label and local interpretability results, PDP plots were
generated for the 2 first ranked features.
5
RESULTS AND DISCUSSION
This section presents and discusses the results of this
empirical study, namely the models performance, and
interpretability results.
5.1
Models Evaluation
The Hyperparameters combination gave 12 MLP
classifiers. Table 3 describes the overall performance
of the 3 first and the last ranked MLP models accord-
ing to the Borda Count method using accuracy, pre-
cision, recall, and AUC as voters. The classifiers are
presented according to their assigned ranks. Accord-
ing to Table 3, the top-ranked model has 25 neurons in
its hidden layer, 64 as a batch size, which determines
the number of training examples utilized in one iter-
ation (Murphy, 2019), and ’sgd’ as a solver, which
specifies the algorithm weight optimization over the
nodes (Fuchs, 2021). To note that this is the classifier
used in the interpretability phase.
5.2
Local Interpretability
To test the trustworthiness of individual predictions,
a group of 3 instances was randomly chosen, Fig-
ure2 shows their relative SHAP’s waterfall plot ex-
planations. The waterfall plot’s purpose is to show the
SHAP values of each feature as well as its im- pact on
the final prediction. The model’s prediction is
represented in the y-axis by f(x), each bar illustrates
how the feature helps to push the model’s output away
from the base value E(x) that indicates the average of
the model output over training data.
The features with a right arrow influence the pre-
diction more in favor of an appropriate habitat for
African elephants, whilst the features with a left arrow
influence the prediction more in favor of the species’
inadequacy for such environments.
Equation (4) demonstrates how f(x) is calculated.
F(x) = E(x) +
∑
SHAPvalues
(1)
The base value in this study is 0.537, it represents
the average of all observations. The model prediction
for instance 2 is 1 meaning that the studied species can
survive in this location, in this case f(x) is obtained
using Equation (4): 0.536 + 0.01 - 0.02 + 0.02 - 0.03
+ 0.03 + 0.03 + 0.03 + 0.04 - 0.05 + 0.05 + 0.06 +
0.06 + 0.11 + 0.12 = 0.996 1, it sums the base value
E(x) with all SHAP values, the same process is true
for the other instances.
Figure 2: Instance 2 SHAP’s waterfall plot.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
694
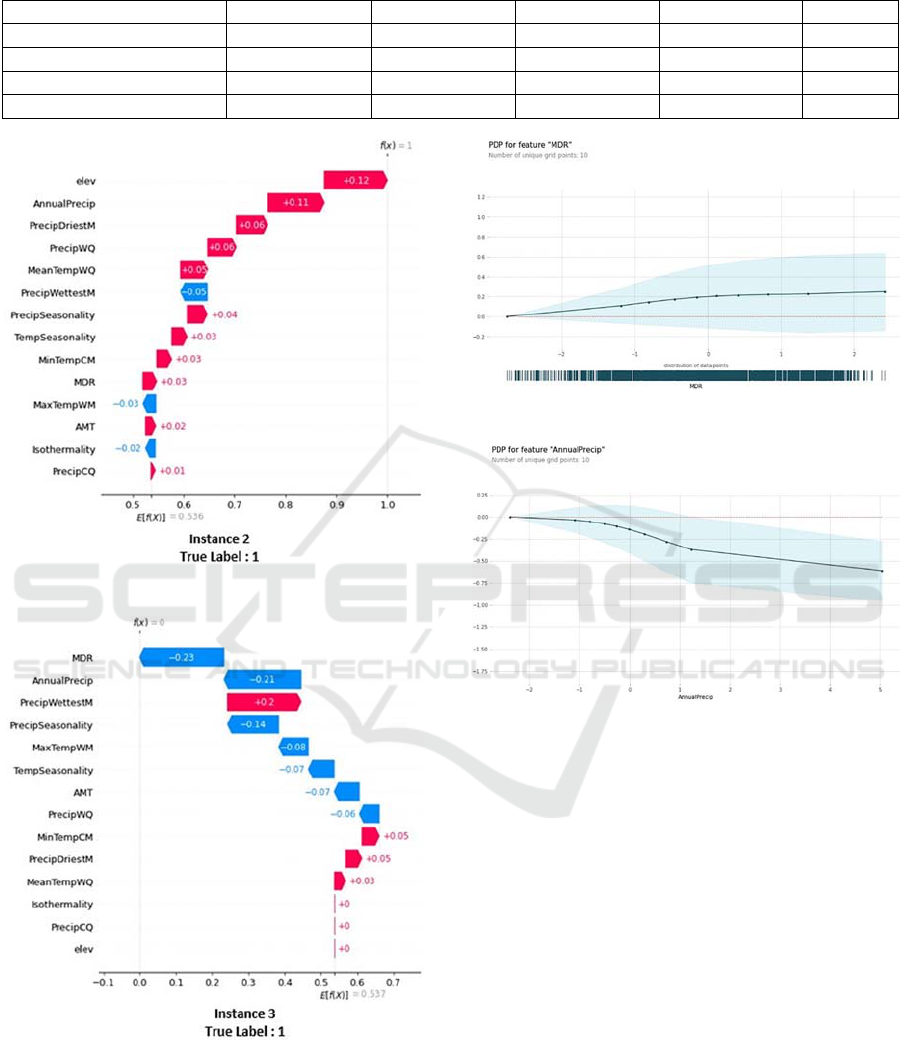
Table 3: Borda Count Results.
MLP Accuracy Precision Recall AUC Ran
k
[(25,),sgd,64] 0.857059 0.8576 0.856452 0.924786
1
[(30,),adam ,64] 0.854046 0.854450 0.853481 0.923254
2
[(30,),sgd,64] 0.851284 0.851420 0.850775 0.920037
3
[(25,),adam,32] 0.815967 0.816859 0.814878 0.889776 12
Figure 3: Instance2 SHAP’s waterfall plot.
Figure 4: Instance 3 SHAP’s waterfall plot.
In the first instance (Figure 2), the classifier pre-
dicts a low habitat suitability for the location in
question which was mainly proven by SHAP since
f(x) =0 and the ’AnnualPrecip’ variable was ranked
the first among all other variables with a SHAP value
Figure 5: ‘MDR’ Partial Dependence Plot.
Figure 6: ‘Annual Precip’ Partial Dependence Plot.
of -0.25. For instance 2 (Figure 3), the MLP classifier
pre-dicts a high habitat suitability f(x)=1.It was
noticed that according to SHAP the 1st ranked
feature was ‘elev’ which increased the base value
with 12% in comparison with its initial value.
Instance 3 (Figure
4) presents a false negative since the
classifier predicts
a low habitat suitability f(x)=0 while
the true label of the instance is 1 which means that the
models’ prediction affirms the absence of the species
in this location while it is not the case in reality.
In order to gain a deeper understanding of this in-
stance and discover the reasons behind this inaccurate
prediction with the point’s true label, PDP plots were
generated for the first and second ranked features.
Normally ’MDR’ ranges from -3.017 to 2.439, in
this case it is equal to -0.95. According to the PDP
plot in Figure 5, the max reached value at this posi-
tion is less than 0.4, which means that the classifier
predicts a low habitat suitability and that agrees well
with SHAP explanations since f(x) was equal to 0.
Explainability of MLP Based Species Distribution Models: A Case Study
695

The ‘AnnualPrecip’ range is between -2.388 and
5.115, its value in the studied position is approxi-
mately 0.82. According to the PDP plot in Figure 6,
the mean reached value in this point is less than -0.25,
which agrees with SHAP results where f(x) is equal to
0, and explains more why the model predicted a false
0 since both values 0.4 and -0.25 in the PDP plots
(Figure 5 and Figure 6) are far from being around 1.
6 THREATS TO VALIDITY AND
CONCLUSION
This work includes limitations that should be taken
into account when evaluating its findings. During the
data retrieval phase, only one occurrence dataset was
used, incorporating additional data types to the used
tabular dataset may generate better results.
Optimizing the built MLP classifier and creating
more black box models, as well as comparing SHAP
with other global and local interpretability techniques
would undoubtedly provide better explanations to the
misclassified instances.
To conclude, several MLP models were used to
study the distribution of the Loxodonta Africana, the
top performing model was used to predict the species’
occurrence and absence values. Based on SHAP’s re-
sults, the ‘AnnualPrecip’ contributed significantly to
the proposed model’s output since the studied species
lives in the African Savanna known with its tropical
wet and dry climate where rain falls in a single sea-
son and the rest of the year is dry.
SHAP allowed the conduction of models analysis
in depth and leads the selection of appropriate fea-
tures making it a suitable explanation technique for
biodiversity experts to consider when drawing critical
decisions.
Future work would attempt to include more black
box models, and compare their performance as well
as their interpretability with the obtained results using
different techniques such as SHAP’s summary plot,
FI, and LIME.
REFERENCES
Abdollahi, A. and Pradhan, B. (2021). Urban vegeta- tion
mapping from aerial imagery using explainable ai (xai).
Sensors.
AI, I. (2020). What is interpretability? https://www.
interpretable.ai/interpretability/what/.
American Museum of Natural History. Biodiversity
informatics. https://www.amnh.org/research/center- for-
biodiversity-conservation/capacity- development/biodi
versity-informatics.
Barbet-Massin, M., Jiguet, F., Albert, C., and Thuiller, W.
(2012). Selecting pseudo-absences for species distri-
bution models: How, where and how many? Methods in
Ecology and Evolution.
B.Daley and R.Kent (2005). P120 Environmental Science
and Management. London.
Brownlee, J. (2017). Gentle introduction to the adam
optimization algorithm for deep learn- ing. https://ma
chinelearningmastery.com/adam- optimization-algorith
m-for-deep-learning/.
Brownlee, J. (2020). Hyperparameter optimiza- tion with
random search and grid search. https://machinelearn
ingmastery.com/hyperparameter- optimization-with-
random-search-and-grid-search/.
Cheng, B. and Titterington, D. M. (1994). Neural Networks:
A
Review from a Statistical Perspective. Statistical
Science, 9(1).
Doran, D., Schulz, S., and Besold, T. (2017). What does
explainable ai really mean? a new conceptualization of
perspectives.
D.Tuia, B.Kellenberger, s.Beery, Blair.R.Costelloe, S.Zuffi,
B.Risse, A.Mathis, W.Mathis, M., Langevelde, F.,
T.Burghardt, H.Klinck, R., M.Wikelski, Horn, G.,
D.Couzin, I., M.Crofoot, V.Stewart, C., and T.Berger-
Wolf (2022). Perspectives in machine learning for
widlife conservation. Nature Communications.
EcoCommons (2022). Data for species distribution models.
https://support.ecocommons.org.au/support/solutions/ ar
ticles/6000255996-data-for-species-distribution- models.
Fuchs, M. (2021). Nn - multi-layer perceptron clas- sifier
(mlpclassifier). https://michael-fuchs-python.netlify.ap
p/2021/02/03/nn-multi-layer- perceptron-classifier-mlp
classifier/.
GBIF (2021). Loxodonta africana (blumenbach, 1797).
https://www.gbif.org/fr/species/2435350.
Gopinath, D. and Kurokawa, D. (2021). The shapley value
for ml models. https://towardsdatascience.com/the-
shapley-value-for-ml-models-f1100bff78d1.
Gurney, K. (1997). An Introduction to Neural Networks.
Hakkoum, H., Idri, A., and Abnane, I. (2021). Assessing
and comparing interpretability techniques for artificial
neural networks breast cancer classification. Com-
puter Methods in Biomechanics and Biomedical En-
gineering: Imaging & Visualization.
Lippman, D.Bordacount. https://courses.lumenlearning.
com/ mathforliberalartscorequisite/chapter/borda-count/.
Liu, M., Han, Z., Chen, Y., Liu, Z., and Han, Y. (2021).
Tree species classification of lidar data based on 3d
deep learning. Measurement, 177:109301.
Lundberg, S. M. and Lee, S.-I. (2017). A unified approach
to interpreting model predictions. In Advances in Neu-
ral Information Processing Systems.
Molnar, C. (2019). Interpretable Machine Learn- ing.
https://christophm.github.io/interpretable-ml- book/.
https://christophm.github.io/interpretable-ml- book/.
Murphy, A. (2019). Batch size (machine learning).
https://radiopaedia.org/articles/batch-size-machine-lea
rning.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
696

Park, S. (2021).A 2021 guide to im- proving cnns-
optimizers: Adam vs sgd. https://medium.com/
geekculture/a-2021-guide-to-improving-cnns-optimize
rs-adam-vs-sgd- 495848ac6008.
Ribeiro, M. T., Singh, S., and Guestrin, C. (2016). ”why
should i trust you?”: Explaining the predictions of any
classifier. In Proceedings of the 22nd ACM SIGKDD
International Conference on Knowledge Discovery
and Data Mining. Association for Computing Ma-
chinery.
Rolnick, D., Donti, P. L., Kaack, L. H., Kochanski, K.,
Lacoste, A., Sankaran, K., Ross, A. S., Milojevic-
Dupont, N., Jaques, N., Waldman-Brown, A., Luc-
cioni, A., Maharaj, T., Sherwin, E. D., Mukkavilli,
S. K., Kording, K. P., Gomes, C., Ng, A. Y., Hassabis,
D.,
Platt, J. C., Creutzig, F., Chayes, J., and Bengio, Y.
(2019).
Tackling climate change with machine learn- ing.
Ryo, M., Angelov, B., Mammola, S., Kass, J., Benito, B.,
and Hartig, F. (2021). Explainable artificial in-
telligence enhances the ecological interpretability of
black-box species distribution models. Ecography, 44.
S.Beery, E.Cole, J.Parker, P.Perona, and K.Winner (2021).
Species distribution modeling for machine learning
practitioners: A review. In ACM SIGCAS Conference on
Computing and Sustainable Societies (COMPASS ’21).
Science, S. D. (2021). Absence and background points.
https://rspatial.org/raster/sdm/3
s
dm
a
bsencebackground
.html.
SEON (2022). Whitebox machine learning. https://seon
.io/resources/dictionary/whitebox-machine- learning/h-
why-choose-whitebox-over-blackbox-machine-learning.
Wen, L. and Hughes, M. (2020). Coastal wetland mapping
using ensemble learning algorithms: A comparative
study of bag- ging, boosting and stacking techniques.
Remote Sensing.
Explainability of MLP Based Species Distribution Models: A Case Study
697
