
Disease Prediction with Heterogeneous Graph of Electronic Health
Records and Toxicogenomics Data
Ji-Hyeong Park
1 a
, Hyun-Soo Choi
2,∗ b
, Sunhwa Jo
3 c
and Jinho Kim
3,∗ d
1
Dept. of Convergence Security, Kangwon Nat’l Univ., Chuncheon, Republic of Korea
2
Dept. of Computer Science and Engineering, Seoul Nat’l Univ. of Science and Technology, Seoul, Republic of Korea
3
Dept. of Computer Science and Engineering, Kangwon Nat’l Univ., Chuncheon, Republic of Korea
Keywords:
Heterogeneous Graph, Node Embedding, Disease Prediction, Electronic Health Records, Toxicogenomics
Data.
Abstract:
Disease prediction is an important technology in the field of medicine. Several studies have been conducted on
disease prediction using electronic health records (EHR). However, existing methods have several limitations,
such as predicting only a single disease and utilizing limited data sources of textual or drug-related data; thus,
they cannot capture the relationship between a patient and a disease, or among diseases. Furthermore, they
suffer from the problem that additional information other than EHR exists only for a limited set of diseases
and cannot be used for a wide range of diseases. To mitigate these problems, we utilize Toxicogenomics
Data (TD) that contains extensive information about most diseases, and analyze this complicated data using a
heterogeneous graph embedding technique. We utilize metapath and graph neural network for graph embed-
ding of heterogeneous relationships in EHR-TD, and then develop a novel disease prediction framework. To
achieve this goal, we first present a process for the collection and processing of EHR and TD data to improve
their reliability. Secondly, we propose a method for efficiently constructing heterogeneous EHR-TD graphs,
and present an embedding model that can be effectively used. Finally, we propose a metapath interaction
encoder that can address the problems of RNN-based encoders in previous models. Thereafter, we validate
the effectiveness of the proposed framework and modules with extensive evaluations of various designs for
disease prediction using EHR and TD data.
1 INTRODUCTION
Recent advancements in medical technology have led
to the generation of large amounts of high-quality data
in hospitals. These data are commonly referred to
as electronic health records (EHRs) and typically in-
clude a wide range of information, such as visit and
hospitalization records, symptoms, and patient infor-
mation. Due to the wealth of valuable information
contained in EHRs, there has been a rapid increase
in the number of data analysis studies utilizing EHRs
for medical applications, such as hospital stay predic-
tion(Gentimis et al., 2017), diagnostic prediction(Ma
et al., 2018), and mortality prediction(DeSalvo et al.,
a
https://orcid.org/0009-0003-7817-1774
b
https://orcid.org/0000-0002-3594-8948
c
https://orcid.org/0000-0002-6696-6276
d
https://orcid.org/0000-0003-1125-3938
∗
Corresponding authors
2006). Among these studies, disease prediction is a
valuable technology that can reduce the cost of med-
ical care. In existing studies on disease prediction,
gene-based approaches have been used, which are
limited in accessibility due to the high cost and re-
source requirements. Therefore, we aim to develop
a disease prediction method that can be used with
reasonable costs and resources by utilizing EHR data
generated by patients visiting a hospital.
Existing disease prediction studies address spe-
cific disease prediction using only EHR (Jin et al.,
2018), which contains textual information, including
user examination records and disease predictions us-
ing drug information databases, such as PubMed and
DrugBank. However, these disease predictions do not
employ sufficient information for disease prediction,
although they utilize additional data, such as images,
text, or drug-related data. Furthermore, these addi-
tional data cannot be consistently used for all diseases
because their presence depends on the characteristics
Park, J., Choi, H., Jo, S. and Kim, J.
Disease Prediction with Heterogeneous Graph of Electronic Health Records and Toxicogenomics Data.
DOI: 10.5220/0012096400003541
In Proceedings of the 12th International Conference on Data Science, Technology and Applications (DATA 2023), pages 97-104
ISBN: 978-989-758-664-4; ISSN: 2184-285X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
97

of each disease. In addition, existing methods do not
capture the relationship between a patient and disease
or among different diseases.
Our primary goal is to achieve a more accurate
disease prediction that is practical, as well as solve
the problem in which the existing methods are avail-
able to limited diseases and show limited accuracy
owing to a lack of information for prediction. To
supplement information about relatively rare diseases,
we introduced Toxicogenomics Data (TD), which in-
cludes various environmental factors affecting health-
care. TD contains a variety of information about
disease-related genes and chemicals, which is useful
for inferring information about the disease of a pa-
tient as well as other diseases similar to a particular
disease. As TD appears in most diseases, it does not
depend on the characteristics of a disease; thus, TD
can be actively utilized.
To understand the complex structure that com-
bines EHR and TD, we adopted a graph-based em-
bedding approach. A graph-based approach can ef-
fectively model the data structure in which each ob-
ject has similar information, and multiple relation-
ships between any two objects, as validated in so-
cial graph analysis(Leskovec and Mcauley, 2012) and
protein-protein graph analysis(Nasiri et al., 2021). In
our work, to cope with the heterogeneous objects of
patients and diseases, we proposed a heterogeneous
graph and heterogeneous node embedding model via
metapath design. In addition, we proposed a metap-
ath interaction encoder that can be utilized in the pro-
posed node-embedding model. Through experimental
evaluations of a couple of graph configurations and
node embedding methods, we suggested a promising
framework for disease prediction for a patient and val-
idated the effectiveness of the proposed framework
and encoder.
The contributions of this study are summarized as
follows.
• We present a novel framework to achieve further
accurate disease prediction that can be applied to
different diseases.
• We construct a reliable heterogeneous graph data
structure that represents the complex data struc-
ture and relationship among objects in EHR and
TD.
• We suggest a heterogeneous graph embedding
model with a metapath interaction encoder to
learn the EHR-TD graph data effectively.
• We achieve outperforming performance to exist-
ing approach without using complex data, such as
images and text.
2 PROPOSED METHOD
In this study, we proposed an EHR-TD combined
graph-utilized disease prediction framework to sup-
plement patient disease information. The proposed
framework consists of four steps: Data Construction,
Graph Configuration, Graph Embedding, and Predic-
tion Process. Here, we introduce each step and de-
scribe how it is performed. Figure 1 illustrates the
overall process of the proposed framework.
2.1 Data Construction
In this study, we extracted information from two
databases: MIMIC-III (Johnson et al., 2016) and CT-
DBase (Davis et al., 2020). This section describes
how the data used in the experiments were extracted
and pre-processed from the two databases.
The ADMISSIONS table in MIMIC-III, which
contains information on patients, such as insurance
status and religion, has been used in addition to the
disease that the patient suffers from. Before extract-
ing patient information, we first used ADMITTIME
and DISCHTTIME in the table to calculate the pa-
tient’s visit time and discharge time to obtain hospi-
talization periods and remove records with hospital-
ization periods of less than 1 d. Thereafter, disease
prediction was performed only for patients who ap-
peared more than twice, based on SUBJECT ID rep-
resenting the patient.
CTDBase includes genes and chemicals associ-
ated with the disease that appear in MIMIC-III. To
collect disease-related genes or chemicals from CT-
DBase, the disease MeshID is required. Therefore,
we first performed text-to-MeshID matching to con-
vert a disease name(text) in the MIMIC-III table into
MeshID in CTDBase. However, the disease pre-
sented in the ADMISSIONS table of MIMIC-III has
not been pre-processed, such as abbreviated words
or containing words that are not commonly used.
For more accurate text-to-MeshID matching, we pre-
processed each disease name, and then matched it
to a MeshID. When a search term(text) is entered,
the Mesh Browser provides approximate information
about the disease and MeshID for the search term,
and slightly corrects the synonyms. The MeshID cor-
responding to the disease name was collected using
Web crawling. Thereafter, text-to-MeshID matching
was performed using a text search. Because the dis-
ease expression between MIMIC-III and CTDBase
is inconsistent, they may not be perfectly matched.
For further accurate matching, two types of text pre-
processing were performed, without using text in-
formation. First, all special characteristics were re-
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
98
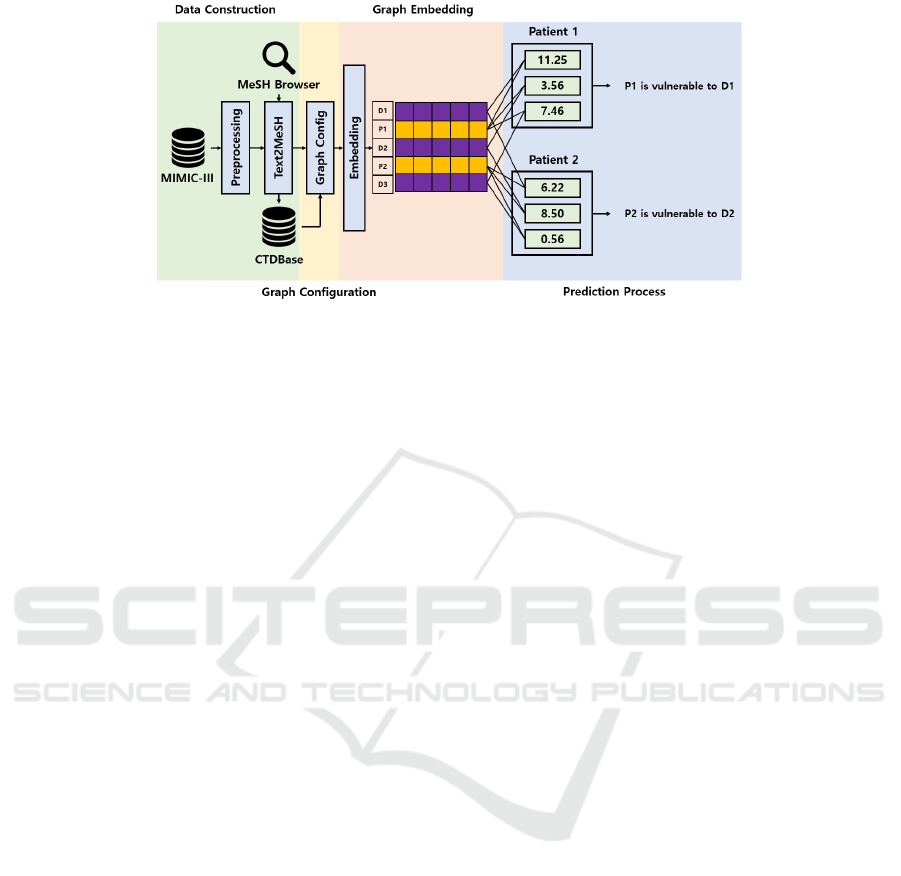
Figure 1: Overall process of the proposed framework.
moved, and second, if there were several diseases in
one column, they were separated.
When processing was completed, we collected
genes and chemicals associated with MeshID. We hy-
pothesized that many genes and chemicals, compared
to the number of diseases, would not be helpful in
disease prediction. Thus, when collecting Gene and
Chemical, only up to 200 genes and chemicals were
collected for each. When there were fewer than 20
associated Genes and Chemicals, we did not collect
them. Additionally, the disease was collected only if
the disease name exactly coincided with one another.
Finally, we added features to the collected ob-
jects. Among the above-described objects, Gene did
not have essential information to be used as a feature;
thus, the feature was assigned by a One-Hot Vector,
and the feature was added as follows for patients, dis-
eases, and chemicals.
• Patient: Patient information in the ADMISSIONS
table was encoded into a multi-hot vector as
a feature vector, depending on the specific pa-
tient information (admission type, admission lo-
cation, discharge location, insurance status, reli-
gion, marital status, and race).
• Disease and Chemical: The words in Definition of
CTDBase were encoded into a multi-hot vector as
a feature. When collecting Definition, the feature
of an object without Definition was assigned by a
zero vector.
2.2 Heterogeneous Graph
Configuration
In the previous section, we described the data col-
lection and processing for EHR and TD, respectively.
This section describes the configuration of the EHR-
TD combined graph required for the proposed frame-
work.
For disease prediction, a graph provides similarity
between a patient’s outbreak disease and other dis-
eases. We define the heterogeneous graph G as fol-
lows: G has two or more node types, including vertex
V , edge E, and type T : G = (V, E,T ). Each node v
in the graph can have heterogeneous neighbor nodes
connected along the edges in E, and the set of neigh-
bor nodes N
v
for v is defined as N
v
= {v
n
|(v, v
n
) ∈
E; v, v
n
∈ V }.
We first add the node types of the patient (P)
and disease (D) to reflect the patient’s outbreak in-
formation in the graph. The similarity between any
two disease nodes can be obtained indirectly from
the disease node features or outbreak information of
similar patients. This indirectly obtained similarity
might degrade the performance of the disease predic-
tion. Therefore, we used TD to provide the similarity
in the graph. We added node types, such as genes
(G) and chemicals (C) in TD to the graph, and the
diseases sharing the types were regarded as similar
diseases to each other. Finally, the type T
v
of each
node v in the heterogeneous graph is defined as T
v
∈
{P,D,G,C}, v ∈ V. Edge type T
e
,e ∈ E for a hetero-
geneous graph is defined as T
e
∈ {O,R
G
,R
C
},e ∈ E,
where O =Occur(P-D) denotes the relationship be-
tween a patient and a disease, R
G
= RelatedGene(D-
G) denotes the relationship between a disease and a
gene, and R
C
= RelatedChemical(D-C) denotes the
relationship between a disease and chemical.
2.3 Heterogeneous Graph Embedding
2.3.1 Metapath Feature Transform
A data sample, such as an image or text, can be eas-
ily vectorized into a point in the Euclidean space.
However, because computers cannot analyze (or cal-
culate) complex non-Euclidean data, such as graph-
structured data, a process of vectorizing graphs is re-
Disease Prediction with Heterogeneous Graph of Electronic Health Records and Toxicogenomics Data
99
(𝒍, 𝟏)&
Kernel Convolutions
𝑿 = [𝐜𝐨𝐧𝐜𝐚𝐭(𝒉
𝟏
, 𝒉
𝟐
, 𝒉
𝟑
)]
𝑻
𝒘
𝑴
(𝒗,𝒖)
𝒌(𝟏, 𝟏)
𝒌(𝟐, 𝟏)
𝒌(𝟑, 𝟏)
𝒉
𝑴
(𝒗,𝒖)
𝟏
Linear Transformation via
Matrix-Vector Multiplication
𝒉
𝑴
(𝒗,𝒖)
%
𝒉
𝑴
(𝒗,𝒖)
&
𝒉
𝑴
(𝒗,𝒖)
.
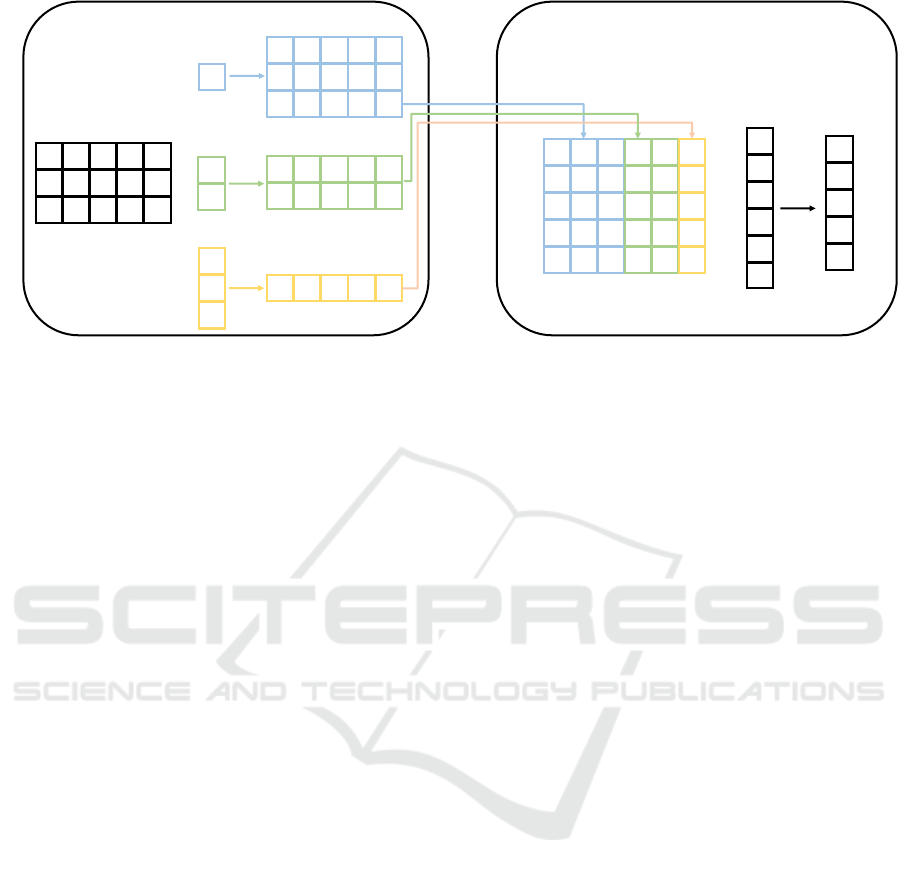
Figure 2: Entire operation of metapath interaction encoder.
quired. In addition, the heterogeneous graph we tar-
get can express more information compared to a ho-
mogeneous graph; however, there are many factors
to be considered when constructing a heterogeneous
graph, such as node types and relations among het-
erogeneous nodes. Therefore, when vectorizing a
graph, diverse methods can be used, depending on the
graph structure. In this study, we designed a hetero-
geneous graph structure and node-embedding scheme
for disease prediction based on MAGNN(Fu et al.,
2020), which uses metapath and graph neural network
(GNN).
In a heterogeneous graph, the shape of the features
of each node is different from that of the other nodes.
For a node v ∈ V of type a ∈ A, encoding is conducted
through a single-layer neural transformation as
h
′
v
= W
a
· x
a
v
, (1)
where x
v
is the input feature vector and W
a
indicates
a trainable matrix for type a ∈ A.
Thereafter, we generated a metapath instance to
apply heterogeneous graph data to an embedding
model. The metapath M is defined by a serial path
of user-specified node types, as follows:
M = {T
v
1
− T
v
2
− ··· − T
v
n
}, T
v
i
∈ T
V
, (2)
where the start node v
1
and end node v
n
are of the
same type. metapath is used in most heterogeneous
graph-embedding models. In our study, we defined
four types of metapath. M
1
,M
2
are defined to re-
flect the patient-disease relationship to embedding,
Patient-Disease-Patient(P-D-P), and Disease-Patient-
Disease(D-P-D). Furthermore, M
3
,M
4
are defined to
reflect the relationship between disease and gene, and
disease and chemical to embedding, disease-gene-
disease (D-G-D), and disease-chemical-disease (D-C-
D), respectively.
In the graph, every metapath instance M
(v,u)
with
the start node v, and end node u has one of four meta-
path types. M
(v,u)
was created by stacking the features
in Eq. (1) as follows:
M
(v,u)
= stack(h
′
v
,{h
′
t
,∀t ∈ {m
M(v,u)
}},h
′
u
),(3)
where m
M
(v,u)
denotes the intermediate node of (v,u).
The metapath instances in (3) are expressed by a ma-
trix that becomes an input to the GNN in MAGNN(Fu
et al., 2020) for embedding. GNN consists of two
stages: intra metapath aggregation and inter metap-
ath aggregation. In our work, instead of intra metap-
ath aggregation in MAGNN(Fu et al., 2020), we sug-
gested a new encoder called metapath interaction en-
coder, and is described in the following subsection.
2.3.2 Metapath Interaction Encoder
The intra-metapath aggregation is a process of col-
lecting information on metapaths of a target node
into one vector. This process uses RNN structure in
MAGNN(Fu et al., 2020). However, the RNN struc-
ture has a problem in that it can not fully utilize all the
information appearing in metapath because of their
sequencial message passing. To mitigate this prob-
lem, we propose a CNN-based encoder called meta-
path interaction encoder. We applied a trainable ker-
nel convolution operation to the input matrix M
(v,u)
in
(3), where the (i, j)
th
element of the filtered result for
M
(v,u)
is expressed as
h
l
M
(v,u)
(i, j) =
l
∑
q=1
M
(v,u)
(i + q − 1, j) k(q, 1), (4)
where k ∈ R
l×1
denotes a trainable kernel, whereas
0 < l ≤ L for L denotes metapath instance’s length.
Subsequently, the filtered matrices h
l
M
(v,u)
,0 < l ≤ L,
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
100

are concatenated and transformed by matrix-vector
multiplication of the concatenated matrix and a train-
able vector w ∈ R
Σ
L
l=1
l×1
as
X := [concat({h
l
M
(v,u)
, 0 < ∀l ≤ L})]
T
, (5)
h
M
(v,u)
= Xw. (6)
The entire operation of metapath interaction encoder
is depicted in Figure 2.
2.3.3 Inter Metapath Aggregration
The vector h
M
(v,u)
in (5) is input to the Inter Metap-
ath Aggregration(IMA) process in MAGNN(Fu et al.,
2020) as
h
(v,u)
= IMA(h
M
(v,u)
), ∀v,u ∈ V, (7)
where v is the target node and u is the node connected
to v in the same cluster type. For embedding, we
adopted the GNN in MAGNN(Fu et al., 2020), and
condisered the original paper for the detailed structure
and hyperparameters. We employed the cross-entropy
loss, which is denoted by
Loss = −
∑
(v,u)∈E
logσ(< h
(v,u)
,h
(v,u)
>)
−
∑
(v
′
,u
′
)∈E
c
logσ(1− < h
(v
′
,u
′
)
,h
(v
′
,u
′
)
>),
(8)
where σ(·) is the sigmoid function, E indicates a set
of connected edges through a metapath instance, and
E
c
denotes the complement of E, that is, a set of un-
connected edges through any metapath instance.
2.4 Prediction Process
The node vectors generated through node embedding
become closer to each other as they become simi-
lar. Therefore, it can be observed that the dot product
value between vectors indicates how similar the vec-
tors are to each other. Through embedding, vectors
are adjusted such that the distance between a patient
and the disease the patient is suffering from or the dis-
tance between a disease and another disease with sim-
ilar genes and chemicals is reduced. These embed-
ding vectors that reflect the graph structure and graph
feature can be used in various applications, such as
link prediction and node classification. In this study,
we used link prediction to predict a patient’s likeli-
hood of an outbreak of a specific disease.
Link prediction predicts the presence or absence
of an edge that already exists or that will be created in
the future in graph G. By predicting the link between
a patient and disease, we can determine the proba-
bility of how risky a patient is for a particular disease.
For a patient node p, its similarity with a disease node
d is obtained by D =< h
p
,h
d
>, p ∈ V
P
,d ∈ V
D
,
where V
P
,V
D
are the patient and disease node sets,
respectively, and < ·,· > denotes the inner product.
The probability that p has disease d is obtained us-
ing the sigmoid function as follows: Prob(p,d) =
1/(1 + e
−D
).
The larger the Prob(p,d), the higher is the proba-
bility that disease d will occur in patient p. To evalu-
ate the link prediction performance, we measured the
prediction errors of the edge existence. In this evalu-
ation, if Prob(p, d) is higher than the threshold η, we
determine that the edge between p and d is connected;
otherwise, it is unconnected.
3 EXPERIMENTS
In this section, we show the validity of the proposed
framework through a performance comparison ac-
cording to three types of graph configurations and six
types of embedding models.
3.1 Experiment Settings
The experimental settings were configured by apply-
ing three graph-configuration methods and six graph-
embedding methods to demonstrate the validity of
the proposed framework. The true-positive and false-
positive rates can differ depending on the threshold
η. Therefore, we used the area under the curve of the
receiver operating characteristic curve (AUROC), and
average precision score(AP).
3.1.1 Types of Graph Configuration
The configurations of the graph types are denoted ac-
cording to the collected data and constructed features
as follows:
• HoEHR-TD Graph: This graph has four node
types (patient, disease, gene, chemical) and
three edge types (patient-disease, disease-gene,
disease-chemical). Although it has several node
types, it is treated as homogeneous graph and
graph embedding method suitable to be em-
ployed.
• BiEHR Graph: This has two node types (patient,
disease) and one edge types (patient-disease). Be-
cause nodes can be divided into two groups, graph
can be classified as bipartite graphs, and an em-
bedding method specialized for bipartite graphs is
used.
• HeEHR-TD Graph (Proposed): This is a hetero-
geneous graph proposed in this study. This graph
Disease Prediction with Heterogeneous Graph of Electronic Health Records and Toxicogenomics Data
101

Table 1: Results of link prediction experiment.
Framework
10% 30% 50% 70%
AUC PR AUC PR AUC PR AUC PR
HoEHR-TD + Deepwalk 0.3903 0.6325 0.4932 0.7059 0.6383 0.7994 0.8632 0.9288
HoEHR-TD + Node2Vec 0.3903 0.6349 0.4893 0.7033 0.6413 0.8013 0.8013 0.9290
BiEHR + BiNE 0.8599 0.8677 0.8806 0.8822 0.8917 0.8839 0.8876 0.8772
BiEHR + BiGI 0.8455 0.8101 0.8618 0.8198 0.8627 0.8220 0.8700 0.8357
HeEHR-TD + Metapath2Vec 0.4142 0.6520 0.5181 0.7236 0.6504 0.8071 0.8629 0.9287
HeEHR-TD + MAGNN 0.9763 0.9740 0.9900 0.9903 0.9907 0.9902 0.9907 0.9907
HeEHR-TD + MAGNN + MIE (Proposed) 0.9870 0.9862 0.9903 0.9900 0.9892 0.9894 0.9922 0.9923
has four node types (patient, disease, gene, chemi-
cal) and three edge types (patient-disease, disease-
gene, disease-chemical). Because it has several
node types and edge types, we designed a spe-
cialized graph embedding model suitable to this
heterogeneous graph.
3.1.2 Type of Graph Embedding Methodologies
Six embedding models were selected based on the
graph configuration. The model selected according
to each graph configuration is described as follows.
In this study, we designed a framework based on the
MAGNN(Fu et al., 2020) model.
• Models for Homogeneous Graph
– Deepwalk(Perozzi et al., 2014): This is a ran-
dom walk-based homogeneous graph embed-
ding method. This is to learn embeddings of
nodes via random walks using skip-gram or c-
bow methods.
– Node2Vec(Grover and Leskovec, 2016): This
is a random walk-based homogeneous graph
embedding method. Similar to DeepWalk, but
when generating a random walk, the probability
of moving to a neighbor is adjusted by parame-
ters p and q to achieve higher performance than
DeepWalk.
• Models for Bipartite Graph
– BiNE(Gao et al., 2018): This is a random walk-
based bipartite graph embedding method. This
attempts to learn the explicit relationship ex-
pressed by an edge, as well as the implicit re-
lationship expressed by a transitive link that is
not observed.
– BiGI(Cao et al., 2021): This is a GCN-based
bipartite graph embedding method. This model
introduces local-global infomax to capture the
global property.
• Models for Heterogeneous Graph
– Metapath2Vec(Dong et al., 2017): This is a
GCN-based bipartite graph embedding method.
This addresses a heterogeneous graph by gen-
erating random walks through the metapath,
which is a list of predefined nodes.
– MAGNN(Fu et al., 2020): This is a model us-
ing both metapath and GCN used in the de-
sign of the proposed framework. The metap-
aths generated from the heterogeneous nodes
are compressed into a single vector, and these
vectors for a metapath are compressed into one
vector, and used as the embedding of the start-
ing node.
– MAGNN + MIE: This is a model that applied
our proposed metapath interaction encoder. As
a metapath interaction encoder, we attempted
to utilize the interaction within metapath that
could not be used in the existing encoders.
3.2 Experiment Result
Based on the experimental settings above, the six
frameworks that were compared in our experiment are
presented as follows:
• HoEHR-TD + (Deepwalk, Node2Vec): we design
(Deepwalk, Node2Vec) for homogeneous EHR-
TD graph.
• BiEHR + (BiNE, BiGI): we design (BiNE, BiGI)
for Bipartite EHR graph.
• HeEHR-TD + (Metapath2Vec, MAGNN): we de-
sign (Metapath2Vec, MAGNN) for heterogeneous
EHR-TD graph.
• HeEHR-TD + MAGNN + MIE: we design
MAGNN using metapath interaction encoder for
heterogeneous EHR-TD graph.
Table 1 lists the results of the link prediction ex-
periment. The edges were divided using four training
data ratios (10%, 30%, 50%, and 70%) and learned
using them. Random walk-based models (Deepwalk,
Node2Vec, BiNE, Metapath2Vec) did not predict the
nodes that did not appear. Thus, when the training
data ratio was low, the performance degradation was
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
102

Table 2: Metapath Comparision in metapath used model.
Metapath Framework
10% 30% 50% 70%
AUC PR AUC PR AUC PR AUC PR
M
1
,M
2
Metapath2Vec 0.3959 0.6349 0.5115 0.7413 0.6417 0.7946 0.7847 0.8738
MAGNN 0.4928 0.4949 0.5059 0.5068 0.4949 0.4967 0.4958 0.5054
M
1
,M
2
,M
3
Metapath2Vec 0.4281 0.6584 0.5298 0.7281 0.6673 0.8155 0.8778 0.9360
MAGNN 0.9795 0.9806 0.9767 0.9757 0.9748 0.9741 0.9571 0.9551
M
1
,M
2
,M
4
Metapath2Vec 0.4229 0.6545 0.5308 0.7288 0.6640 0.8132 0.8689 0.9313
MAGNN 0.9627 0.9642 0.9472 0.9472 0.9551 0.9573 0.9503 0.9533
M
1
,M
2
,M
3
,M
4
Metapath2Vec 0.4213 0.6539 0.5299 0.7276 0.6673 0.8153 0.8839 0.9392
MAGNN 0.9850 0.9864 0.9868 0.9869 0.9634 0.9623 0.9865 0.9883
large compared to that of the graph neural network-
based models (BiGI and MAGNN). In addition, the
model using our proposed metapath interaction en-
coder(MAGNN + MIE) showed better performance
than before.
Comparing the results according to the graph con-
figuration, it can be observed that the performance is
roughly in the order of HeEHR-TD, HoEHR-TD, and
BiEHR. BiEHR embedding models (BiNE and BiGE)
do not reflect the proposed TD. HoEHR-TD embed-
ding models (Deepwalk, Node2Vec) considered EHR
and TD to be of the same type; therefore, the benefit
from the added TD was relatively small. As shown in
the table, we can observe that HeEHR-TD embedding
models exhibit high performance by fully utilizing
TD. In particular, the proposed framework, HeEHR-
TD + MAGNN, outperformed the other frameworks.
Table 2 lists the performance variation depending
on the design of metapaths. As shown in the table, the
design of metapath significantly affected the predic-
tion performance. The results in Table 2 show that the
result using plenty of metapaths through the disease,
gene, and chemical nodes is significantly better than
that using only patient-disease metapaths. In metap-
ath2vec, similar to the results in Table 1, the perfor-
mance is similar at a low training data ratio. How-
ever, it can be observed that the higher the training
data ratio, the better the results of using the metap-
ath reflecting TD. In MAGNN, the results ({M
1
,M
2
})
show worse performance than the others, implying
that simple metapaths cannot reflect sufficient rela-
tionships among heterogeneous node types. How-
ever, other results ({M
1
,M
2
,M
3
}, {M
1
,M
2
,M
4
}, and
{M
1
,M
2
,M
3
,M
4
}) show that adding many relation-
ships between a disease and gene, and a disease and
chemical can contribute to heterogeneous node em-
bedding.
3.3 Discussion
This study aimed to present a new framework for im-
proving disease prediction performance by compos-
ing EHR-TD heterogeneous graphs on the relation-
ships among patients, diseases, and genes/chemicals
related to diseases that cannot be captured by existing
disease prediction studies. This implies TD data can
be helpful for disease prediction.
One of our contributions is that, by using toxi-
cogenomics data, specific diseases as well as all dis-
eases that appear in MIMIC-III (Johnson et al., 2016)
are covered. As introduced in the section on data con-
struction and graph embedding, the probability can
be calculated for all diseases in MIMIC-III because
the embedding vectors are created with edges for all
diseases that have emerged through TD between dis-
eases. Maximum prediction of diseases, rather than
considering a single disease, will provide greater ben-
efits to users.
4 CONCLUSION
In this study, we proposed and introduced an EHR-TD
combined with a heterogeneous graph-based disease
prediction framework to further improve disease pre-
diction. Through the proposed framework, we aimed
to maximally predict diseases, rather than the existing
single disease prediction, to show that the combined
heterogeneous data can improve disease prediction
performance and present a heterogeneous graph struc-
ture that is effective for improving disease prediction
performance. The proposed framework consists of
data construction, which collects and pre-processes
data, graph configuration, graph embedding, which
creates representations for each node with constructed
graphs, and a prediction process that uses representa-
tions of generated nodes. As contributions, we sug-
gested a new heterogeneous graph representing EHR-
Disease Prediction with Heterogeneous Graph of Electronic Health Records and Toxicogenomics Data
103

TD data, and designed a heterogeneous graph embed-
ding model along with metapath design. The pro-
posed frameworks were validated through a compari-
son with possible frameworks using combinations of
graph configurations and embedding models. In addi-
tion, through ablation experiments, we demonstrated
the usefulness of TD for disease prediction, and ef-
fects of the metapath design were investigated. Al-
though the proposed framework shows outstanding
performance compared to existing embedding mod-
els, further study for an enhanced embedding model
specific to our EHR-TD data can be conducted in the
future. We expect that the proposed framework will
contribute to more accurate disease prediction and
disease management in patients.
ACKNOWLEDGEMENTS
This work was supported by the National Research
Foundation of Korea(NRF) grant funded by the Korea
government(MSIT) (No. 2021R1F1A1059255).
REFERENCES
Cao, J., Lin, X., Guo, S., Liu, L., Liu, T., and Wang, B.
(2021). Bipartite graph embedding via mutual in-
formation maximization. In Proceedings of the 14th
ACM International Conference on Web Search and
Data Mining, WSDM ’21, page 635–643, New York,
NY, USA. Association for Computing Machinery.
Davis, A. P., Grondin, C. J., Johnson, R. J., Sci-
aky, D., Wiegers, J., Wiegers, T. C., and Mat-
tingly, C. J. (2020). Comparative Toxicogenomics
Database (CTD): update 2021. Nucleic Acids Re-
search, 49(D1):D1138–D1143.
DeSalvo, K. B., Bloser, N., Reynolds, K., He, J., and Munt-
ner, P. (2006). Mortality prediction with a single gen-
eral self-rated health question. Journal of General In-
ternal Medicine, 21(3):267–275.
Dong, Y., Chawla, N. V., and Swami, A. (2017). Meta-
path2vec: Scalable representation learning for het-
erogeneous networks. In Proceedings of the 23rd
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining, KDD ’17, page
135–144, New York, NY, USA. Association for Com-
puting Machinery.
Fu, X., Zhang, J., Meng, Z., and King, I. (2020). Magnn:
Metapath aggregated graph neural network for hetero-
geneous graph embedding. In Proceedings of The Web
Conference 2020, WWW ’20, page 2331–2341, New
York, NY, USA. Association for Computing Machin-
ery.
Gao, M., Chen, L., He, X., and Zhou, A. (2018). Bine: Bi-
partite network embedding. In The 41st International
ACM SIGIR Conference on Research & Development
in Information Retrieval, SIGIR ’18, page 715–724,
New York, NY, USA. Association for Computing Ma-
chinery.
Gentimis, T., Alnaser, A. J., Durante, A., Cook, K.,
and Steele, R. (2017). Predicting hospital length
of stay using neural networks on mimic iii data.
In 2017 IEEE 15th Intl Conf on Dependable,
Autonomic and Secure Computing, 15th Intl
Conf on Pervasive Intelligence and Comput-
ing, 3rd Intl Conf on Big Data Intelligence and
Computing and Cyber Science and Technology
Congress(DASC/PiCom/DataCom/CyberSciTech),
pages 1194–1201.
Grover, A. and Leskovec, J. (2016). node2vec: Scal-
able feature learning for networks. In Proceedings
of the 22nd ACM SIGKDD international conference
on Knowledge discovery and data mining, pages 855–
864.
Jin, B., Che, C., Liu, Z., Zhang, S., Yin, X., and Wei, X.
(2018). Predicting the risk of heart failure with ehr
sequential data modeling. IEEE Access, 6:9256–9261.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). Mimic-
iii, a freely accessible critical care database. Scientific
Data, 3(1):160035.
Leskovec, J. and Mcauley, J. (2012). Learning to dis-
cover social circles in ego networks. In Pereira, F.,
Burges, C., Bottou, L., and Weinberger, K., editors,
Advances in Neural Information Processing Systems,
volume 25. Curran Associates, Inc.
Ma, F., You, Q., Xiao, H., Chitta, R., Zhou, J., and Gao, J.
(2018). Kame: Knowledge-based attention model for
diagnosis prediction in healthcare. In Proceedings of
the 27th ACM International Conference on Informa-
tion and Knowledge Management, CIKM ’18, page
743–752, New York, NY, USA. Association for Com-
puting Machinery.
Nasiri, E., Berahmand, K., Rostami, M., and Dabiri, M.
(2021). A novel link prediction algorithm for protein-
protein interaction networks by attributed graph em-
bedding. Computers in Biology and Medicine,
137:104772.
Perozzi, B., Al-Rfou, R., and Skiena, S. (2014). Deep-
walk: Online learning of social representations. In
Proceedings of the 20th ACM SIGKDD International
Conference on Knowledge Discovery and Data Min-
ing, KDD ’14, page 701–710, New York, NY, USA.
Association for Computing Machinery.
DATA 2023 - 12th International Conference on Data Science, Technology and Applications
104
