
A Machine Learning Approach to Digitize Medical History and Archive
in a Standard Format
Mohamed Mehfoud Bouh
a
, Forhad Hossain
b
and Ashir Ahmed
c
Faculty of Information Science and Electrical Engineering, Kyushu University, Fukuoka, Japan
Keywords:
Machine Learning, Electronic Health Records, Portable Health Clinic, Data Digitization, Data
Standardization, Data Integration.
Abstract:
Thanks to the advancement of information technology and the wide adoption of smartphone-based apps, an
enormous amount of medical information is being produced worldwide. However, most of the medical records
are yet to be standardized. Small clinics in developing countries generate only handwritten medical documents.
Our past medical history is not digitized. Machine learning approaches applied to predict disease are quite
common. But it will need sufficient past medical records to analyze. However, we do not have past medical
records in digital form. This research aims to generate standard Electronic Health Records (EHRs) from
paper-based documents. The major research tasks will be to investigate (1) the commonalities and differences
of current unstructured paper-based medical documents, (2) the best technology to convert the paper-based
documents into unstructured data, and (3) Extracting structured data from the unstructured data, (4) Integrating
the structured into EHR databse using FHIR-based or OpenEHR Type System. This will produce standard
medical history. Once medical histories are available in a standard format, it will be possible to predict
personalized health status more accurately.
1 INTRODUCTION
In developing countries, access to healthcare can be
limited due to a lack of infrastructure, resources, and
trained medical professionals. Portable Health Clinic
(PHC) has been proposed as a solution to this prob-
lem, as it is designed to be easily transported to re-
mote and under-served areas. (Ahmed et al., 2014).
In the field of healthcare, the use of paper-based
documents is still prevalent despite the increasing
adoption of Electronic Health Records (EHRs) and
Electronic Medical Records (EMRs)(Rigatti, 2017).
While many healthcare organizations have imple-
mented EMRs and EHRs, some still rely on paper-
based systems, particularly in smaller clinics and ru-
ral areas (Tolera et al., 2022). However, paper docu-
ments can be difficult to manage and access, leading
to inefficiencies in patient care and increased risk of
errors. Additionally, older adults often have multi-
ple chronic conditions and take multiple medications,
which means they have a lot of medical information
a
https://orcid.org/0000-0002-7716-7007
b
https://orcid.org/0000-0002-3593-0860
c
https://orcid.org/0000-0002-8125-471X
to keep track of. This can be difficult for them to re-
member. One solution to these problems is the use
of document scanning technology to digitize paper-
based medical documents and integrate them into
EHRs using FHIR(Fast Healthcare Interoperability)
or OpenEHR(Open Electronic Health Record). FHIR
and OpenEHR are both standards for electronic health
records that aim to improve interoperability and data
sharing between EHR systems, but they differ in their
approach, focus, and adoption(Saripalle et al., 2019).
This process allows for the conversion of physical
documents into digital formats, which can then be
easily stored, shared, and accessed by healthcare pro-
fessionals. This paper will investigate the technical
aspects of scanning paper-based medical documents
and integrating them into EHRs in a standard format.
It will also outline a system architecture with the goal
of providing systems such as PHC and SHGC (Smart
Health Gantt Chart) that our lab is developing (Hos-
sain et al., 2022) with the appropriate data collection
mechanism from paper-based documents in order to
predict patient’s health status more accurately.
230
Bouh, M., Hossain, F. and Ahmed, A.
A Machine Learning Approach to Digitize Medical History and Archive in a Standard Format.
DOI: 10.5220/0011986400003476
In Proceedings of the 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2023), pages 230-236
ISBN: 978-989-758-645-3; ISSN: 2184-4984
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
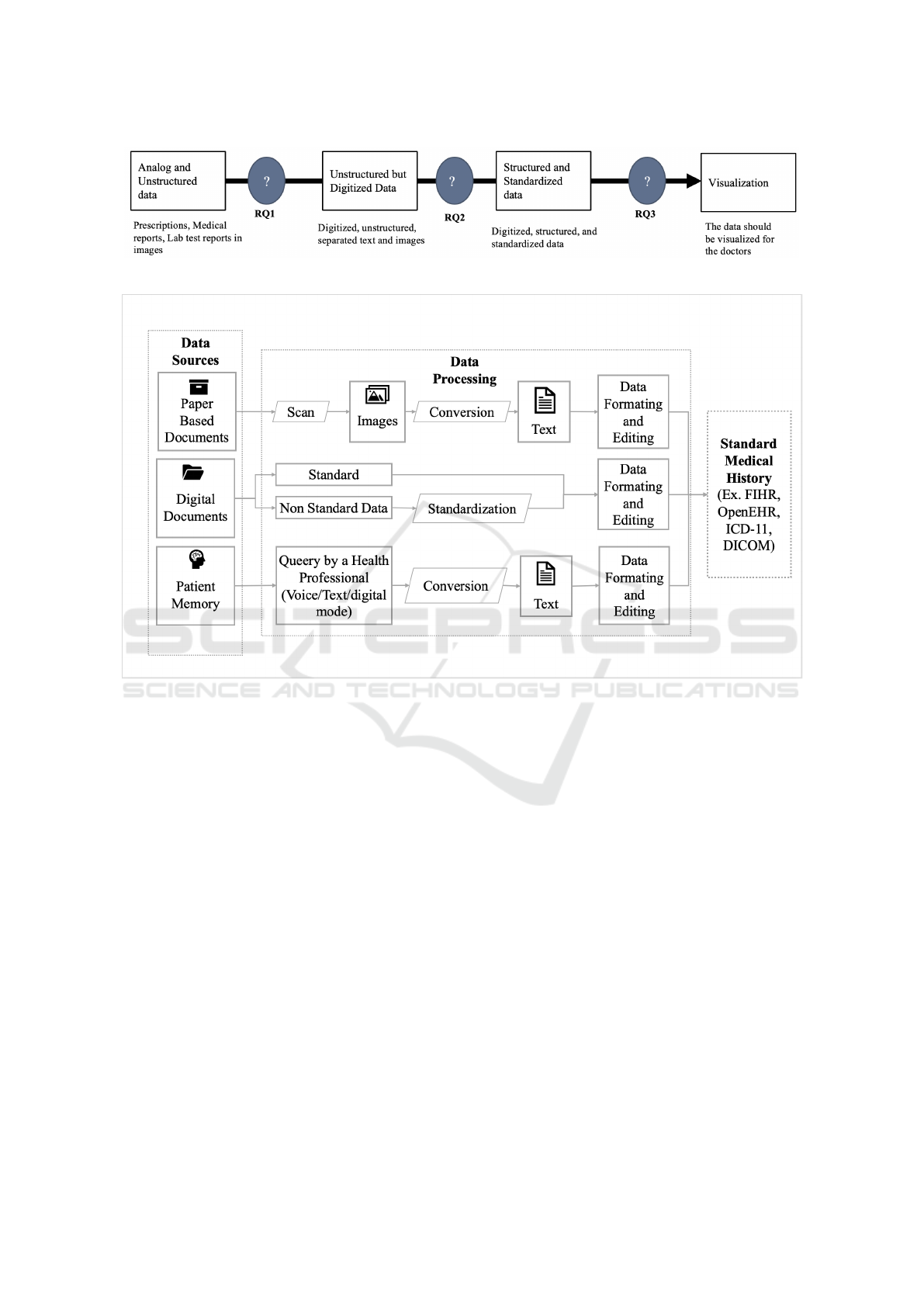
Figure 1: Research Questions.
Figure 2: Data Collection Mechanism for the Smart Health Gantt Chart (Hossain et al., 2022).
2 NEED FOR DIGITIZATION
AND STANDARDIZATION OF
MEDICAL HISTORY
• Digitization: Digitization of medical history can
improve the efficiency and accessibility of health-
care. EMRs can be easily shared between health-
care providers, reducing the need for patients to
repeat medical tests and information. This can
also reduce errors and improve continuity of care.
EMRs can also be accessed remotely, which can
be especially beneficial in rural or underserved
areas. Additionally, digitized medical records
can be analyzed to improve population health
and research(Kaneko et al., 2018). Most medical
records are not digitized. Without digitized medi-
cal records, it is difficult for researchers to predict
future health events.
• Standardization: Different systems and devices
can communicate and share information with ease
thanks to standardization. This can reduce the risk
of errors and improve the continuity of care.
Currently, many hospitals are generating digitized
medical records but they are not shared with indi-
vidual patients. Personal healthcare management re-
quires healthcare records to be managed by individu-
als. This research will enable patients to manage and
control their health records. By enabling better data
management, data sharing, and regulatory compli-
ance, digitizing medical records can raise care qual-
ity, increase patient engagement, and reduce costs.
This research aims to generate standard EHRs from
scanned paper-based documents.
3 RESEARCH MOTIVATION AND
OBJECTIVES
To increase the effectiveness and precision of EHR
systems, this research is going to study and pro-
A Machine Learning Approach to Digitize Medical History and Archive in a Standard Format
231

Figure 3: Proposed System Architecture.
pose a machine learning approach to extract medi-
cal data from scanned documents and standardize it
using FHIR/OpenEHR. By using machine learning
techniques to extract this data and standardize it us-
ing FHIR/OpenEHR, healthcare providers can more
easily access and use this information to make clini-
cal decisions and improve patient care.
3.1 Research Questions
Figure 1 illustrates the research questions that this re-
search will seek to answer in further studies in the
future.
1. RQ1: How to accurately convert Analog and
Unstructured medical data into machine-readable
text?
2. RQ2: How to accurately extract structured data
from unstructured and then standardize it?
3. RQ3: How to accurately integrate the data into a
visualization system?
3.2 Research Objectives
The objective of this research is to develop and eval-
uate a machine learning-based system for extracting
medical data from scanned documents and standard-
izing it using FHIR/OpenEHR.
1. To investigate the current state of medical history
digitization and standardization practices.
2. Develop a comprehensive framework for digitiz-
ing and standardizing medical histories that can
be implemented in a variety of healthcare settings.
3. To evaluate the effectiveness of digitization and
standardization of medical history in improving
the performance of digital healthcare systems.
4. To identify the best practices for digitizing and
standardizing medical history and how they can
be implemented in digital healthcare systems.
5. To investigate the role of FHIR and OpenEHR in
enabling the sharing and interoperability of medi-
cal history across different digital healthcare sys-
tems.
As shown in Figure 2, There will be 3 data collection
mechanisms for the Smart Health Gantt Chart.
• Paper-based medical documents: They contain in-
formation about a patient’s medical history, cur-
rent conditions, treatments, and test results. The
purpose of this research is to study the commonal-
ities and differences of current paper-based medi-
cal documents and propose a system for digitizing
and storing them in a standardised way, so they
can be used for the benefit of patients, doctors,
and researchers.
• Digital Documents: Medical history can be col-
lected as digital data from a variety of sources,
including EHRs, EMRs, and Internet of Things
(IoT) devices. EHRs and EMRs typically include
information such as patient demographics, med-
ical history, lab results, medications, and visit
notes. EHRs are primarily used by hospitals and
large healthcare systems, while EMRs are used by
individual providers, such as doctors and clinics.
IoT devices, such as wearable fitness trackers and
smartwatches, can also collect and transmit med-
ical data, such as heart rate, sleep patterns, and
activity levels. This data can be integrated with
EHRs and EMRs to provide a more complete pic-
ture of a patient’s health (Kodali et al., 2015).
• Patient Memory: This can be a limitation in the
collection of accurate and complete medical his-
tory when relying on digital data. Patients may
not remember all of the relevant information about
their medical history, such as past illnesses, surg-
eries, and medications (Kessels, 2003). Overall,
while digitization of medical history can improve
the efficiency and accessibility of healthcare, it
is important to also consider patient memory and
other limitations in the collection of accurate and
complete medical history. All medical histories of
a patient may not be in paper nor in digital doc-
uments, especially in developing countries, and
studies have shown that age can affect memory
(Rhodes et al., 2019).
Figure 3 demonstrates the architecture of the ap-
proach that will be studied further in the future. This
architecture can be divided into 5 main components:
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
232

1. Analog data: Three different types of data can be
identified (ACCERN, 2022).
• Structured Data: Data that has been organized
in a specific format, such as a database table
or an Excel spreadsheet. This form of data has
a predetermined structure, making it simple to
search, sort, and analyze.
• Unstructured data: Data that does not have a
specific format or structure such as text docu-
ments, images, videos, and audio recordings.
This type of data is more difficult to analyze
and process because it lacks a clear structure.
Our research will be conducted on this type of
data, especially on medical scanned documents
that contain text such as medical notes, reports,
and measurements that can be used to assess a
person’s health. Moreover, computer-generated
text will be our interest. The theories and tools
for data extraction from this type of data will be
investigated.
• Semi-structured data: Type of data that has
some structure, but not as much as structured
data such as XML and JSON files. This type of
data is easier to process than unstructured data,
but not as easy as structured data.
2. Digital Transformation and Data Extraction: The
significance of turning scanned documents into
usable data has increased dramatically as society
is trying to move away from paper and handwrit-
ing in favor of digital documents for convenience.
To suggest a well-designed tool to perform, text
tool conversion from scanned documents, and
data formatting, this study will assess the cur-
rent state of each of these processes. A pipeline
of techniques used to process the scanned med-
ical documents will be proposed. This pipeline
will include OCR approach to convert the scanned
images to machine-readable text followed by an
NLP concept such as NER, Text-classification
(Miro
´
nczuk and Protasiewicz, 2018), and/or in-
formation extraction using pre-trained language
models such as BioBERT (Lee et al., 2019), Clini-
calBERT (Pawar et al., 2022), MedBERT (Rasmy
et al., 2021), etc. This step will produce structured
data.
3. Standardization: The ability for the medical his-
tory documents to be stored in a standard for-
mat will enable them to be shared between EHRs
and EMRs more easily. Thus, this research will
standardize the medical history collected from the
medical scanned documents based on FHIR or
OpenEHR. This step will make the structured data
mapped to FHIR or OpenEHR resources.
4. EHR(Electronic health Records): The EHR sys-
tem will be built based on the FHIR/OpenEHR
standards. The performance of the proposed ar-
chitecture will be evaluated, and different ma-
chine learning techniques will tested in order to
choose the best performing one. This may involve
using metrics such as accuracy, recall, precision
and F1-score to evaluate the performance of the
system, and testing different machine learning al-
gorithms such as Support Vector Machine (Pisner
and Schnyer, 2020), Logistic Regression (LaVal-
ley, 2008), Random Forest (Rigatti, 2017) , Neural
Networks (Bishop, 1994) etc. Additionally, The
proposed architecture will be evaluated using real-
world datasets and different scenarios, which will
help to identify any potential limitations and areas
for improvement.
4 EXISTING RESEARCH AND
THEIR LIMITATIONS
Data extraction from scanned medical documents has
attracted a lot research interest. The goal of this re-
search is to develop automated methods for extract-
ing structured data from unstructured one in scanned
documents and standardizing it which will allow the
data to be more accessible and to be used for research
and healthcare operations. Some common techniques
which are being used include, but not exclusive to:
• Optical character recognition (OCR) to convert
scanned images of text into machine-readable text
(Mithe et al., 2013)
• Natural language processing (NLP) (Chowdhary,
2020) tasks such as Named Entity Recogni-
tion(NER) (Mohit, 2014) to extract structured
data from the text. For instance, patient’s demo-
graphics, medical history, and test results.
• Machine Learning(ML) traditional algorithms to
train models to automatically identify and extract
specific information from the documents, such as
diagnoses, medications, and lab results.
• BERT(Bidirectional Encoder Representation
from Transformers) (Rogers et al., 2020) mod-
els which are pre-trained models that can be
fine-tuned for a wide range of natural language
processing tasks, including NER and can be
used for extracting structured information from
scanned medical documents.
When it comes to extracting structured data from un-
structured data, researchers have been using:
A Machine Learning Approach to Digitize Medical History and Archive in a Standard Format
233
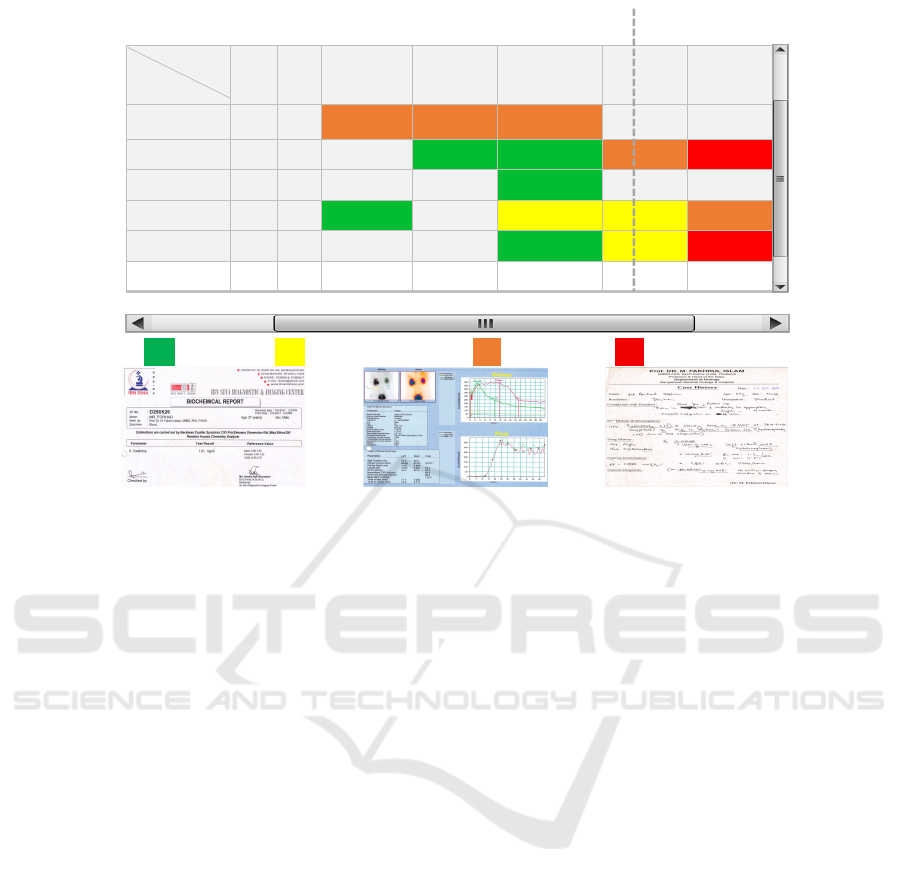
Health
Events
1 2 10 20 30 40 50
Problems
Pain-left
stomach
Pain-left
stomach
Nephrolithiasis-
Left Kidney
Surgery
Urethroplasty
SWL
Medication
xyz
Dr Visit
xyz
Event X
xyz
1987
1988
1997 2007 2017 2027 2037
Cured
Not cured but not so risky
Risky
Emergency
Age
Health Events
Ye a r
Past medical history collected from different sources Prediction
Figure 4: Smart Health Gantt Chart.
• Rule-based methods: Simple yet powerful tech-
niques such as Regular Expressions to extract
structured data from scanned medical documents
(Aggarwal et al., 2018). However, Regular ex-
pressions have some limitations:
– Variability in formatting: Medical documents
can vary widely in terms of formatting and
structure, making it difficult to create a single
regular expression that can match all relevant
information.
– Regular expressions can be difficult to maintain
and update, especially for complex patterns.
• Combination of different and complex techniques
techniques such OCR, NLP, ML, and BERT mod-
els. (Hsu et al., 2022). This combination is
quite effective in extracting structured data from
unstructured data in scanned documents, as each
technology has its own strengths. However, it’s
important to note that the performance of these
technologies will depend on the quality of the data
and models which are used. So, it’s important to
conduct thorough testing and evaluation on a rep-
resentative dataset to measure the performance of
the combination of these technologies before de-
ploying it in a real-world scenario. Moreover, us-
ing all of these technologies can be computation-
ally intensive, which can require a large amount
of computational resources such as memory, CPU
and GPU.
5 APPLICATIONS
There can be a number of potential applications for
exploiting the medical data once it has been extracted
and standardized. Some examples include:
• Smart Health Gantt Chart: Figure 4 represents the
concept of this Gantt Chart. Basically, the doc-
tor will be able to see all of the patient’s medi-
cal history into one window (Hossain and Ahmed,
2021).
• Predictive modeling: Predictive models can be
created using the extracted data to help identify
patients who are at risk of developing specific
health issues, such as chronic diseases.
6 DISCUSSION
Data collection from scanned documents is a crucial
stage in this process, but it can be difficult due to the
variety of formats and layouts that may be encoun-
tered. The studies are limited in application to a spe-
cific clinical domain. To the best of our knowledge, a
generic integration approach for extracting structured
data from scanned documents, modeling EHR data
with the FHIR/OpenEHR data model has not yet been
well studied. For the sake of its effectiveness, an ar-
chitecture that is based on several steps and combine
various techniques such as ML, NLP, OCR, etc was
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
234

proposed, and it will be further investigated through
academic research in the future. This architecture will
enable medical data to be extracted from scanned doc-
uments, standardized using FHIR/OpenEHR and then
stored in EHRs.
7 CONCLUSION
In recent years, there has been an increasing interest
in using the quantity of data found in scanned medical
documents. Healthcare delivery could be revolution-
ized by using scanned medical documents to predict
patient outcomes and utilizing them in this way has
the potential to improve patient outcomes, save doc-
tors’ times, and save costs for the healthcare systems.
Extracting structured data from scanned documents
using technologies such as OCR and NLP, and ML
models to extract useful information standardizing it
in a common format like FHIR or OpenEHR, and us-
ing methods like ML and BERT models to generate
predictions is a relatively new field. As healthcare or-
ganizations look for ways to use the enormous quan-
tity of data included in scanned medical documents to
enhance patient outcomes and reduce costs, this ap-
proach has gained increasing attention in recent years.
However, it is important to note that the process for
collecting, standardizing, and analyzing the data can
be challenging, time-consuming, and expensive. Nev-
ertheless, the advantages of using scanned documents
in this way make it a worthwhile endeavor for health-
care researchers.
REFERENCES
ACCERN (2022). Differences between
structured, unstructured, and semi-
structured data. https://accern.com/blog/
structured-vs-semi-structured-vs-unstructured-data/.
Aggarwal, A., Garhwal, S., and Kumar, A. (2018). Hedea:
a python tool for extracting and analysing semi-
structured information from medical records. Health-
care informatics research, 24(2):148–153.
Ahmed, A., Rebeiro-Hargrave, A., Nohara, Y., Kai, E.,
Ripon, Z. H., and Nakashima, N. (2014). Targeting
morbidity in unreached communities using portable
health clinic system. IEICE Transactions on Commu-
nications, E97.B(3):540–545.
Bishop, C. M. (1994). Neural networks and their applica-
tions. Review of scientific instruments, 65(6):1803–
1832.
Chowdhary, K. R. (2020). Natural Language Processing,
pages 603–649. Springer India, New Delhi.
Hossain, F. and Ahmed, A. (2021). Visualization of health-
care data for busy doctors in developing countries to
make efficient clinical decisions. In 10th Social Busi-
ness Academia Conference.
Hossain, F., Islam, R., Ahmed, M. T., and Ahmed, A.
(2022). Technical requirements to design a personal
medical history visualization tool for doctors. In Pro-
ceedings of the 8th International Conference on Hu-
man Interaction and Emerging Technologies. IHIET,
https://ihiet. org.
Hsu, E., Malagaris, I., Kuo, Y.-F., Sultana, R., and Roberts,
K. (2022). Deep learning-based nlp data pipeline for
ehr-scanned document information extraction. JAMIA
open, 5(2):ooac045.
Kaneko, K., Onozuka, D., Shibuta, H., and Hagihara, A.
(2018). Impact of electronic medical records (emrs)
on hospital productivity in japan. International jour-
nal of medical informatics, 118:36–43.
Kessels, R. P. (2003). Patients’ memory for medical in-
formation. Journal of the Royal Society of Medicine,
96(5):219–222.
Kodali, R. K., Swamy, G., and Lakshmi, B. (2015). An im-
plementation of iot for healthcare. In 2015 IEEE Re-
cent Advances in Intelligent Computational Systems
(RAICS), pages 411–416. IEEE.
LaValley, M. P. (2008). Logistic regression. Circulation,
117(18):2395–2399.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2019). BioBERT: a pre-trained biomed-
ical language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Miro
´
nczuk, M. M. and Protasiewicz, J. (2018). A recent
overview of the state-of-the-art elements of text clas-
sification. Expert Systems with Applications, 106:36–
54.
Mithe, R., Indalkar, S., and Divekar, N. (2013). Optical
character recognition. International journal of recent
technology and engineering (IJRTE), 2(1):72–75.
Mohit, B. (2014). Named Entity Recognition, pages 221–
245. Springer Berlin Heidelberg, Berlin, Heidelberg.
Pawar, Y., Henriksson, A., Hedberg, P., and Naucler, P.
(2022). Leveraging clinical bert in multimodal mortal-
ity prediction models for covid-19. In 2022 IEEE 35th
International Symposium on Computer-Based Medi-
cal Systems (CBMS), pages 199–204. IEEE.
Pisner, D. A. and Schnyer, D. M. (2020). Support vector
machine. In Machine learning, pages 101–121. Else-
vier.
Rasmy, L., Xiang, Y., Xie, Z., Tao, C., and Zhi, D. (2021).
Med-bert: pretrained contextualized embeddings on
large-scale structured electronic health records for dis-
ease prediction. NPJ digital medicine, 4(1):1–13.
Rhodes, S., Greene, N. R., and Naveh-Benjamin, M.
(2019). Age-related differences in recall and recog-
nition: A meta-analysis. Psychonomic Bulletin & Re-
view, 26(5):1529–1547.
Rigatti, S. J. (2017). Random forest. Journal of Insurance
Medicine, 47(1):31–39.
Rogers, A., Kovaleva, O., and Rumshisky, A. (2020). A
primer in bertology: What we know about how bert
works. Transactions of the Association for Computa-
tional Linguistics, 8:842–866.
A Machine Learning Approach to Digitize Medical History and Archive in a Standard Format
235

Saripalle, R., Runyan, C., and Russell, M. (2019). Using hl7
fhir to achieve interoperability in patient health record.
Journal of Biomedical Informatics, 94:103188.
Tolera, A., Oljira, L., Dingeta, T., Abera, A., and Roba,
H. S. (2022). Electronic medical record use and asso-
ciated factors among healthcare professionals at pub-
lic health facilities in dire dawa, eastern ethiopia: A
mixed-method study. Frontiers in Digital Health, 4.
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
236
