
QFLS: A Cloud-Based Framework for Supporting Big Healthcare Data
Management and Analytics from Big Data Lakes: Definitions,
Requirements, Models and Techniques
Alfredo Cuzzocrea
1,2 a
and Selim Soufargi
1 b
1
iDEA Lab, University of Calabria, Rende, Italy
2
Department of Computer Science, University of Paris City, Paris, France
Keywords:
Big Healthcare Data, Big Data Management, Big Data Analytics, Cloud-Based Frameworks.
Abstract:
This paper introduces definitions, requirements, models and techniques of QUALITOP Federated Big Data
Analytics Learning System (QFLS), a Cloud-based framework for supporting big healthcare data management
and analytics from big data lakes. QFLS anatomy and main functionalities are described, along with the main
software solutions proposed with the framework.
1 INTRODUCTION
Nowadays, big data management and analytics play
a disruptive role in modern database research (e.g.,
(Chen et al., 2013; Zhang et al., 2015; Chaudhuri,
2012; Cuzzocrea et al., 2014; Campan et al., 2017;
Balbin et al., 2020)), with a special touch on both
theory and systems. When applied to healthcare do-
mains (e.g., (Dash et al., 2019; Sun and Reddy, 2013;
Patil and Seshadri, 2014)), relevant challenges arise,
starting from big data representation to big data in-
dexing, from big data understanding to big data an-
alytics, and so forth. These open issues have been
recently highlighted by authoritative proposals in the
scientific field (e.g., (Ghayvat et al., 2022; Ding et al.,
2022; Parimanam et al., 2022)).
Indeed, healthcare domains dictate special re-
quirements, among that the need for supporting ad-
vanced analytics while ensuring the privacy of in-
put big healthcare datasets is a first-class point (e.g.,
(Onesimu et al., 2022; Abbasi and Mohammadi,
2022; Singh et al., 2022)). This is one of the main
goal of the EU H2020 project QUALITOP (QUALI-
TOP, 2023). QUALITOP aims at devising a collec-
tion of models, techniques and algorithms for sup-
porting prediction and recommendation for the qual-
ity of life of cancer patients treated with the modern
immunotherapy treatment (e.g., (Vinke et al., 2023;
Beaulieu et al., 2022)).
a
https://orcid.org/0000-0002-7104-6415
b
https://orcid.org/0009-0000-5476-9403
One of the most distinctive characteristics of
QUALITOP is represented by the so-called QUALI-
TOP big data lake, a big data lake (e.g., (Cuzzocrea,
2021)) oriented to collect anonymized data/informa-
tion from a reference data federation built at the med-
ical premises, and to support big data analytics and
big data predictive analytics over such datasets. This,
with the final goal of supporting analysis and recom-
mendations of immunotherapy-treated cancer patients
during their post-trauma life. The big data lake re-
lies within the general QUALITOP software platform
(e.g., (Elgammal and Kr
¨
amer, 2021)).
In order to fulfill the requirements super-imposed
by such scenarios, we propose the QUALITOP Feder-
ated Big Data Analytics Learning System (QFLS), a
Cloud-based federated framework for supporting big
healthcare data management and analytics from big
data lakes. Among various results, QFLS introduces
the so-called Tree-Like Analytical Query (TLAQ)
model, a powerful analytical model for healthcare
analytics. Given a TLAQ Q, every node n ∈ Q is
equipped by query node Q
n
, modeled as follows:
Q
n
= ⟨A
n
, Σ
A
n
, P
n
⟩, such that: (i) A
n
is the target query
attribute; (ii) Σ
A
n
is a constraint over A
n
; (iii) P
n
is an
aggregate operator over A
n
. Another relevant charac-
teristic of the TLQA model is that, given two query
nodes Q
n
and Q
m
, such that Q
n
≺ Q
m
, there not ex-
ists a strict hierarchical relation between Q
n
and Q
m
,
meaning that A
m
̸⊂ A
n
. This special feature is particu-
larly suitable to support precision medicine processes.
When executed, TLAQ generate a tree-like analytical
422
Cuzzocrea, A. and Soufargi, S.
QFLS: A Cloud-Based Framework for Supporting Big Healthcare Data Management and Analytics from Big Data Lakes: Definitions, Requirements, Models and Techniques.
DOI: 10.5220/0012092700003541
In Proceedings of the 12th International Conference on Data Science, Technology and Applications (DATA 2023), pages 422-428
ISBN: 978-989-758-664-4; ISSN: 2184-285X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

data structure that indexes anonymized datasets, made
available for several privacy-preserving big data ana-
lytics purposes (e.g., (Lin et al., 2016)).
In more details, QFLS is a data federation solu-
tion that is based on the big data processing frame-
work Hadoop cluster, to which all queries are dis-
patched, In addition, specialized Apache-Spark-based
servers that run remotely within each of the data fed-
eration sources are introduced. QFLS is developed
and deployed on top of the iDEA Lab Cloud, located
at University of Calabria, Rende, Italy, which is com-
posed of 21 VMs equipped with Windows Server Es-
sentials 2019 OS, each having 32GB of memory, 8-
Core CPUs, 32GB RAM and 60GB HDD.
It goes without saying that it is of best interest to
run Spark jobs on the Hadoop cluster in a MapRe-
duce manner, especially for larger datasets, and to
be able to do that, fine tuning Spark job submission
onto the cluster was necessary. Using full potential
of the Hadoop cluster is a key to bring effectiveness
and efficiency during query executions. Specifically,
the server runs on Hadoop (through YARN resource
manager) with fine-tuned worker instances, worker
memory and CPU, and driver memory and CPU (e.g.,
(Chen et al., 2017; Gounaris and Torres, 2018)).
The latter sub-system represents the QFLS Core
component of QFLS, which is the Cloud-based en-
gine that runs everything. QFLS encompasses more
two components, namely QFLS-ADPT and QFLS-
ADAT. The QUALITOP Anonymized Dataset Popula-
tion Tool (QFLS-ADPT) is a web-based tool for man-
aging the anonymized dataset at the federation node.
The QUALITOP Anonymized Dataset Analytics Tool
(QFLS-ADAT) is a web-based tool for supporting big
healthcare analytics and predictive analytics over the
anonymized datasets stored in the federation, via the
TLAQs.
2 QFLS ANATOMY
Figure 1 shows the main blueprint of the big data pro-
cessing flow supported by QFLS. Here, we introduce
an example scenario where the following two nodes
occur: one QFLS federated node, located in France,
and one QFLS Core node, located in Italy. There-
fore, the French node provides (anonymized) health-
care data, and the Italian node support big data ana-
lytics and predictive analytics over these data.
As shown in Figure 1, at the French node (1),
where the target healtcare dataset D is located, med-
ical operators generate an anonymized version of
D, denoted by D
′
(3), according to specific med-
ical guidelines (2), yet compliant with the GDPR.
Then, the analytics tools located at the Italian node
(4) execute aggregate queries, defined by the input
TLAQ (8), which, in turn, is driven by target big
data analytics tools defined within the QUALITOP
big data lake, over the remote French node, via feder-
ated query algorithms based on Apache Spark, and
the anonymized representation of the result is so-
obtained (5). The final TLAQ analytics answer is
shaped as a tree (9) such that each node indexed a
proper anonymized healthcare dataset, empowered by
the Cloud computing potentialities (e.g., distribution,
indexing, load balancing, mirroring, and so forth).
The latter data structure is accessed by medical de-
cision makers (10) who provide the final big data an-
alytics and predictive analytics (11).
It should be noted some relevant characteristics of
QFLS, which we summarize as follows:
1. Data Federation. While QFLS Core is entirely
Cloud-based, the main processing of QFLS hap-
pens over data federation nodes, according to
severe data privacy protection guidelines. This
scenario fully converges with real-life systems,
where data federation participants are interested
in participating to the federation (for data analyt-
ics purposes) but are not willing to unveil their
data. The latter is the common case of medical
centers.
2. Privacy-Preserving Advanced Analytical Mod-
els. QFLS encompasses the TLAQ analytical
model, a powerful privacy-preserving analyti-
cal model particularly target to support preci-
sion medicine processes that, by definition, are
built upon lazy aggregations. For instance, at
first, physicians may be interested in analyzing
COVID-19 data about female patients who live in
Canada and whose age in between the range [25-
50], then, within this range, they may want to an-
alyze COVID-19 data about female people of age
30 who are resident in Toronto ans have also been
diagnosed with Tuberculosis.
3. Big Data Analytics & Big Data Predictive Ana-
lytics. QFLS not only supports big data analytics,
mostly driven by the TLAQ model, but also the
so-called big data predictive analytics. Indeed, re-
trieved TLAQ analytics can be combined in a mul-
tidimensional fashion, and so-derived summary
data can be used to develop fortunate big multidi-
mensional analytics metaphors (e.g., (Cuzzocrea
et al., 2004)).
4. Flexible Big Data Processing Tools Integration.
QFLS fully relies on the smoothly integration of
several big data processing tools, mostly within
the eco-system defined by Hadoop, such Apache
QFLS: A Cloud-Based Framework for Supporting Big Healthcare Data Management and Analytics from Big Data Lakes: Definitions,
Requirements, Models and Techniques
423

Figure 1: QFLS Main Blueprint.
Spark, YARN, Hive, MongoDB, etc. This en-
sures high data-availability, high scalability and,
moreover, a full tendency to be further integrated
within more complex big data stack architectures.
3 RELATED WORK
The problem of supporting big healthcare data man-
agement and analytics has been of great interest re-
cently, even due to the COVID-19 outbreak (e.g., (Liu
et al., 2023b; Dimitsaki et al., 2023)). In this Section,
we provide a brief overview of most relevant propos-
als in this scientific area.
(Ghayvat et al., 2022) focuses on Internet of
Things (IoT)-based Healthcare services, which are
becoming more widespread today, continuously gen-
erate huge amounts of big data. Due to the data mag-
nitude, data intricacy, privacy preservation, data in-
tegrity and identity verification requirements, indeed
novel research challenges and issues in healthcare big
data service management arise. To overcome these
problems, author propose a scalable computing sys-
tem that provides verifiable data access mechanism
for IoT-enabled health data analytics in the big data
ecosystem. There are two primary sub-architectures
in the proposed architecture, namely a big data an-
alytics tracking system and a derived blockchain-
based data storage/access system. This approach
leverages big data systems and blockchain architec-
ture to analyze, and securely store data from IoT-
enabled devices and allow verified access to the stored
data. The zero-knowledge protocol is used to ensure
that no information is accessible to unauthenticated
users alongside avoiding data linkability. The results
demonstrate the effectiveness of the proposed method
to solve the problems of big data analytics and privacy
issues in healthcare.
(Ding et al., 2022) proposes two differentially
private algorithms, i.e., Output Perturbation with
aGM (OPERA) and Gradient Perturbation with aGM
(GRPUA) for empirical risk minimization, a useful
method to obtain a globally optimal classifier, by
leveraging the analytic Gaussian mechanism (aGM)
to achieve privacy preservation of sensitive medical
data in a healthcare system. Authors theoretically an-
alyze and prove utility upper bounds of proposed al-
gorithms and compare them with prior algorithms in
the literature. The analyses show that in the high pri-
vacy regime, the proposed algorithms can achieve a
tighter utility bound for both settings: strongly convex
and non-strongly convex loss functions. Besides, the
proposed private algorithms are evaluated against five
benchmark datasets. The simulation results demon-
strate that these approaches can achieve higher accu-
racy and lower objective values compared with exist-
ing ones in all three datasets while providing differ-
ential privacy guarantees.
(Parimanam et al., 2022) notices that, current
health information systems, when coupled big data
trends, fail to maintain a highly organized analysis
and processing of health data for analytics purposes.
In addition, it collects issues that play an impor-
tant role in determining quality. In this article, au-
thors propose a Hybrid Optimization based Learn-
ing technique for Multi-Disease analytics (HOL-
MD) from healthcare big data using optimal pre-
processing, clustering, and classifier. First, authors
introduce a capuchin search based optimization al-
gorithm for pre-processing which removes the un-
wanted artifacts to enhance the detection accuracy.
Second, a modified Harris Hawks Optimization based
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
424
Clustering (MHHOC) technique is introduced used
to select optimal features among multiple features
which discovers the subgroups and reduce the dimen-
sionality issues. Performance of proposed HOL-MD
technique is evaluated using standard US healthcare-
organization SUSY and HIGGS datasets, and it turns
that existing state-of-art techniques are outperformed
in terms of accuracy, precision, recall and F-measure,
respectively.
4 QFLS: REQUIREMENTS AND
MAIN FUNCTIONALITIES
This Section highlights, mostly from a software-
engineering and component-architecture point-of-
view, the requirements and the main functionalities
of QFLS.
4.1 Data Ingestion and Storage: The
QADPT Component
In order to enable data ingestion into QFLS, feder-
ated nodes are enabled with a client web component
that ensures data can be referred by QFLS, and can be
processed for later analytics, according to FAIR prin-
ciples (Findable, Accessible, Interoperable and Re-
peatable).
In order to ingest the (anonymized) data, current
QADPT provides an interface to select data from lo-
cal file system as CSV files. The data are then stored
into Hive table and QADPT notifies QFLS of the
ingested data through updating the data setting of
QFLS federated nodes. After updating the QFLS sys-
tem configuration, users are allowed to see the added
datasets in the respective node and use them for sub-
mitting their TLAQ (federated query answering). To
note that, at ingestion time, data are cleansed and
transformed to meet QFLS standards and constraints
(date format, column values encoding issues, and so
forth). On the other hand, it should be noted that the
QFLS-based anonymization mechanism ensures that
users cannot never access the original data, but only
privacy-preserving summarized versions of them.
QADPT is also capable or removing a dataset
from QFLS federated node or also list existing ones in
a intuitive browsable interface. QADPT implements
a role-based access control, and allows authentication
of medical operator users solely.
QADPT is a client web component implemented
using Spring v. 5.3 and deployed over a Tomcat v.
8.5 instance. Main functionalities of QADPT is to
load ingested data into Hive. The insertion into Hive
is performed through a JDBC client which takes care
of loading the dataset (then, a CSV file) into a cor-
responding table in Hive, it also, as mentioned, up-
sert the associated (dataset) information into a HDFS-
based configuration system file (.ini) depending on
whether the node is previously existent in QFLS (up-
date) or the node is newly created (insert).
4.2 Data Analytics: The QADAT
Component
In order to enable data scientists and medical oper-
ators with insightful recommendation on the health-
care data, QFLS provides QADAT, a client web com-
ponent that offers numerous tools to be used, all in
a matter of a deep analysis of data and to eventually
provide recommendations, or, better, predictive ana-
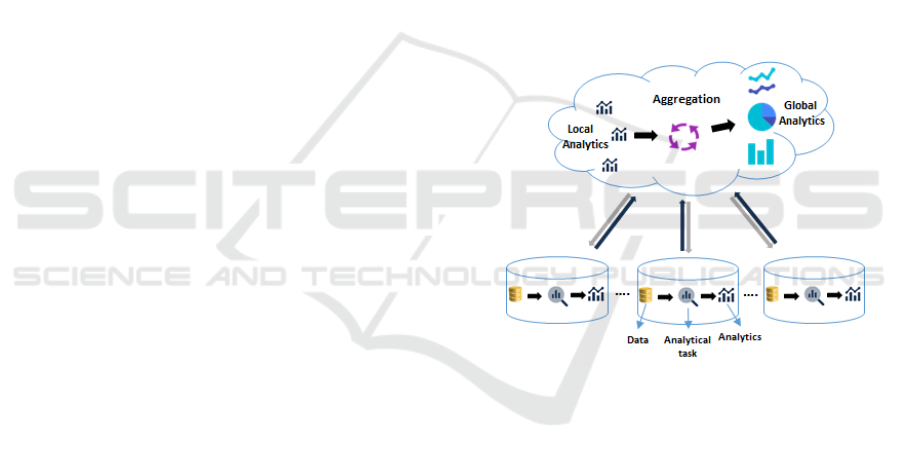
lytics, upon these global analysis. Figure 2 shows the
main workflow for predictive analytics supported by
QFLS.
Figure 2: QFLS Workflow for Predictive Analytics.
In more details, Figure 2 shows the interaction be-
tween the Cloud-based QFLS framework QFLS Core
and the federated nodes. At each federated node, data
are processed as partial analytical tasks, via the TLAQ
queries, and (partial) results are sent back to the Cloud
for aggregation with other partial results similarly ob-
tained from other nodes. The partial results are then
processed and structured as needed and global analyt-
ics are then derived. QFLS produces from the TLAQ-
shaped global analytics a visual dashboard consisting
of predictive analytics tools. These can be in many
forms: clustering, ROC areas, confusion matrices,
and so forth. These predictive analytics tools serve
for recommendation purposes by the competent med-
ical operators for the patients. It is also worth notic-
ing that the aforementioned QFLS system set-up pre-
serves data locality which would increase anonymity
and produce more tailored analytics. All of the data
QFLS: A Cloud-Based Framework for Supporting Big Healthcare Data Management and Analytics from Big Data Lakes: Definitions,
Requirements, Models and Techniques
425
processing logic is executed on the QFLS Core, in a
MapReduce fashion.
QADAT is a client web component implemented
using Spring v. 5.3 from which users can analyze
the data stored in all linked federated nodes, in an
anonymized manner. Like for the case of QADPT,
the choice of Spring was made because it is compliant
with the employed JDK version 8, and, with our set
of technological solutions such as those used for the
server (e.g., Spark) and for deployment (e.g., Tom-
cat), that was the most conforming solution among
all the available ones. JWT authentication (Spring se-
curity) along with role based access were also part
of our Spring application. Indeed, data analyst role
has full components access whereas medical opera-
tor modules access is restricted. For instance, a med-
ical operator user cannot use some SQL-based mod-
ules. This general idea could be further extended by
adopting adaptive metaphors developed in the context
of web information systems (e.g., (Cannataro et al.,
2001; Cannataro et al., 2002)).
In order to test our Spring application, we used
JUnit v. 5 to perform unit tests over the developed
code base. In addition, deployment of each of the
client applications was achieved thanks to Tomcat ap-
plication server, like for QADPT.
Finally, given the need to store information about
the user (such as their credentials) and other infor-
mation related to the implemented functionalities, a
MongoDB database was installed and used for the
purpose.
4.2.1 A Cloud-Based, Client-Server Solution
In order to reach the data at each of the federated
nodes, QADAT employs Java RMI technology to
transport result data (i.e., aggregations) from one ma-
chine to another. In addition, and since data could
potentially be large, conveying them over the net-
work requires the usage of specialized libraries such
as RMIIO to enable large data to be transported over
the network in a convenient way. More specifically,
a Spark running instance server receives the remote
method invocation request from the client through
an RMI call, processes the dataset accordingly (in
a MapReduce fashion) over the Hadoop cluster, and
then returns the results to the client. The results are
then gathered, structured and displayed to users.
4.3 Main Components and
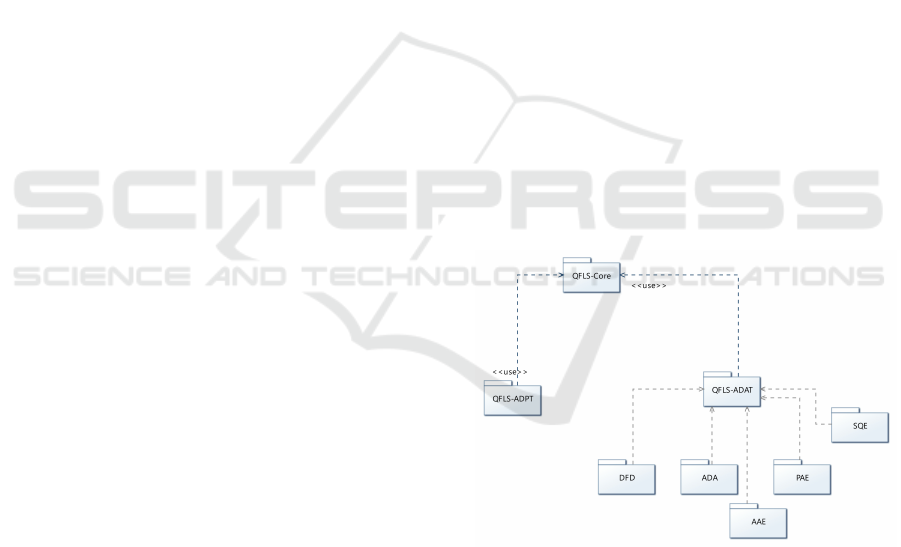
Functionalities
QFLS is intended to satisfy basic analytical needs by
performing analytical computations over diverse data
sources in a federated context. First, QFLS enables
data exploration throughout its Data Federation dis-
covery (DFD). DFD exposes, based on the configura-
tion file, the content of each of the federated node in
terms of datasets. Second, the Anonymized Dataset
Analysis (ADA) component provides an analysis of
each of the target anonymized dataset, by providing
details on its level of anonymization of the attributes,
and other statistics such as the minimum value, the
maximum value, the type. Third, the Anonymized
Analytics Environment (AAE) allows users to sub-
mit TLAQ queries onto the remote target datasets and
to retrieve analytics in a tree-like shaped analytical
structure through. QFLS is also capable of deriv-
ing a dashboard over the query answers in order to
enable recommendations based on the obtained ana-
lytics through the Predictive Analytical Environment
(PAE). Furthermore, QFLS enables SQL queries to
be submitted and executed on a target dataset, and
to retrieve the corresponding result set in an intu-
itive interface, easily navigable by non-medical per-
sons, through the SQL Query Environment (SQE).
Each submitted tree-like analytical query is stored in
a database. All of the aforementioned modules use
Java RMI to request and retrieve results from the re-
mote Spark-instance running server.
Figure 3 shows the UML package diagram of
QFLS, as to highlight the inter-dependencies among
the various components.
Figure 3: UML Package Diagram of QFLS.
Since, within the scope of the QUALITOP project,
medical data at the federated nodes cannot be ac-
cessed, all of the data processing logic occurs on
server side (on each federated node) and only ag-
gregate information is returned back to the QADAT
component. This ensures the necessary level of
anonymization, as per the requirements of the QUAL-
ITOP project.
To better understand the data access constraint, we
adopt the strategy of retrieving distinct values within
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
426

dataset columns, which are necessary at various sub-
components of QADAT (e.g., AAE component). In
this case, Spark processing yields only distinct val-
ues through adequately processing the anonymized
dataset on the corresponding node, while data are
never returned raw, nor are fully-processed at the
client side. Indeed, even though data are anonymized
on each federated node, data access may still expose
certain sensitive information such as patient names or
their diseases (e.g., (Sweeney, 2002)). In addition to
this measure, identifier columns are removed at the
time of processing the dataset at the federated node,
as to reinforce anonymity of data and to provide op-
timal security guarantees over the privacy of patients
and data owners.
Finally, QFLS is obviously extendable to a fed-
erated machine learning system (e.g., (Yang et al.,
2019; Fu et al., 2022)), where the processing at each
node level would be based on a machine learning
model rather than a data aggregation model, thus re-
sulting in a global machine model. Under this as-
sumption, SparkML would be our best bet to employ
for embedding further capabilities to our framework
(e.g., (Omran et al., 2021; Mohamed et al., 2021)).
5 CONCLUSIONS AND FUTURE
WORK
This paper has described in details definitions, re-
quirements, models and techniques of QFLS, a
Cloud-based framework for supporting big health-
care data management and analytics from big data
lakes. QFLS anatomy and main functionalities have
been described, along with the main software solu-
tions proposed with the framework.
Concepts and guidelines deriving from the pro-
posed framework also opens the door to emerging
research challenges for the future. Among those,
an interesting research line consists in extending our
framework as to deal with advanced machine learn-
ing techniques, such as those experimented in other
research efforts (e.g., (Cuzzocrea et al., 2017; Leung
et al., 2019; Coronato and Cuzzocrea, 2022; Liu et al.,
2023a; Adeoye et al., 2023; Tan et al., 2023)).
ACKNOWLEDGEMENTS
This research has been funded by the EU H2020
QUALITOP research project - Call Reference: H2020
- SC1-DTH-01-2019; Project Number: 875171.
REFERENCES
Abbasi, A. and Mohammadi, B. (2022). A clustering-
based anonymization approach for privacy-preserving
in the healthcare cloud. Concurr. Comput. Pract. Exp.,
34(1).
Adeoye, J., Koohi-Moghadam, M., Choi, S., Zheng, L., Lo,
A. W. I., Tsang, R. K., Chow, V. L. Y., Akinshipo,
A., Thomson, P., and Su, Y. (2023). Predicting oral
cancer risk in patients with oral leukoplakia and oral
lichenoid mucositis using machine learning. J. Big
Data, 10(1):39.
Balbin, P. P. F., Barker, J. C. R., Leung, C. K., Tran,
M., Wall, R. P., and Cuzzocrea, A. (2020). Predic-
tive analytics on open big data for supporting smart
transportation services. Procedia Computer Science,
176:3009–3018.
Beaulieu, E., Spanjaart, A., Roes, A., Rachet, B., Dalle, S.,
Kersten, M. J., Maucort-Boulch, D., and Jalali, M. S.
(2022). Health-related quality of life in cancer im-
munotherapy: a systematic perspective, using causal
loop diagrams. Qual Life Res., 31(8).
Campan, A., Cuzzocrea, A., and Truta, T. M. (2017). Fight-
ing fake news spread in online social networks: Actual
trends and future research directions. In IEEE Big-
Data, 2017, pages 4453–4457. IEEE Computer Soci-
ety.
Cannataro, M., Cuzzocrea, A., and Pugliese, A. (2001). A
probabilistic approach to model adaptive hypermedia
systems. In Proceedings of the First International
Workshop on Web Dynamics, WebDyn@ICDT 2001,
London, UK, January 3, 2001, pages 50–60.
Cannataro, M., Cuzzocrea, A., and Pugliese, A. (2002).
XAHM: an adaptive hypermedia model based on
XML. In Proceedings of the 14th international con-
ference on Software engineering and knowledge engi-
neering, SEKE 2002, Ischia, Italy, July 15-19, 2002,
pages 627–634.
Chaudhuri, S. (2012). What next?: a half-dozen data man-
agement research goals for big data and the cloud. In
Benedikt, M., Kr
¨
otzsch, M., and Lenzerini, M., edi-
tors, ACM SIGMOD-SIGACT-SIGART PODS, 2012,
pages 1–4. ACM.
Chen, D., Chen, H., Jiang, Z., and Zhao, Y. (2017). An
adaptive memory tuning strategy with high perfor-
mance for spark. Int. J. Big Data Intell., 4(4):276–
286.
Chen, J., Chen, Y., Du, X., Li, C., Lu, J., Zhao, S., and
Zhou, X. (2013). Big data challenge: a data manage-
ment perspective. Frontiers Comput. Sci., 7(2):157–
164.
Coronato, A. and Cuzzocrea, A. (2022). An innovative risk
assessment methodology for medical information sys-
tems. IEEE Trans. Knowl. Data Eng., 34(7):3095–
3110.
Cuzzocrea, A. (2021). Big data lakes: Models, frameworks,
and techniques. In IEEE BigComp, 2021, pages 1–4.
IEEE.
Cuzzocrea, A., Furfaro, F., Mazzeo, G. M., and Sacc
`
a, D.
(2004). A grid framework for approximate aggregate
QFLS: A Cloud-Based Framework for Supporting Big Healthcare Data Management and Analytics from Big Data Lakes: Definitions,
Requirements, Models and Techniques
427

query answering on summarized sensor network read-
ings. In OTM 2004 Workshops, 2004, volume 3292,
pages 144–153.
Cuzzocrea, A., Leung, C. K., and MacKinnon, R. K. (2014).
Mining constrained frequent itemsets from distributed
uncertain data. Future Gener. Comput. Syst., 37:117–
126.
Cuzzocrea, A., Martinelli, F., Mercaldo, F., and Vercelli,
G. V. (2017). Tor traffic analysis and detection via
machine learning techniques. In IEEE BigData, 2017,
pages 4474–4480. IEEE Computer Society.
Dash, S., Shakyawar, S. K., Sharma, M., and Kaushik, S.
(2019). Big data in healthcare: management, analysis
and future prospects. J. Big Data, 6:54.
Dimitsaki, S., Gavriilidis, G. I., Dimitriadis, V. K., and Nat-
siavas, P. (2023). Benchmarking of machine learning
classifiers on plasma proteomic for COVID-19 sever-
ity prediction through interpretable artificial intelli-
gence. Artif. Intell. Medicine, 137:102490.
Ding, J., Errapotu, S. M., Guo, Y., Zhang, H., Yuan, D.,
and Pan, M. (2022). Private empirical risk minimiza-
tion with analytic gaussian mechanism for healthcare
system. IEEE Trans. Big Data, 8(4):1107–1117.
Elgammal, A. and Kr
¨
amer, B. J. (2021). A reference ar-
chitecture for smart digital platform for personalized
prevention and patient management. In Next-Gen
Digital Services. A Retrospective and Roadmap for
Service Computing of the Future - Essays Dedicated
to Michael Papazoglou on the Occasion of His 65th
Birthday and His Retirement, volume 12521, pages
88–99.
Fu, X., Zhang, B., Dong, Y., Chen, C., and Li, J. (2022).
Federated graph machine learning: A survey of con-
cepts, techniques, and applications. SIGKDD Explor.,
24(2):32–47.
Ghayvat, H., Pandya, S., Bhattacharya, P., Zuhair, M.,
Rashid, M., Hakak, S., and Dev, K. (2022). CP-
BDHCA: blockchain-based confidentiality-privacy
preserving big data scheme for healthcare clouds and
applications. IEEE J. Biomed. Health Informatics,
26(5):1937–1948.
Gounaris, A. and Torres, J. (2018). A methodology for
spark parameter tuning. Big Data Res., 11:22–32.
Leung, C. K., Cuzzocrea, A., Mai, J. J., Deng, D., and
Jiang, F. (2019). Personalized deepinf: Enhanced so-
cial influence prediction with deep learning and trans-
fer learning. In IEEE BigData, 2019, pages 2871–
2880. IEEE.
Lin, C., Song, Z., Song, H., Zhou, Y., Wang, Y., and
Wu, G. (2016). Differential privacy preserving in big
data analytics for connected health. J. Medical Syst.,
40(4):97:1–97:9.
Liu, C., Yao, Z., Liu, P., Tu, Y., Chen, H., Cheng, H., Xie,
L., and Xiao, K. (2023a). Early prediction of MODS
interventions in the intensive care unit using machine
learning. J. Big Data, 10(1):55.
Liu, X., Hasan, M. R., Ahmed, K. A., and Hossain, M. Z.
(2023b). Machine learning to analyse omic-data for
COVID-19 diagnosis and prognosis. BMC Bioinform.,
24(1):7.
Mohamed, M. A., El-Henawy, I. M., and Salah, A.
(2021). Usages of spark framework with different ma-
chine learning algorithms. Comput. Intell. Neurosci.,
2021:1896953:1–1896953:7.
Omran, N. F., Ghany, S. F. A., Saleh, H., and Nabil, A.
(2021). Breast cancer identification from patients’
tweet streaming using machine learning solution on
spark. Complex., 2021:6653508:1–6653508:12.
Onesimu, J. A., Karthikeyan, J., Eunice, J., Pomplun, M.,
and Dang, H. (2022). Privacy preserving attribute-
focused anonymization scheme for healthcare data
publishing. IEEE Access, 10:86979–86997.
Parimanam, K., Lakshmanan, L., and Palaniswamy, T.
(2022). Hybrid optimization based learning technique
for multi-disease analytics from healthcare big data
using optimal pre-processing, clustering and classifier.
Concurr. Comput. Pract. Exp., 34(17).
Patil, H. K. and Seshadri, R. (2014). Big data security and
privacy issues in healthcare. In IEEE Congress on Big
Data, 2014, pages 762–765. IEEE Computer Society.
QUALITOP (2023). The QUALITOP project.
https://h2020qualitop.liris.cnrs.fr/wordpress/index.
php/project/.
Singh, S., Rathore, S., Alfarraj, O., Tolba, A., and Yoon,
B. (2022). A framework for privacy-preservation
of iot healthcare data using federated learning and
blockchain technology. Future Gener. Comput. Syst.,
129:380–388.
Sun, J. and Reddy, C. K. (2013). Big data analytics for
healthcare. In ACM SIGKDD KDD, 2013, page 1525.
ACM.
Sweeney, L. (2002). k-anonymity: A model for protecting
privacy. Int. J. Uncertain. Fuzziness Knowl. Based
Syst., 10(5):557–570.
Tan, Q., Xu, X., and Liang, H. (2023). Physiological big
data mining through machine learning and wireless
sensor networks. Int. J. Distributed Syst. Technol.,
14(2):1–12.
Vinke, P. C., Combalia, M., de Bock, G. H., Leyrat, C.,
Spanjaart, A. M., Dalle, S., da Silva, M. G., Essongue,
A. F., Rabier, A., Pannard, M., Jalali, M. S., Elgam-
mal, A., Papazoglou, M., Hacid, M.-S., Rioufol, C.,
Kersten, M.-J., van Oijen, M. G., Suazo-Zepeda, E.,
Malhotra, A., Coquery, E., Anota, A., Preau, M., Fau-
vernier, M., Coz, E., Puig, S., and Maucort-Boulch,
D. (2023). Monitoring multidimensional aspects of
quality of life after cancer immunotherapy: protocol
for the international multicentre, observational quali-
top cohort study. BMJ Open, 13(4).
Yang, Q., Liu, Y., Chen, T., and Tong, Y. (2019). Federated
machine learning: Concept and applications. ACM
Trans. Intell. Syst. Technol., 10(2):12:1–12:19.
Zhang, H., Chen, G., Ooi, B. C., Tan, K., and Zhang, M.
(2015). In-memory big data management and pro-
cessing: A survey. IEEE Trans. Knowl. Data Eng.,
27(7):1920–1948.
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
428
