
Personalized Semantic Annotation Recommendations on Biomedical
Content Through an Expanded Socio-Technical Approach
Asim Abbas
1 a
, Steve Mbouadeu
1 b
, Tahir Hameed
2 c
and Syed Ahmad Chan Bukhari
1 d
1
Division of Computer Science, Mathematics and Science St. John’s University, Queens NY 11439, U.S.A.
2
Girard School of Business, Merrimack College, North Andover, Massachusetts, U.S.A.
Keywords:
Annotation Recommendation, BERT, Semantic Annotation Optimization, Biomedical Semantics, Biomedical
Content Authoring, Peer-to-Peer, Annotation Ranking, Structured Data.
Abstract:
There are huge on-going challenges to timely access of accurate online biomedical content due to exponential
growth of unstructured biomedical data. Therefore, semantic annotations are essentially required with the
biomedical content in order to improve search engines’ context-aware indexing, search efficiency, and preci-
sion of the retrieved results. In this study, we propose a personalized semantic annotation recommendations
approach to biomedical content through an expanded socio-technical approach. Our layered architecture gen-
erates annotations on the users’ entered text in the first layer. To optimize the yielded annotations, users can
seek help from professional experts by posing specific questions to them. The socio-technical system also
connects help seekers (users) to help providers (experts) employing the pre-trained BERT embedding, which
matches the profile similarity scores of users and experts at various levels and suggests a run-time compati-
ble match (of the help seeker and the help provider). Our approach overcomes previous systems’ limitations
as they are predominantly non-collaborative and laborious. While performing experiments, we analyzed the
performance enhancements offered by our socio-technical approach in improving the semantic annotations in
three scenarios in various contexts. Our results show overall achievement of 89.98% precision, 89.61% recall,
and an 89.45% f1-score at the system level. Comparatively speaking, a high accuracy of 90% was achieved
with the socio-technical approach whereas the traditional approach could only reach 87% accuracy. Our novel
socio-technical approach produces apt annotation recommendations that would definitely be helpful for vari-
ous secondary uses ranging from context-aware indexing to retrieval accuracy improvements.
1 INTRODUCTION
Efficient practices for accessing biomedical publica-
tions are crucial to timely information transfer from
the scientific research community to other peer in-
vestigators and healthcare practitioners. This ex-
plosive growth in the biomedical domain has intro-
duced several access-level challenges for researchers
and practitioners. Due to the lack of machine-
interpretable metadata (semantic annotations), this
valuable information is available in the contents ac-
cessible on the web but still opaque to information
retrieval and knowledge extraction search engines.
Search engines require the metadata to correctly in-
dex contents in a context-aware fashion for the pre-
a
https://orcid.org/0000-0001-6374-0397
b
https://orcid.org/0000-0002-9137-407X
c
https://orcid.org/0000-0002-6824-6803
d
https://orcid.org/0000-0002-6517-5261
cise search of biomedical literature and to foster sec-
ondary activities such as automatic integration for
meta-analysis (Bukhari, 2017). Including machine-
interpretable semantic annotations in biomedical in-
formation at the pre-publication stage (during first
drafting) is desirable and will significantly benefit
the larger semantic web vision (Warren et al., 2008).
However, these processes are complex and require
deep technical and/or domain knowledge. Therefore,
a state-of-the-art, freely accessible biomedical seman-
tic content authoring framework would be a game-
changer.
Semantic Content Authoring is manual and/or semi-
automatic composing of textual content with an ex-
plicit semantic structure. The main components of
the semantic content authoring process are ontolo-
gies, annotators, and user interface(UI). Similarly,
semantic annotators are designed to facilitate tag-
ging/annotating the related ontology concepts with
638
Abbas, A., Mbouadeu, S., Hameed, T. and Bukhari, S.
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach.
DOI: 10.5220/0011926700003414
In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023) - Volume 5: HEALTHINF, pages 638-648
ISBN: 978-989-758-631-6; ISSN: 2184-4305
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

pre-defined terminologies in a manual, automatic, or
hybrid way(Abbas et al., 2021). As a result, users pro-
duce semantically richer content than traditional com-
posing processes, e.g., using a word processor (Ab-
bas et al., 2022). We categorized the current se-
mantic content authoring approaches into two stages
such as bottom-up and top-down. In the bottom-up
approach, semantic annotations or semantic markup
techniques, the textual contents of a document are
annotated using a set of ontologies such as Seman-
tic MediaWiki (Laxstr
¨
om and Kanner, 2015), Sweet-
Wiki (Buffa et al., 2008), and Linkator (Araujo et al.,
2010) are a few examples of bottom-up tools for creat-
ing semantic content. The current tools have a few no-
table shortcomings, though: The first drawback is that
it is an offline, non-collaborative, application-centric
way of content development. Second, because it was
created more than eleven years ago, it is incompatible
with the most recent version of MS Word. Likewise,
top-down methods were developed to add semantic
information to existing ontologies, each ontology be-
ing extended or filled using a particular template de-
sign. So this method is also known as an ontology
population method of content authoring. However,
Top-down approaches do not take the non-semantic
contents of a given text and uplift their quality by an-
notating them with the appropriate ontology terms.
Instead, it begins with the ontology to use the par-
ticular ontology concepts as fillers while authoring
content. Examples of top-down approaches are On-
toWiki (Auer et al., 2006), OWiki (Iorio et al., 2012),
and RDFAuthor(Tramp et al., 2010).
The development of a biomedical semantic annota-
tor has received significant support from the scien-
tific community due to the importance of the seman-
tic annotation process in biomedical informatics re-
search and retrieval. The biomedical annotators can
be further divided into a) general-purpose annotators
for biomedical, which assert to cover all biomedical
subdomains, and b) use case-specific annotators for
biomedical, which are created for a specific subdo-
main or to annotate specific entities like genes and
mutations in a given text. Whereas the general pur-
pose non-biomedical semantic annotators combine
technologies such as NLP (Natural Language Pro-
cessing), ontologies, semantic similarity algorithms,
machine learning (ML) models, and graph manip-
ulation techniques(Jovanovi
´
c and Bagheri, 2017).
Biomedical annotators predominantly use term-to-
concept matching with or without machine learning-
based methods. Biomedical annotators such as NO-
BLE Coder (Tseytlin et al., 2016), Neji (Campos
et al., 2013), and Open Biomedical Annotator(Shah
et al., 2009) use machine learning and annotate text
with an acceptable processing speed. However, they
lack a robust capacity for disambiguation or the abil-
ity to pick out the right biological concept for a
particular text from numerous competing notions.
Whereas NCBO Annotator(Jonquet et al., 2009) and
MGrep services are pretty slow, RysannMd annotator
claims to balance speed and accuracy in the annota-
tion process. However, on the flip side, its knowl-
edge base is limited to particular ontologies available
in UMLS (Unified Medical Language System) and
does not provide full coverage of all biomedical sub-
domains(Mbouadeu et al., 2022).
To address the above limitation and problem, we
proposed and developed ”Semantically Knowledge
Cafe” a freely accessible interactive system that en-
ables individuals at different expertise levels in the
biomedical domain to collaboratively author biomed-
ical semantic content. Finding the proper semantic
annotations in real-time during content authoring is
quite challenging because one semantic annotation is
frequently available in many biomedical ontologies
with various texts or implications. The main research
issue is to balance speed and accuracy. Therefore, we
proposed a state-of-the-art socio and personalized se-
mantic annotation recommendation approach to de-
velop a biomedical semantic content authoring sys-
tem that balances the speed and accuracy of avail-
able biomedical annotators while involving the orig-
inal author throughout the process. Additionally, our
infrastructure enables users to export their content in
various online interoperable formats for hosting and
sharing in a decentralized manner. To demonstrate
the usefulness of the proposed system, we conduct a
set of experiments on biomedical research articles ac-
quired from Pubmed.org(Macleod, 2002). The find-
ings of the proposed system indicate a significant re-
duction in annotation costs by achieving a higher ac-
curacy compared to the existing approaches used for
the same task in the past. Furthermore, our system is
unique by applying a novel Socio-technical and per-
sonalized approach to develop a biomedical semantic
content authoring to enhance the FAIRness(Jonquet
et al., 2009) of the published research.
2 PROPOSED APPROACH
In this section, we present an enhanced approach
to recommend semantic annotation on biomedical
content by developing a web-based free accessible
biomedical semantic content authoring system for an
individual with a different level of expertise in the
biomedical domain. The end user is facilitated with
an authoring interface similar to the MS Word editor
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach
639

Figure 1: Workflow of Personalized Semantic Annotation recommendations on Biomedical Contents.
to type/write biomedical semantic content such as re-
search papers, clinical notes, and biomedical reports.
To fetch the first layer of semantic annotation, we
leveraged Bioportal (Jonquet et al., 2009) endpoint
APIs and automated the configuration process for au-
thors. Henceforth, the annotated biomedical terms
are highlighted for the user. Besides, a ”Semantically
Knowledge Cafe” social-collaborative environment is
proposed to aid authors in getting help from an ex-
pert for accurate annotation of a particular term or
the entire text. Furthermore, the author is aided in
receiving personalized quality annotation recommen-
dations while directly communicating with an expert.
The proposed methodology is further distributed in
three sections: the first layer of annotations, personal-
ized expert recommendation, and annotation recom-
mendation through the Socio-Technical Approach as
shown in Figure.1.
2.1 First Layer Annotation
A biomedical annotator is a crucial component of
biomedical content semantic annotation or enrich-
ment (Mbouadeu et al., 2022). These biomedical an-
notators leverage publicly accessible biomedical on-
tologies, such as Bioportal (Jonquet et al., 2009) and
UMLS (Abbas et al., 2019), to assist researchers in
the biomedical community in structuring and anno-
tating their data with ontology notions to enhance in-
formation retrieval and indexing. However, the se-
mantic annotation and augmentation process is time-
consuming and entails expert curators. Therefore, we
automate the semantic annotations assignment proce-
dure with our designed solutions. To do that, we used
the NCBO Bioportal web-service resources (Jonquet
et al., 2009) to evaluate the original text and anno-
tate it with the proper biomedical ontology terms. As
shown in Figure.1(a), by pressing the ”Annotate” but-
ton, users can generate the First layer of annotations
without the need for any technical knowledge. In the
beginning, authors have two options: pre-existing im-
port content from research papers, clinical notes, and
biological reports, or begin typing in the semantic text
editor. The user’s free text is accepted by our sys-
tems, which then feed it into a concept recognition en-
gine as input. The machine follows a string-matching
approach to locate pertinent acronyms, definitions,
ontologies, and hyperlinks for specific terminologies
that best fit based on the context. This semantic infor-
mation is displayed in our system’s annotation panel
for human interpretation and understanding. Mean-
while, authors can modify the generated semantic in-
formation dependent on their knowledge and experi-
ence. For instance, they can choose an appropriate
ontology from the list, pick relevant acronyms, re-
move semantic information, annotate for explicit ter-
minology, etc. While more experienced users may use
complex features to regulate the semantic annotation
and authoring process competently, individuals with
a technical background can easily use a simplified in-
terface.
2.2 Personalized Expert
Recommendation
In the proposed study, the author can obtain personal-
ized semantic annotation recommendations by either
posting questions in the semantic content authoring
environment or by a system that can make recommen-
dations automatically see Figure.1(b). We used a pro-
file similarity-based method to match the authors with
experts Figure.1(c). A pre-trained BERT (Bidirec-
tional Encoder Decoder Transformer) (Devlin et al.,
2018) NLP deep learning model is utilized to gen-
erate contextual word embeddings of the author pro-
file and expert E
i
profile. So far, various algorithms
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
640

have been used by the NLP research community for
text similarity matching, such as cosine similarity, Eu-
clidian distance, and Jaccard similarity (Han et al.,
2006). We utilized the cosine similarity index be-
tween author and expert E
i
contextual word embed-
ding in the proposed approach. We chose the top five
experts E
i
to the author of a similar profile. There-
after, each of the top five recommended experts’ E
i
previously available profile scores is catered from the
expert repository. The product of the profile similar-
ity score and the expert’s previously available score
is calculated to find a weighted score for each top-
selected expert E
i
. Finally, as shown in Figure.1(d),
we used a filter to rank and recommend the expert E
i
with the highest weighted score among the top five.
Hereafter an author set up personalized communica-
tion with the recommended expert for semantic an-
notation suggestions. In the below section, a detailed
process of expert recommendation is presented.
2.3 Personalized Expert
Recommendation Scenario
This section illustrates a case study of expert, per-
sonalized recommendations to the author in a seman-
tic optimization environment. We have designed a
web-based interface called ”Semantically Knowledge
Cafe” for an author to search and communicate with
an optimal expert for a precise and quality seman-
tic annotation recommendation. Initially, an author
can pose questions manually to seek help from the
community domain expert. As shown in Figure.1(i),
our system stores author profile information and other
users’ U
i
profile information in the users’ repository.
This profile information is collected during first-time
registration, consisting of research interest, qualifica-
tion, profession, organization and experience in an
unstructured format. First, the BERT model is used to
generate a contextual word embedding of author and
user U
i
profile text (see Figure.1(j)). Then, as shown
in Figure.1(k), a cosine similarity index is used to de-
termine the contextual profile similarity between the
author and user U
i
. As Equ.1 is presented, the cosine
similarity mathematical representation.
ψ = similarity(a,u) = cos(θ) =
⃗a.⃗u
∥⃗a ∥∥⃗u ∥
(1)
Where, cos θ represent the angle between author and
users U
i
embedding vectors, whereas ⃗a represent the
author embedding vectors and ⃗u denoted the User U
i
embedding vectors.
After all identifying the similarity score between
author and other users U
i
profile, the users U
i
profile
is sorted in descending order based on obtained
similarity. Finally the top high profile similarity score
expert E
i
is selected Figure.1(i). As discussed above
during first time registration we filled the question-
naire from the users U
i
related to their profession,
qualification, experience and research interest. Each
section of the questionnaire consists of subsections as
shown in the scoring table Table.1, where a suitable
score is randomly assigned to each category. Finally
we recorded the average mean subsequently filling
the questionnaire as shown in Equ.2. Where X
i
is
the score of each category in a scoring table and N
is the sum of all category scores. After all, the final
mean score stored into the repository named as initial
profile scoring.
Mean
Score
= µ =
n
∑
i=0
X
i
!
/N (2)
Subsequently profile similarity, a weighted score
is calculated for each top five selected expert E
i
as shown in Equ.3. Whereas weighted score is the
product of mean average score (µ) and user profile
similarity score (ψ) as shown in Equ.3
Weighted
score
= φ
i
= µ
i
∗ ψ
i
(3)
Where, φ
i
represents the weighted score of an expert
E
i
and µ
i
is the mean score calculated during first time
registration from a questionnaire and ψ
i
is the each
top five user profile similarity score. Afterward the
experts are sorted in the ascending order and filter is
applied to choose only top weighted score among. Fi-
nally the top weighted score expert is recommended
to the author for a personalized recommendation and
communication.
2.4 Proficient Annotation
Recommendation
Succeeding in obtaining initial/base level semantic
annotation, “Semantically Knowledge Cafe” provides
an out-of-the box socio-technical environment where
the author is allowed to communicate and get recom-
mendation from peer review for a correct and high
quality annotation. The proposed approach is eval-
uated while taking three types of scenario as shown
in Table.2. Following these scenarios we collected
the features as shown in Table.3, then applied a sta-
tistical approach to rank and recommend the correct
annotation to the author. The environment is called
“Semantically Knowledge Cafe” where the author can
post their query, peers or domain expert can reply for
the author post with some self confidence score, other
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach
641
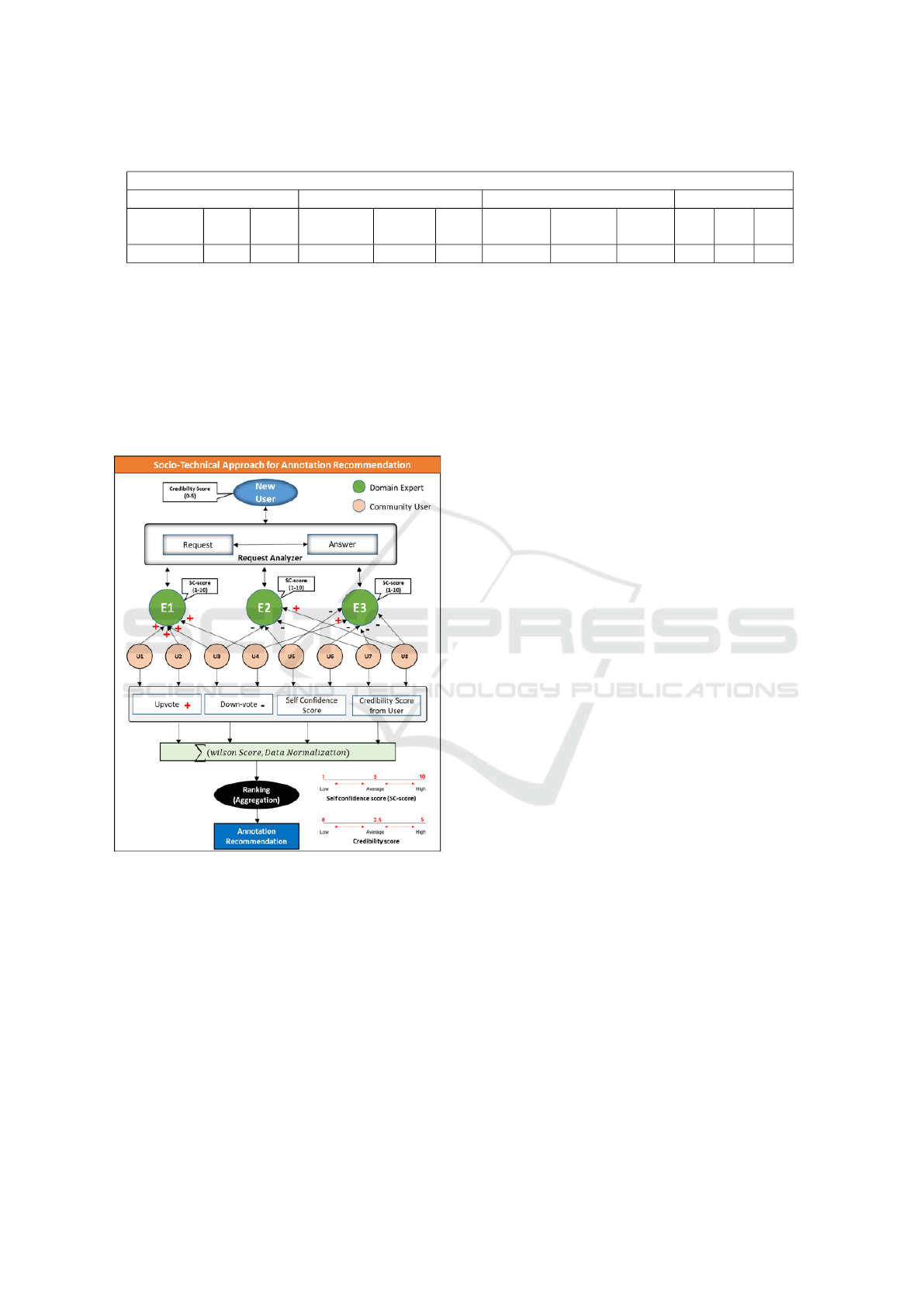
Table 1: Preliminary Users Profile Scoring Table.
Scoring Table
Research Interest Qualification Profession Experience
Semantic
Web
DL
/ML
NLP Bachelor Master PhD
Develo-
per
Resear-
cher
Profe-
ssor
1-2 3-5 >5
5 5 5 3 4 5 4 4 5 3 4 5
community users can credit the expert reply by up-
vote and down-vote in a collaborative mode. Finally
the author receives the notification for their post with
recommended annotation. Meanwhile the author is
allowed to accept the recommended annotation or re-
ject the recommendation and the ultimate results are
stored in the database. In this section we have pre-
sented the statistical process of the annotation recom-
mendation as shown in Figure.2.
Figure 2: Socio-technical annotation recommendation eval-
uation process.
In Table.2 we have listed three different scenarios
to evaluate the proposed socio-technical annotation
optimization approach. These scenarios are related to
the query regarding semantic annotation for medical
content. In addition to scrutinizing the existing ques-
tion answering platform such as Stackoverflow, such
kind of scenarios or query can be found. An example
is presented for a Scenario:1 below:
Suppose the author is required to find correct on-
tology annotations from experts for the biomedical
term “Coronary artery disease”. The “Semantically
Knowledge Cafe” provides an interface where user
can put their query.
For Example:
”Which ontology should I use for ”Coronary
artery disease”?
Following submission of the aforementioned query, it
appeared as a new post on the ”Semantically Knowl-
edge Cafe” forum for expert E
i
responses where E
i
=
e
1
,e
2
,e
3
,...e
n
. Similarly, the ”Semantically Knowl-
edge Cafe” provides an interface to the expert E
i
to smooth the response process, allowing an expert
to describe the suggested annotation, provide a self-
confidence score for their recommendation, and eas-
ily search for a correct ontology using the NCBO on-
tology tree widget tool. When the expert E
i
responds
to the author’s post for Scenario 1, other community
users U
i
= u
1
,u
2
,u
3
,....u
n
respond with an upvote as
+V and a downvote as -V, to the expert reply as shown
in Figure.2. A statistical measure is taken for ex-
pert self-confidence score, upvote +V and downvote
-V, and credibility score from the author by applying
the Wilson formula and data normalization process as
shown in Figure.2. Finally, an optimal recommenda-
tion of annotation is generated for the author.
2.5 Semantically Recommendation
Features(SR-FS)
Finding an optimal and quality annotation recommen-
dation, we have addressed several features in the pro-
posed socio-technical approach as shown in Table.3.
These features are generated by and collected from
the community users, who actively participated in the
socio-technical environment. We presented features
such as upvote (+V), downvote (-V), expert confi-
dence score, and author credibility score as accepting
or rejecting in the following way.
2.5.1 Upvote and Downvote (SR-f1)
In the social network environment, upvotes +V and
downvotes -V plays a crucial role, whereas +V indi-
cates the usefulness or quality of a response or answer
while -V points to irrelevance or low quality. This fea-
ture measures the quality of domain expert response
to the post for suggesting annotation by achieving
high numbers of up-votes and low numbers of down-
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
642

Table 2: Semantically Annotation Recommendation Scenarios.
Scenario. No Scenario Description
Scenario:1 Which Ontology should I use?
Scenario: 2 What is the suitable ontology vocabulary?
Scenario: 3 Does this Ontology best describe this terminology?
Table 3: Semantically Recommendation Features(SR-f) Descriptions.
Features. No Features Name Feature Descriptions
SR.f1
Upvotes(+) and
Downvotes(-)
Up-votes and down-votes from
community users.
SR.f2
Self Confidence
Score
Score from expert for their response
to post.
SR.f3
Credibility Score
from Author
Credibility score from the author to
expert annotation recommendation.
votes. Wilson’s formula takes these feature score con-
fidence intervals and applies them to a Bernoulli pa-
rameter (see Equ.4) to determine the expert-suggested
annotation quality score.
Wilson
score
=
ˆp +
Z
2
α/2
2n
± Z
α/2
r
j
ˆp(1 − ˆp) + Z
2
α/2
/4n
k
/n
/
1 + Z
2
α/2
/n
!
(4)
Where,
b
p =
N
∑
n=1
+V
!
/(n) (5)
n =
N
∑
i=0
M
∑
j=0
(+V
i
,−V
j
) (6)
and, z
α
2
is the
1 −
α
2
quantile o f the standard
normal distribution (7)
In Equ.4. ˆp is the sum of upvotes (+V) of a com-
munity user’s U
i
to the Expert E
i
response for a post
from an author for correct annotation divided by over-
all votes (+V,-V). Likewise, n is the sum of the num-
ber of upvotes and downvotes (+V,-V), and α is the
confidence refers to the statistical confidence level:
pick 0.95 to have a 95% chance that our lower bound
is correct. However, the z-score in this function never
changes.
2.5.2 Self Confidence Score (SR-f2)
In Psychology, self-confidence refers to an individ-
ual’s trust in their abilities, capacities, and judgments
that they can successfully make. In the proposed ap-
proach, we allow the Expert to give a confidence score
for their decision-making for an annotation recom-
mendation. As shown in Figure.3, experts can rate
how they feel about recommended annotations by as-
signing a self-confidence score between 1 and 10. Use
a number between 1 and 10 to accurately describe the
expert confidence response. For example, if an ex-
pert feels slightly above average for their recommen-
dation, rate them a six score, but if an expert feels
more confident, rate them an eight score.
Figure 3: Self Confidence Score Selection Level.
All the features (SR-f1, SR-f2, and SR-f3)
are equally contributed and deeply correlated with
each other for the final annotation recommendation.
Though the final output of SR-f1 is between 0 and
1, we normalize the self-confidence score (SR-f2) be-
tween 0 and 1 using Equ.8 to keep the process consis-
tent and feature dependent.
z
i
= (x
i
− min(x))/(max(x) − min(x)) ∗ Q (8)
Where, z
i
is the i
th
normalized value in the dataset.
Where x
i
is the i
th
value in the dataset, e.g., the user
confidence score. Similarly, min(x) is the minimum
value in the dataset, e.g the minimum value between
1 and 10 is 1, so the min(x)=1 and max(x) is the max-
imum value in the dataset, e.g the maximum value
between 1 and 10 is 10, so the max(x)=10. Finally,
Q is the maximum number wanted for a normalized
data value, e.g. we normalized the confidence score
between 0 and 1, and the maximum value between 0
and 1 for Q is 1.
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach
643

2.5.3 Credibility Score from an Author (SR-f3)
Credibility is deemed to be the quality of being be-
lieved or accepted as true and accurate. As an at-
tribute, credibility is crucial because it helps to influ-
ence domain expert knowledge, experience, and pro-
file. Therefore if a domain expert profile is not credi-
ble, others are less likely to believe what is being said
or recommended. Subsequently, annotation recom-
mendations are received by the author from an expert,
the author is allowed to either accept or reject the rec-
ommended annotation with some credibility score be-
tween 0 and 5 as shown in Figure.4. Whenever an au-
thor agrees with the recommended annotation, a cred-
ibility score between 2 and 5 is added to the expert
or help provider profile by the author. Also to pre-
vent repetitive questions on the ”Semantically Knowl-
edge Cafe” forum, the author’s credit score effectively
adds value to automatic annotation recommendations
to other community users for relevant questions or ter-
minologies. As discussed in the above section SR-f2,
all the features (SR-f1,SR-f2 and SR-f3) are equally
contributed and deeply correlated with each other for
final annotation recommendation. Therefore, we ap-
ply Equ.8 on SR-f3, to transform values between 0
and 1.
Figure 4: Credibility Score from an Author Selection Level.
Finally, Equ.9 is used to compute and aggregate
the SR-FS (Semantically Ranking Feature Score) for
each expert’s E
i
recommended annotations.
Sr − Fs =
m
∑
j=1
n
∑
i=1
p
∑
k=0
(F
j
,E
i
,A
k
) (9)
f inal − score = argmax[
N
∑
i=1
(Sr − Fs)] (10)
Where F
j
is feature score for Expert E
i
and Annota-
tion A
k
. The final decision or ranking happens based
on maximum feature scoring gained by the Expert E
i
response to the author’s post or query see Equ.10.
2.6 Annotation Recommendation
Experimental Environment
As shown in Figure.5, we have designed three pecu-
liar studies to evaluate the proposed socio-technical
approach for annotation recommendation. To do that,
we process individual levels of features (SR-f1, SR-
f2, and SR-f3) for ranking annotations. In this sec-
tion, we presented an experimental practices environ-
ment for Scenarios in Table.2 by evaluating features
statistically.
As shown in the Figure.5. An author posted a
query on a ”Semantically Knowledge Cafe” forum
such as ”Which Ontology should I use for medical
content ’Coronary artery disease?. As a result, an au-
thor receives reply for their post from a domain ex-
pert E
i
presented as ”Reply-post” and suggests the
correct annotation for a required biomedical content
as ”Social Annotation”. In the study, four experts
participated, and each Expert suggested the annota-
tion as (”NCIT”,” MESH”, ”ADO” and ”LPRO”).
Meanwhile expert E
i
also provides a self confidence
score of (4,7,3 and 2) as “Expert Confidence Level”
between 1 and 10 for the suggested annotation. Now
community users U
i
are allowed to give their feed-
back in the form of upvotes (10,15,4,5) and down-
votes (2,3,8,11) to the Expert recommended annota-
tion, and finally, a sum of upvotes and downvotes is
calculated as ”Total Votes”. Similarly, whenever the
user accept suggested annotation from experts E
i
, a
credibility score of (1,4,1,1) is gain by the experts
E
i
. As previously discussed in section E(1), a sta-
tistical approach Wilson score see Equ.4 is applied
on upvotes and downvotes for expert E
i
to calculate
final estimation as (0.552,0.608, 0.138 and 0.142).
Likewise, a data normalization formula sees Equ.8 is
employed on the Expert confidence score and author
credibility score to downstream the value between 0
and 1. Consequently a mean ˆx =
1
N
∑
N
i=0
x
i
applied on
Wilson score, normalize self confidence and author
credibility score of expert E
i
suggested annotation as
“Aggregate score” of (0.295, 0.675,0.12 and 0.084).
Finally argmax(x
i
) function is applied on the aggre-
gate score to obtain the maximum score earned by the
expert E
i
annotation which is (0.675). So that even-
tually, the highly proficient and ranking annotation is
recommended to the author as ”Mesh” and ”Reply-
post=2” for the biomedical content ”Coronary artery
disease”. The same process is applied for another
biomedical content ”burst of atrial fibrillation” but
the scenario or query can be changed.
3 Experiments
3.1 Datasets
In the evaluation, a total of 30 persons took part in
the proposed methodology. We recruited people by
making a social media call asking them to partici-
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
644
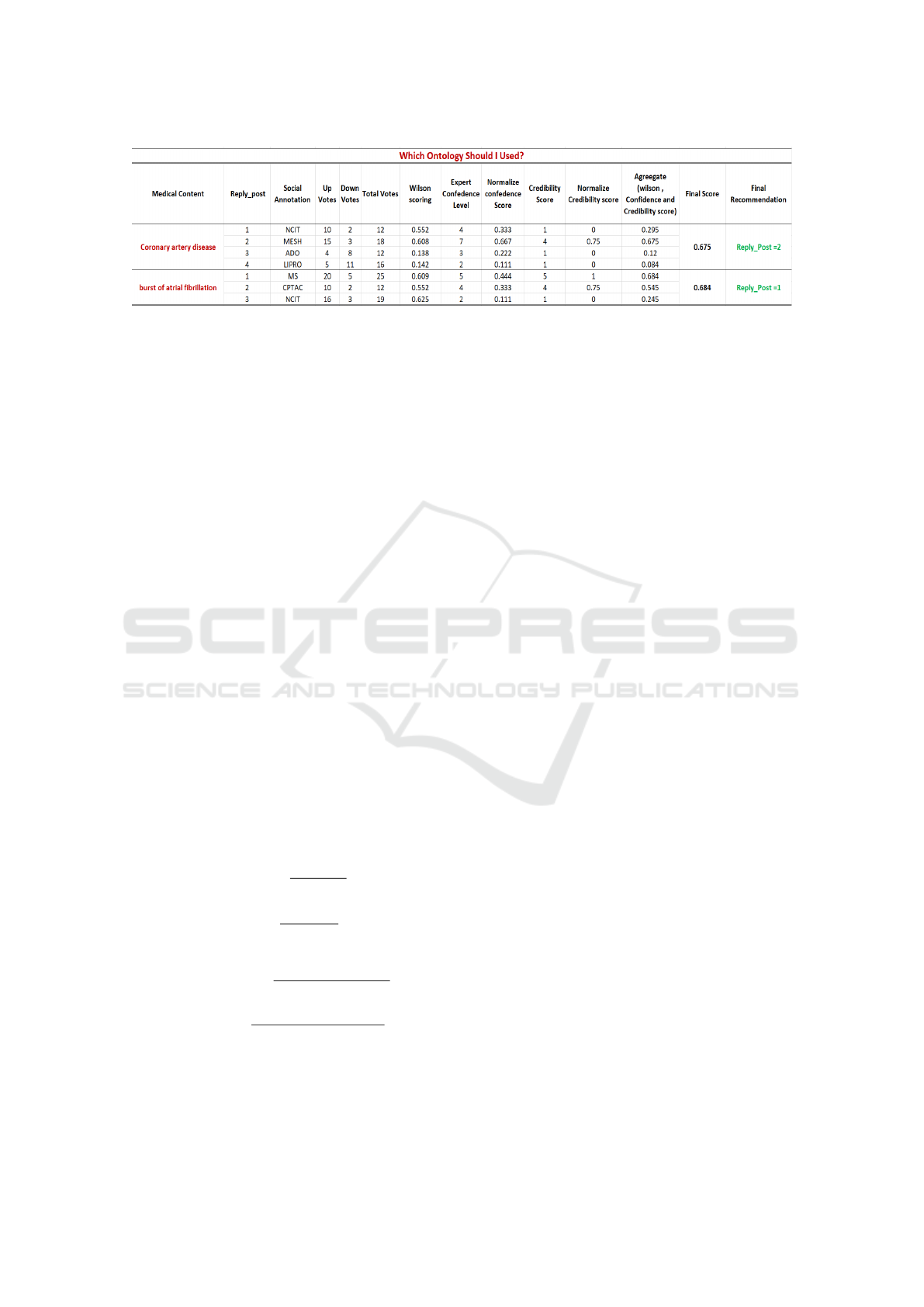
Figure 5: A statistical evaluation representation of Annotation Recommendation.
pate in the study. We classify participants as pri-
marily graduate-level students with computer and bi-
ological sciences backgrounds. In addition, we ran-
domly assigned individuals a batch of 30 papers from
PubMed.org(Macleod, 2002). We provided a user
manual of systems along with a pre-recorded video
about system usage. We asked each participant to cre-
ate a query on ”Semantically Knowledge Cafe” about
the biomedical content annotation they like to ask for
help from an Expert. Collectively, our participants
post 140 questions to the system. All the participants
have also recorded confidence scores between 0 and
1 from the suggestions they received as a satisfaction
score.
3.2 Results and Analysis
The effectiveness and performance of the suggested
system or approach are often assessed using four
indexes Precision, Recall, f1-Score, and Accuracy.
Since recall counts the number of valid examples in
the targeted class of instances, precision counts the
number of valid instances in the set of all retrieved in-
stances. Similar to the modified f1-score, it is the har-
monic mean of precision and recall, where accuracy
is the proportion of true positives and true negatives to
all positive and negative observations. The following
formulae can be used to calculate the measurements:
Precision =
T P
T P + FP
(11)
Recall =
T P
T P + FN
(12)
f 1 − score = 2 ∗
Precision ∗ Recall
Precision + Recall
(13)
Accuracy =
T P + T N
T P + FN + T N + FP
(14)
3.2.1 Users of Similar Profile
As mentioned above, thirty users actively participated
in the proposed methodology evaluation. Initially,
we received user profile information about the user’s
background, knowledge, and experience in a particu-
lar domain. A word embedding similarity approach
is employed to identify similar users that help in
expert recommendations with equivalent knowledge
and experience in a specific field. A BERT (Bidi-
rectional Encoder Representation of Transformer) an
NLP model is utilized to generate the user profile em-
beddings. A cosine similarity algorithm is used to find
the embedding similarity score between users. It has
been analyzed all the user’s profiles are at least 50%
similar to one another. The lowest similarity between
user profiles is found 0.51 (51%), the highest similar-
ity is 0.98 (98%), and the average users lie in the range
of 0.60 (60%) and 0.80 (80%) as shown in Figure.6.
3.2.2 Inter Annotator Agreement(IAA)
In the socio-technical approach, when the author re-
ceived recommended annotations from a community
expert through ”Semantically Knowledge Cafe”, we
also evaluated these annotations from domain experts
employing the IAA (Inter Annotator Agreement) ap-
proach.
The Inter-Annotator Agreement (IAA), a measure
of how well multiple annotators can make the same
annotation decision for a certain category. It is a vi-
tal part of both the validation and reproducibility of
annotation results. There were three domain expert
participated to evlauted the socio-technical generated
semantic annotations. We take two measurment into
account for evaluation purpose:i) Cohen’s Kappa and
ii) Fleiss’ Kappa. In this way Cohen’s(κ) is the mea-
sures of agreement between two annotators annotat-
ing each instance into mutually exclusive categories.
Whereas Fleiss(κ) is the measuremnt, where the num-
ber of annotator can be more than two.
Where, In Equ.15 p
o
is the relative observed agree-
ment among annotators(similar to the accuracy), and
p
e
is the hypothetical probability of chance agree-
ment. To interpret Cohen’s kappa results, refer the
following study (Landis and Koch, 1977). However,
if the annotators are in complete agreement then κ =
1 and perfect agreement. If there is no agreement
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach
645
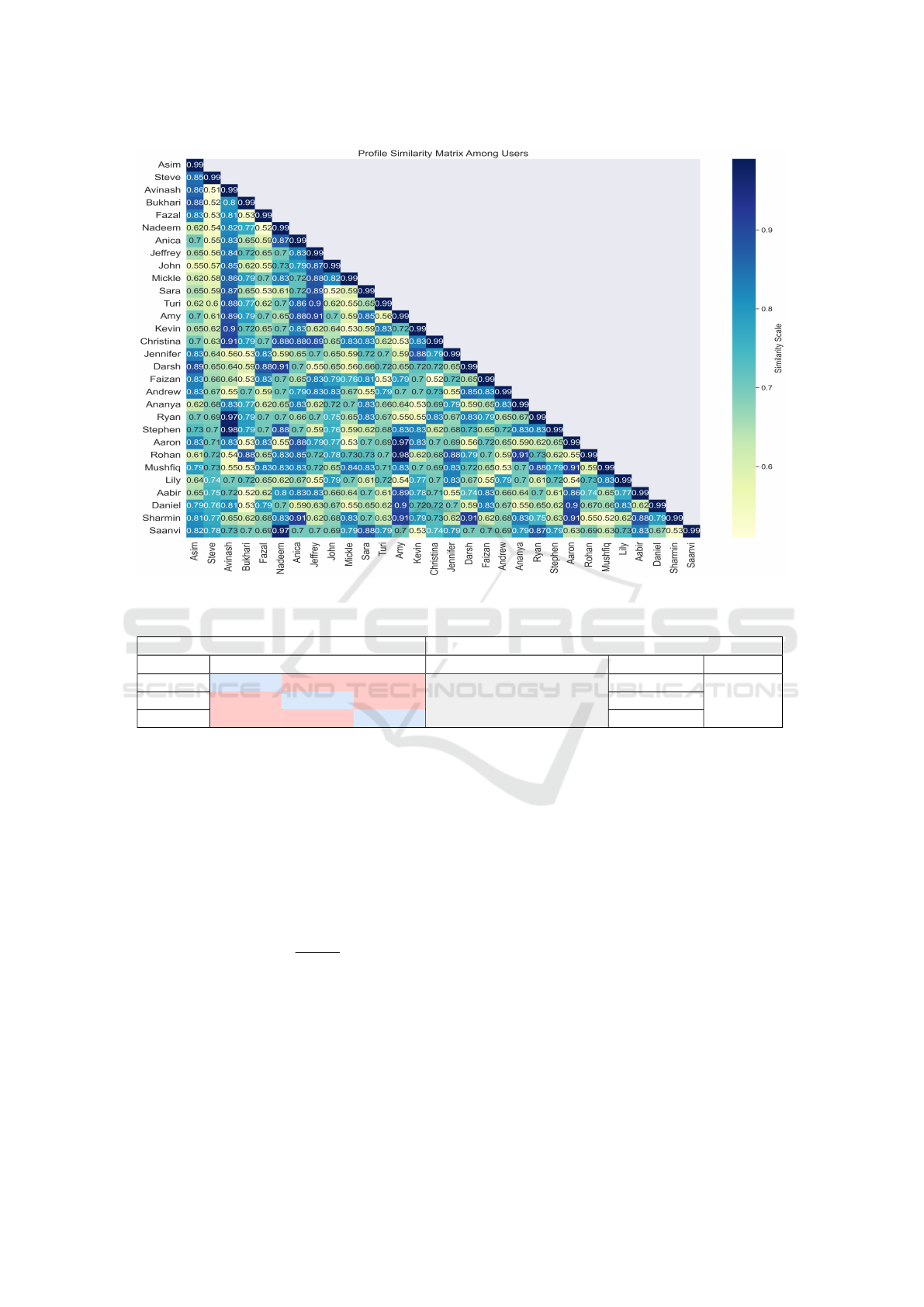
Figure 6: User’s Having Similar Profile by Employ BERT based Embedding Similarity score.
Table 4: Inter Annotator Agreement(IAA) Results Among Domain Experts.
% of Agreement Cohen’s and Fleiss Kappa Value
Expert1 Expert2 Expert3 IAA between two Expert Cohen’s(κ ) Fleiss(κ )
Expert1 95.56% 95.3% Expert1, Expert2 0.88
Expert2 95.56% 95.04% Expert1, Expert3 0.87
Expert3 95.3% 95.04% Expert2, Expert3 0.87
0.88
among the annotators then κ ≤ 0 or slight agree-
ment as shown in the Table.4. As per guidlines in
the study (Landis and Koch, 1977), we obtained al-
most perfect agreeemnt among three domain epx-
ert(Annotators) for our porposed socio-technical ap-
proach where all agreement value is placed more than
90%. Similarly perfect Cohen’s and Fleiss kappa
value of more than 85% is gain by domain experts for
socio-technical annotation recommendation as shown
in Table.4.
kappa(κ) =
P
o
− P
e
1 − P
e
(15)
3.2.3 System Level Performance
The outcomes of the scenario-level calculations are
averaged to get a system-level performance. For each
case, we first discover the findings at the document
level. We then average the outcomes at the sys-
tem level after combining the findings at the scenario
level.
After gathering the results for each scenario, a
professor-level domain expert from the academic
community is engaged to manually assess these out-
comes using their expertise. Then, to determine
the system’s effectiveness, we manually compare the
outcomes of domain experts to those of the socio-
technical approach and use precision, recall, and f1-
score. As a result, the system has demonstrated a
nearly identical performance of 90% for an annota-
tion recommendation in a socio-technical setting, as
illustrated in Figure.7. Additionally, we use both a
non-sociotechnical and a socio-technical approach to
measure the system’s performance utilizing Equ.14.
Since it was founded, high accuracy is gained after
a socio-technical approach, as shown in Figure.8. A
document’s level of accuracy is identified without a
socio-technical approach and with a socio-technical
approach process. The Figure.8 shows the number
of 30 documents processed on the X-axis, while the
Y-axises on the left and right, respectively, show the
accuracy levels with and without socio-technical ap-
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
646
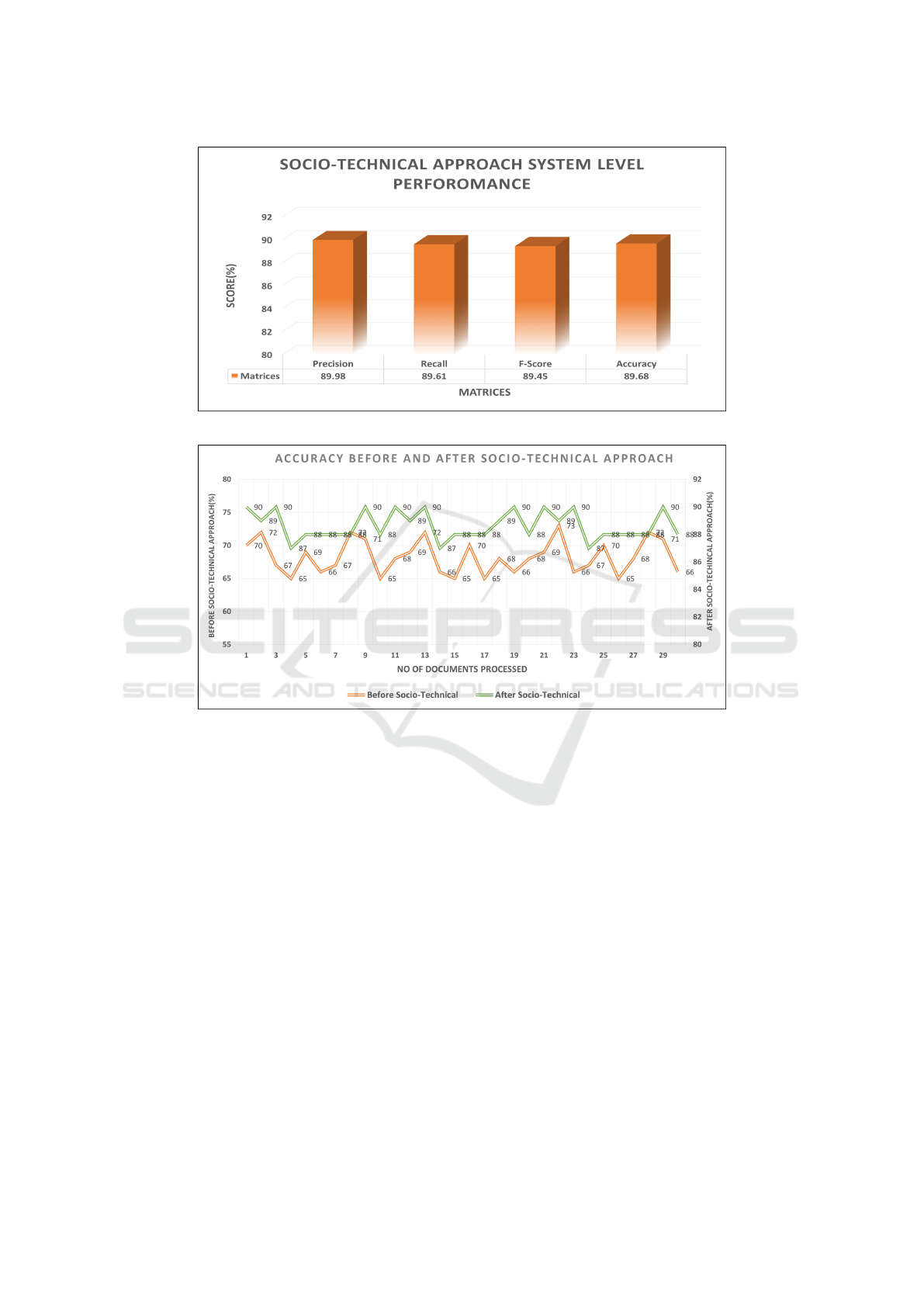
Figure 7: Presented the System Level Performance of a Socio-technical approach.
Figure 8: Presented the Accuracy of the system without and with Socio-technical approach.
proaches. As a result, analyzing a system’s findings
with a socio-technical approach is more effective than
it is without a socio-technical at the document level.
Nine documents achieved an accuracy of 90%, three
documents achieved an accuracy of 87%, and a max-
imum of documents achieved an accuracy of between
87% and 90% using a socio-technical method see
Figure.8. Similarly, high accuracy of 73% is yielded
by a single document and low accuracy of 65% is
gained by five documents and the maximum number
of documents gained accuracy in the range of 65% to
73% with a socio-technical approach. Overall the pro-
posed socio-technical approach remains the winner
by obtaining high precision of 89.98%, recall 89.61%,
and f1-score 89.45%.
4 CONCLUSIONS
This work aims to develop a publicly available sys-
tem that allows users with various levels of biomed-
ical expertise to produce correct semantic annotation
for a biomedical content. To balance speed and ac-
curacy, we present a hybridized approach for seman-
tic annotation optimization method to create correct
biomedical semantic content by involving the original
author throughout the process. We utilized Bioprtal
end-point web services to cater initial level seman-
tic information and automate the configuration pro-
cess for the authors. Similarly, ”Semantically Knowl-
edge Cafe” is designed where the author can commu-
nicate to the experts similar to their profile and get
personalized semantic annotation recommendations.
A pre-trained NLP Model BERT is utilized to rec-
ommend a proficient expert to the author based on
the similar profile employed contextual word embed-
Personalized Semantic Annotation Recommendations on Biomedical Content Through an Expanded Socio-Technical Approach
647

dings similarity approach. Similarly, ”Semantically
Knowledge Cafe” is created so that authors may pub-
lish their queries and receive appropriate annotation
recommendations. While peers or domain experts re-
view the author’s post, other community members can
appreciate the expert reply by up- and down-voting in
a collaborative way. Finally, the author receives the
notification for their post with recommended annota-
tion. The author can accept the recommended annota-
tion or reject the recommendation, and the final find-
ings are recorded in the database. The semantically
system is available at https://gosemantically.com
ACKNOWLEDGEMENTS
This work is supported by the National Science Foun-
dation grant ID: 2101350.
REFERENCES
Abbas, A., Afzal, M., Hussain, J., Ali, T., Bilal, H. S. M.,
Lee, S., and Jeon, S. (2021). Clinical concept extrac-
tion with lexical semantics to support automatic an-
notation. International Journal of Environmental Re-
search and Public Health, 18(20):10564.
Abbas, A., Afzal, M., Hussain, J., and Lee, S. (2019).
Meaningful information extraction from unstructured
clinical documents. Proc. Asia Pac. Adv. Netw, 48:42–
47.
Abbas, A., Mbouadeu, S. F., Keshtkar, F., DeBello, J., and
Bukhari, S. A. C. (2022). Biomedical scholarly article
editing and sharing using holistic semantic uplifting
approach. In The International FLAIRS Conference
Proceedings, volume 35.
Araujo, S., Houben, G.-J., and Schwabe, D. (2010). Linka-
tor: Enriching web pages by automatically adding
dereferenceable semantic annotations. In Interna-
tional Conference on Web Engineering, pages 355–
369. Springer.
Auer, S., Dietzold, S., and Riechert, T. (2006). Ontowiki–
a tool for social, semantic collaboration. In Inter-
national Semantic Web Conference, pages 736–749.
Springer.
Buffa, M., Gandon, F., Ereteo, G., Sander, P., and Faron, C.
(2008). Sweetwiki: A semantic wiki. Journal of Web
Semantics, 6(1):84–97.
Bukhari, S. A. C. (2017). Semantic enrichment and simi-
larity approximation for biomedical sequence images.
PhD thesis, University of New Brunswick (Canada).
Campos, D., Matos, S., and Oliveira, J. L. (2013). A mod-
ular framework for biomedical concept recognition.
BMC bioinformatics, 14(1):1–21.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Han, J., Kamber, M., and Pei, J. (2006). Data mining: con-
cepts and techniques morgan kaufmann. San Fran-
cisco.
Iorio, A. D., Musetti, A., Peroni, S., and Vitali, F. (2012).
Owiki: enabling an ontology-led creation of seman-
tic data. In Human–Computer Systems Interaction:
Backgrounds and Applications 2, pages 359–374.
Springer.
Jonquet, C., Shah, N., Youn, C., Callendar, C., Storey, M.-
A., and Musen, M. (2009). Ncbo annotator: seman-
tic annotation of biomedical data. In International
Semantic Web Conference, Poster and Demo session,
volume 110. Washington DC, USA.
Jovanovi
´
c, J. and Bagheri, E. (2017). Semantic annota-
tion in biomedicine: the current landscape. Journal
of biomedical semantics, 8(1):1–18.
Landis, J. R. and Koch, G. G. (1977). The measurement of
observer agreement for categorical data. biometrics,
pages 159–174.
Laxstr
¨
om, N. and Kanner, A. (2015). Multilingual semantic
mediawiki for finno-ugric dictionaries. In Septentrio
Conference Series, volume 2, pages 75–86.
Macleod, M. (2002). Pubmed: http://www. pubmed. org.
Journal of Neurology, Neurosurgery & Psychiatry,
73(6):746–746.
Mbouadeu, S. F., Abbas, A., Ahmed, F., Keshtkar, F.,
De Bello, J., and Bukhari, S. A. C. (2022). Towards
structured biomedical content authoring and publish-
ing. In 2022 IEEE 16th International Conference on
Semantic Computing (ICSC), pages 175–176. IEEE.
Shah, N. H., Bhatia, N., Jonquet, C., Rubin, D., Chiang,
A. P., and Musen, M. A. (2009). Comparison of con-
cept recognizers for building the open biomedical an-
notator. In BMC bioinformatics, volume 10, pages 1–
9. Springer.
Tramp, S., Heino, N., Auer, S., and Frischmuth, P. (2010).
Rdfauthor: Employing rdfa for collaborative knowl-
edge engineering. In International Conference on
Knowledge Engineering and Knowledge Manage-
ment, pages 90–104. Springer.
Tseytlin, E., Mitchell, K., Legowski, E., Corrigan, J., Cha-
van, G., and Jacobson, R. S. (2016). Noble–flexible
concept recognition for large-scale biomedical natural
language processing. BMC bioinformatics, 17(1):1–
15.
Warren, P., Davies, J., and Brown, D. (2008). The semantic
web-from vision to reality. ICT futures: Delivering
pervasive, real-time and secure services, pages 55–66.
CCH 2023 - Special Session on Machine Learning and Deep Learning for Preventive Healthcare and Clinical Decision Support
648
