
Barycentre Averaging for the Move-Split-Merge Time Series Distance
Measure
Christopher Holder
1 a
, David Guijo-Rubio
1,2 b
and Anthony Bagnall
3 c
1
School of Computing Sciences, University of East Anglia, Norwich, U.K.
2
Department of Computer Science and Numerical Analysis, University of C
´
ordoba, C
´
ordoba, Spain
3
School of Electronics and Computer Science, University of Southampton, Southampton, U.K.
Keywords:
Time Series Distances, Time Series Clustering, Move Split Merge, Barycentre Averaging, Dynamic
Barycentre Averaging, MSM Barycentre Averaging, DBA, MBA.
Abstract:
Distance functions play a core role in many time series machine learning algorithms for tasks such as cluster-
ing, classification and regression. Time series often require bespoke distance functions because small offsets
in time can lead to large distances between series that are conceptually similar. Elastic distances compensate
for misalignment by creating a path through a cost matrix by warping and/or editing time series. Time series
are most commonly clustered with partitional algorithms such as k-means and k-medoids using elastic distance
measures such as Dynamic Time Warping (DTW). The distance is used to assign cases to the closest cluster
representative. k-means requires the averaging of time series to find these representative centroids. If DTW is
used to assign membership, but the arithmetic mean is used to find centroids, k-means performance degrades
significantly. An averaging technique specific to DTW, called DTW Barycentre Averaging (DBA), overcomes
the averaging problem however, can only be used with DTW. As such alternative distance functions such as
Move-Split-Merge (MSM) are forced to use the arithmetic mean to compute new centroids and suffer similar
degraded performance as k-means-DTW without DBA. To address this we propose a averaging method for
MSM distance, MSM Barycentre Averaging (MBA) and show that when used to find centroids it significantly
improves MSM based k-means and is better than commonly used alternatives.
1 INTRODUCTION
Machine learning applied to time series data is a ex-
tensively studied area within the literature. One of
the key factors contributing to its advancement is the
improved ability to collect temporal data using cost-
effective sensor technology across various scientific
disciplines. Specifically, time series data can be sub-
jected to different tasks, each with its own objective.
Among these tasks, Time Series Classification (TSC)
(Bagnall et al., 2017; Middlehurst et al., 2023) has
gained widespread attention, involving the prediction
of a discrete output value for a given time series. Fur-
thermore, if the aim is to predict a continuous output
value instead of a discrete one, the task is referred
to as Time Series Extrinsic Regression (TSER) (Tan
et al., 2021; Guijo-Rubio et al., 2023). Concerning
a
https://orcid.org/0000-0001-9571-3764
b
https://orcid.org/0000-0002-8035-4057
c
https://orcid.org/0000-0003-2360-8994
unsupervised tasks, Time Series Clustering (TSCL)
(Liao, 2005; Aghabozorgi et al., 2015) is particularly
well-known, aiming to group time series without the
need of labelled data. TSCL is the primary focus of
this work.
TSCL is used across multiple different diciplines.
It has been used to identify anomalous amplitude and
shape patterns in disease outbreak data (Li et al.,
2021) Climate researchers used TSCL to discover
common patterns preceding important paleoclimate
events (Nikolaou et al., 2015). TSCL has been used
extensivly in bioinformatics to group gene expression
patterns (McDowell et al., 2018). These applications
of TSCL demonstrate the versatility of this task across
several disciplines.
With respect the TSCL taxonomy, it can be ap-
proached from two main points of view: 1) extract-
ing features from the time series; and 2) employing
a standard clustering approach along with a time se-
ries distance measure. The majority of existing TSCL
techniques fall into the second category. These ap-
Holder, C., Guijo-Rubio, D. and Bagnall, A.
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure.
DOI: 10.5220/0012164900003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 1: KDIR, pages 51-62
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
51

proaches critically depend on the choice of an appro-
priate distance measure, combined with the clustering
approach used.
Measuring distance between time series is a prim-
itive operation that can be used for a range of
tasks such as classification, clustering, regression and
query. Time series require bespoke distance func-
tions because small offsets between series can lead
to large distances between series that are conceptu-
ally similar. Elastic distances compensate for mis-
alignment by creating a path through a cost ma-
trix through either warping or editing time series.
The most common elastic distance is Dynamic Time
Warping (DTW) (Ratanamahatana and Keogh, 2005),
however, there have been numerous others proposed.
A recent comparison study that evaluated 9 com-
monly used elastic distances found that the MSM
distance measure was the best performing distance
function for the k-means clustering algorithm (Holder
et al., 2022). Unlike DTW, MSM satisfies the mathe-
matical conditions of a metric, meaning it can, for ex-
ample, exploit the triangle inequality for fast distance
calculations. Figure 1 shows the method for finding
the MSM distance between two series.
In addition to computing similarity, many TSCL
approaches must also choose or synthesise exemplars
to represent clusters. One of the most common tech-
niques to do this is averaging. k-means (Lloyd, 1982)
is one of the most commonly used TSCL approaches
in the literature and requires both to compute the dis-
tance between time series and the creation of syn-
thetic time series (by averaging) that represent a clus-
ter. Various methods have been proposed to compute
the average of a collection of time series (Brill et al.,
2019; Holznigenkemper and Seeger, 2023).
Our contribution is to propose a method for av-
eraging time series using MSM. Our method, MSM
Barycentre Averaging (MBA) is based on the approx-
imate barycentre averaging method which was made
popular for DTW using DTW Barycentre Averag-
ing (DBA) (Petitjean et al., 2011). We show that,
when integrated with k-means clustering, MBA sig-
nificantly outperforms clusters created using either
DTW or MSM with arithmetic mean averaging and
DBA.
We have conducted all experiments with the aeon
time series machine learning toolkit
1
and we demon-
strate how to reproduce all our experiments with the
associated experimental code and notebook
2
.
The rest of this paper is structured as follows.
1
https://github.com/aeon-toolkit/aeon
2
https://github.com/time-series-machine-learning/tsml
-eval/blob/main/tsml eval/publications/y2023/distance bas
ed clustering/MBA.ipynb
In Section 2 we provide background into elastic dis-
tance functions and time series averaging. Section 2.3
describes our approach to averaging using MSM,
known as MSM Barycentre Averaging (MBA). In
Section 3.1, we present related works in TSCL litera-
ture. Our results are presented in Section 4 before we
conclude in Section 5.
2 BACKGROUND
Distance-based time series machine learning has been
a popular theme in time series classification and clus-
tering research. There have been numerous exper-
imental evaluations of distance-based classification,
such as (Ding et al., 2008; Lines and Bagnall, 2014).
For many years, the received wisdom was that DTW
was the best choice. For example, the first sentence of
(Petitjean et al., 2016) is: “The last decade has seen
increasing acceptance that the nearest neighbour al-
gorithm with dynamic time warping as the distance
measure is the technique of choice for most time se-
ries classification problems”. However, recent exper-
imental papers (Lines et al., 2018; Paparrizos et al.,
2020; Holder et al., 2022) have identified that MSM
is more effective for both classification and cluster-
ing. Nevertheless, DTW is still by far the most widely
used elastic distance measure. Hence, we limit our
focus to DTW and MSM, as well as the standard Eu-
clidean Distance (ED), and direct the interested reader
to (Holder et al., 2022; Shifaz et al., 2023) for more
detailed background on elastic distances.
2.1 Time Series Elastic Distance
Functions
Suppose we want to measure the distance between
two time series (assumed to be equal lengths and uni-
variate), a = {a
1
,a
2
,...,a
m
} and b = {b
1
,b
2
,...,b
m
}.
The ED, d
ED
is the L2 norm between series,
d
ED
(a,b) =
s
m
∑
i=1
(a
i
− b
i
)
2
. (1)
d
ED
puts no priority on the ordering of the series.
Elastic distance measures allow for possible misalign-
ment by attempting to optimally align two series. This
is done by either distorting indices or by editing the
series to add or remove values.
2.1.1 Dynamic Time Warping (DTW)
DTW mitigates distortions in the time axis by re-
aligning (also known as warping) the series to best
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
52
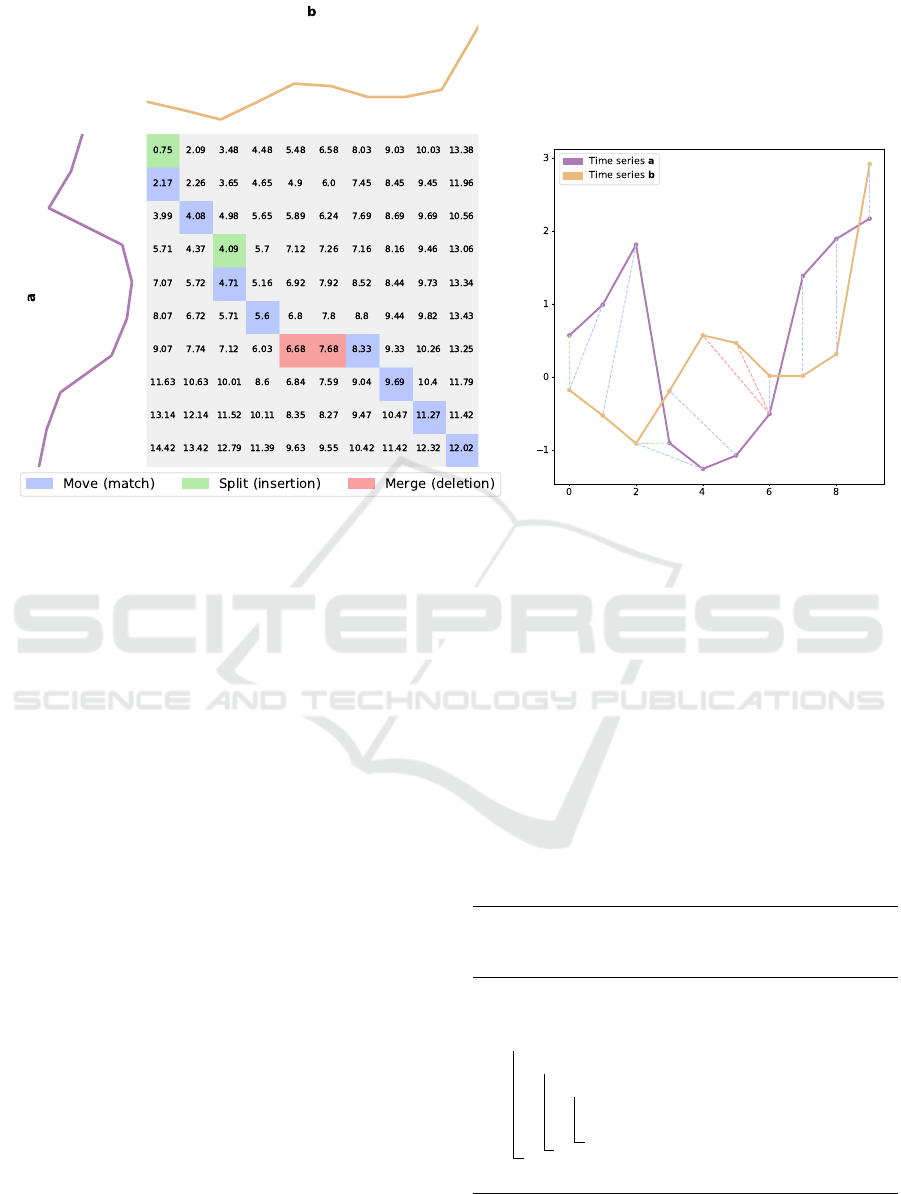
Figure 1: An example of MSM distance: cost matrix (left) to align series (right).
match each other. Let M(a,b) be the m × m point-
wise distance matrix between series a and b, where
M
i, j
= (a
i
− b
j
)
2
. A warping path
P =< (e
1
, f
1
),(e
2
, f
2
),...,(e
s
, f
s
) >
is a set of pairs of indices that define a traversal of
matrix M. A valid warping path must start at location
(1,1), end at point (m,m) and not backtrack, i.e. 0 ≤
e
i+1
− e
i
≤ 1 and 0 ≤ f
i+1
− f
i
≤ 1 for all 1 < i < m.
The DTW distance between series is the path through
M that minimises the total distance. The distance for
any path P of length s is
D
P
(a,b,M) =
s
∑
i=1
M
e
i
, f
i
. (2)
If P is the space of all possible paths, the DTW path
P
∗
is the path that has the minimum distance, hence
the DTW distance between series is
d
DTW
(a,b) = D
P∗
(a,b,M). (3)
The optimal warping path P
∗
can be found exactly
through the dynamic programming formulation de-
scribed in Algorithm 1. This can be a time-consuming
operation, and it is common to put a restriction on the
amount of warping allowed.
2.1.2 Move-Split-Merge (MSM)
At any step, elastic distances can use one of three
costs: diagonal, horizontal or vertical, in forming an
alignment. The alignment path is a series of moves
across the cost matrix. DTW assigns no explicit
penalty for moving off the diagonal. Instead, it uses
an implicit penalty (long paths have longer total dis-
tance) and a hard cut off on window size to stop large
warpings. An alternative family of distance func-
tions are based on the concept of edit distance (e.g.
Edit distance with Real Penalty (ERP) (Chen and Ng,
2004)). An edit distance, such as MSM, considers a
diagonal move as a match, a vertical move as an in-
sertion and an horizontal move as a deletion. MSM
(Algorithm 2) follows this structure, where move is
a match (diagonal), split is a insertion (vertical) and
merge is deletion (horizontal).
Algorithm 1: DTW (a,b, (both series of length m), w (win-
dow proportion, default value w ← 1), M (pointwise dis-
tance matrix)).
1 Let C be an (m + 1) × (m + 1) matrix
initialised to zero, indexed from zero.
2 for i ← 1 to m do
3 for j ← 1 to m do
4 if |i − j| < w · m then
5 C
i, j
← M
i, j
+
min(C
i−1, j−1
,C
i−1, j
,C
i, j−1
)
6 return C
m,m
Figure 1 shows the method for finding the MSM
distance between two time series. As can be ob-
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure
53

served, the three operations (match/insert/deletion)
are identified with different colours (blue/green/red)
and using the specific terminology of MSM distance
(move/split/merge).
The move operation in MSM uses the absolute dif-
ference rather than the squared euclidean distance for
matching in DTW. The cost of the split operation is
given by cost function C (Equation 4) with a call to
C(a
i
,a
i−1
,b
j
,c). If the value being inserted, b
j
, is be-
tween the two values a
i
and a
i−1
being split, the cost
is a constant value c. If not, the cost is c plus the
minimum deviation from the furthest point a
i
and the
previous point a
i−1
or b
j
. The delete/merge is given
by C(b
j
,b
j−1
,a
i
,c), which is simply the same oper-
ation as split but applied to the second series. Thus,
the cost of splitting and merging values depends on
the value itself and adjacent values.
C(a
i
,a
i−1
,b
j
,c) =
c if a
i−1
≤ a
i
≤ b
j
c if a
i−1
≥ a
i
≥ b
j
c + min(|a
i
− a
i−1
|,|a
i
− b
j
|)
otherwise.
(4)
Algorithm 2 describes how to calculate the MSM
distance between two time series a and b. MSM sat-
isfies triangular inequality and is a metric. In Algo-
rithm 2 the first return value is the MSM distance
between a and b, the second is the cost matrix used
to compute the MSM distance (this is used in Algo-
rithm 5).
Algorithm 2: MSM(a (of length m), b (of length m), c (min-
imum cost)).
1 Let CM be an m × m matrix initialised to zero.
2 CM
1,1
= |a
1
− b
1
|
3 for i ← 2 to m do
4 CM
i,1
= CM
i−1,1
+C(a
i
,a
i−1
,b
1
,c)
5 for i ← 2 to m do
6 CM
1,i
= CM
1,i−1
+C(b
i
,a
1
,b + i − 1,c)
7 for i ← 2 to m do
8 for j ← 2 to m do
9 move ← CM
i−1, j−1
+ |a
i
− b
j
|
10 split ← CM
i−1, j
+C(a
i
,a
i−1
,b
j
,c)
11 merge ← CM
i, j−1
+C(b
j
,b
j−1
,a
i
,c)
12 CM
i, j
← min(move,split, merge)
13 return CM
m,m
,CM
2.2 Time Series Averaging Methods
Finding a consensus (average) representation of a set
of sequences has been described as the Holy Grail by
(Gusfield, 1997). One of the main issues with averag-
ing sequences is that it is subjective. As such to de-
velop an algorithm to solve this problem it is normally
formulated as an optimisation problem (Brill et al.,
2019; Holznigenkemper and Seeger, 2023). If we are
not concerned with offset, then the simple arithmetic
average over time points will minimise the ED be-
tween time series. For algorithms such as k-means
this is the default approach. However, when using
this approach with elastic distances (i.e. cluster mem-
bership is assigned based on an elastic distance mea-
sure), the arithmetic mean centroid may misrepresent
the elements of a cluster. Simple averaging will tend
to blur the underlying series and result in worse k-
means clustering performance (Petitjean et al., 2016).
Similarly, if series of the same class are condensed
through arithmetic averaging, it is unlikely these ex-
emplars will be useful for classification or regression.
As such many methods to average time series us-
ing DTW have been proposed. Initially NonLinear
Alignment and Averaging Filters (NLAAF) (Gupta
et al., 1996) was proposed. This method applies
a tournament scheme whereby sequences are paired
and averaged together step by step until only one fi-
nal sequence remains. When two sequences are av-
eraged, the DTW alignment path between the two
series is computed and the point to point average is
taken using this alignment path. The main drawback
of this approach is it leads to a large growth in the av-
erage sequence produced (as every use of the averag-
ing method can lead to the length of the sequence al-
most doubling) (Petitjean et al., 2011). Another issue
with NLAAF is error propagation. Niennattrakul and
Ratanamahatana (2009) proposed Prioritised Shape
Averaging (PSA) which employs a hierarchical av-
eraging method reducing error propagation but also
leading to the same large sequence length growth as
NLAAF. Due to this growth of sequence, both are im-
practical for time series data (given complexities of
algorithms employed, such as DTW). Petitjean et al.
(2011) addresses both of these problems by propos-
ing DTW Barycentre Averaging (DBA) that uses the
optimal warping path to compute a series of the same
length while accounting for alignment of time series.
This will be explained further in Section 2.3.
2.3 Barycentre Averaging
DBA (Petitjean et al., 2011) was proposed to over-
come the limitations of other averaging methods for
time series outlined in Section 2.2. DBA uses a
heuristic strategy to compute a new series that min-
imises the DTW distance to cluster members rather
than the ED. The DBA process begins with an initial
centre, which is typically the medoids of the time se-
ries collection to be averaged. For each time series
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
54

in the collection, the optimal DTW warping path is
computed to the centre. Using these warping paths a
new centre is computed by determining which points
warped onto each element of the initial centre. The
mean is then calculated for each time point warped to
each other time point, taking into account how often
each time point was warped to. This process contin-
ues iteratively until there is no significant change in
the sum of squared DTW distances to the centre, in-
dicating that the optimum centre has been achieved.
While the original proposal for DBA was only
for DTW, assuming an optimal alignment path can
be obtained from an elastic distance (and it aims to
minimise the dissimilarity between two series), any
distance could be used with (Petitjean et al., 2011)
barycentre averaging approach. Holder et al. (2022)
reviewed 9 different elastic distance measures for
TSCL and found the MSM distance significantly out-
performed DTW across multiple clustering metrics.
As such this paper seeks to expand on these finding
by also using the best performing elastic distance in
the averaging stage of computation.
3 MSM BARYCENTRE AVERAGE
(MBA)
To change the distance measure used in the origi-
nal DBA, the optimal warping path needs to be re-
trieved from the distance computation, necessitating
the MSM algorithm to construct and return a cost ma-
trix, as seen in Algorithm 2. This optimal path is iden-
tified by backtracking through the cost matrix and fol-
lowing the trajectory that minimises the total distance,
as outlined in Algorithm 3. Given these traits, MSM
can feasibly be paired with barycentre averaging.
With this prerequisites the DBA algorithm has
been adapted and outlined in Algorithm 4 and 5. This
new algorithm computes the MSM Barycentre Aver-
age (MBA). Each step in the MBA algorithm will now
be outlined.
MSM medoids = arg min
x
m
∈C
∑
x
c
∈C
MSM(x
c
,x
m
,cost).
(5)
Algorithm 4 takes a collection of time series, a num-
ber of max iterations and a minimum cost for MSM as
parameters and computes the MBA. Firstly an initial
centre is computed (line 1). This is done using the
MSM medoids given in Eq 5. Using this initial cen-
tre, a number of iterations are executed (max iters)
to refine the centre (line 3). This is done using
Algorithm 5. Algorithm 5 begins by defining two
values: num war ps to, which is an array that tracks
Algorithm 3: Compute path(CM (of size n × m)).
1 Let alignment be a list.
2 Let i = n − 1 and j = m − 1.
3 while i > 0 or j > 0 do
4 Append (i, j) to alignment.
5 if i == 0 then
6 j ← j − 1.
7 else if j == 0 then
8 i ← i − 1.
9 else
10 min index =
arg min((CM
i−1, j−1
,CM
i−1, j
,CM
i, j−1
)
11 if min index == 0 then
12 i ← i − 1, j ← j − 1.
13 else if min index == 1 then
14 i ← i − 1.
15 else
16 j ← j − 1.
17 Append (0,0) to alignment.
18 return alignment reversed.
Algorithm 4: MSM barycentre average(X (collection of
time series), max iters (max iterations before stop) c (min-
imum cost)).
1 centre ← MSM medoids(X )
2 for i ← 1 to max iters do
3 centre ← MSM BA update(centre,X, c)
4 return centre.
Algorithm 5: MSM BA update(centre (time series of size
m), X (collection of time series of size n × m), c (minimum
cost for MSM computation)).
1 Initialise num warps to as a zeros array of
size m.
2 Initialise alignment as a zeros array of size m.
3 for i ← 1 to n do
4 dist, CM ← MSM(X
i
, centre, c)
5 curr alignment ← compute path(CM)
6 for each ( j, k) in curr alignment do
7 alignment
k
← alignment
k
+ X
i, j
8 num warps to
k
← num warps to
k
+1
9 new centre ← alignment/num warps to
10 return new centre
how many times a point is warped to (due to how
the optimal warping path is computed one point can
be warped to multiple times in a single path), and
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure
55

alignment, which is an array of length m which will
store the values of the new centre (line 1 and 2). Next,
for each value in the collection of time series X, it
computes the MSM Cost Matrix (CM) (Algorithm 2)
between X
i
and the centre (line 4). Using the com-
puted CM, the optimal alignment path can be found
in the form (line 5):
curr alignment =< (e
1
, f
1
),(e
2
, f
2
),...,(e
s
, f
s
) > .
Each tuple in the optimal alignment path is then
looped over (line 6) and the j
th
value in X
i
is added
to the value in alignment
k
(i.e. value j is warped to
the index k) (line 7). Furthermore num war ps to
k
is
incremented (line 8). Once each time series in the col-
lection X has been warped to, the new centre can be
computed by taking the mean of the alignments us-
ing num warps to, which tracked the number of time
each index was warped to (line 9). The new computed
centre is then returned (line 10).
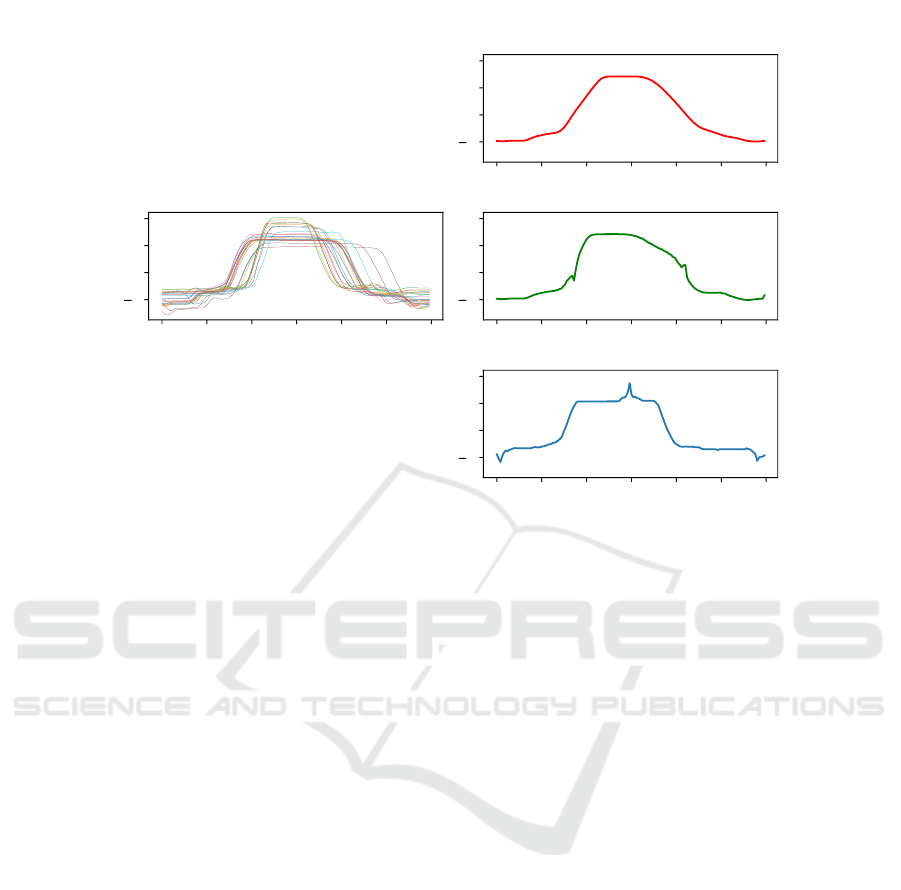
The result of this adaptation, while subtle, leads to
a much different resulting average as shown in Figure
2. This figure shows the average time series of the
whole set of time series with class 1 for the GunPoint
dataset. The discriminatory features are the small
peaks before and after the main peak. These represent
the moment when the actor is drawing and replacing
the gun. As can be observed, MSM is able to clearly
identify these discriminatory features, whereas both
arithmetic averaging or DTW do not.
3.1 Alternative TSCL Algorithms
To put the results of MBA into context we also ex-
plore alternative TSCL approaches. Most of the de-
velopments in TSCL are domain-specific, not being
focused on TSCL as a whole. Nevertheless, there
are a few approaches tackling TSCL as a whole.
The first one is the U-shapelets technique (Zakaria
et al., 2012), which, instead of computing pairwise
distances, considers only relevant subsequences of
time series. This technique shares similarities with
its classification counterpart (Hills et al., 2014). First
of all, subsequences are extracted from the data and
ranked based on their utility, which reflects their dis-
criminative power. This value attempts to maximise
the separation gap between two subsets of time se-
ries: one subset comprises time series with subse-
quences similar to the shapelet being evaluated, while
the other subset consists of the remaining time se-
ries. The subsequences with high utility, referred to as
shapelets, are retained. Once the final set of shapelets
is achieved, a transformation matrix is built with cells
representing distances between U-shapelets and time
series. Finally, the standard k-means method is ap-
plied to the transformation matrix. The main advan-
tages of this approach are that U-shapelets mitigates
the sensitivity to noise and other irrelevant data, and
their ability to provide additional insights into the
data.
Another well-known approach is the Two-step
Time series Clustering (TTC) (Aghabozorgi et al.,
2014). This method firstly reduces the size of the
dataset using the concept of affinity. For this, time
series are grouped according to similarity in time and
then applying an affinity search technique. Subse-
quently, for each cluster a prototype is defined ac-
cording to the affinity of the time series belonging to
it. The second step of this approach involves comput-
ing the DTW distances between the subclusters pro-
totypes. This distance measurement aims to repre-
sent the dissimilarity between the subgroups, in such
a way that the complexity is reduced as much as possi-
ble. Finally, similar subclusters are merged by means
of the k-medoids standard clustering method.
In addition, one of the approaches in the state-of-
the-art of TSCL is the well-known k-shapes (Paparri-
zos and Gravano, 2015). It is a partitional clustering
algorithm that aims to create homogeneous and well-
separated clusters through an iterative process. In a
similar way to k-means, k-shapes also performs two
main processes: 1) the assignment step; and 2) the re-
finement step. For the first one, k-shapes employs an
efficient adaptation of the cross-correlation measure
known as Shape-Based Distance (SBD). In the refine-
ment step, the centroids of the clusters are recom-
puted by solving an optimisation problem that min-
imises (or maximises) the sum of squared distances
(or squared similarities) to all the time series, found
significantly better than computing the average time
series. A key advantage of k-shapes is that it groups
time series based on their shape similarity, regardless
of differences in amplitude and phase. Thus, it pre-
serves the shapes of the time series while measuring
the distance between them.
Finally, a range of deep learning approaches have
been recently compared and analysed in (Lafabregue
et al., 2022). This study represents the first explo-
ration of deep learning techniques in TSCL. Hence,
three components have been studied: architecture,
clustering loss, and pretext loss. Through separate
assessments of each component it has been deter-
mined that a simple autoencoder architecture using a
reconstruction-based pretext loss is the best combina-
tion. Interestingly, the results also indicated that the
incorporation of clustering losses did not lead to a per-
formance increase. Therefore, its addition is not jus-
tified. Finally, authors discussed that more research is
required for improving the performance of these ap-
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
56
0 25 50 75 100 125 150
1
0
1
2
Arithmetic averaging
0 25 50 75 100 125 150
1
0
1
2
Original time series
0 25 50 75 100 125 150
1
0
1
2
MBA
0 25 50 75 100 125 150
1
0
1
2
DBA
GunPoint - Class 1
Figure 2: Visual differences between arithmetic averaging, the MBA approach and the DBA technique.
proaches.
4 COMPARISON OF OUR
METHOD
This section details the experimental settings used as
well as the results obtained, highlighting some of the
most important aspects to be analysed.
4.1 Experimental Settings
We compare MBA to alternative approaches using the
112 univariate, equal-length time series in the UCR
archive (Dau et al., 2019) for clustering. We use the
provided default train/test splits for all experiments.
We z-normalise all datasets prior to clustering. In
addition each model takes a value of k as a param-
eter. We set the value of k equal to the number of
unique class labels for each dataset. To measure per-
formance, we use three measures of the cluster labels:
CLustering ACCuracy (CL-ACC), is calculated
by dividing the number of correct predictions by the
total number of cases, similar to classification accu-
racy. To do this, once each value has been assigned to
a cluster the maximum accuracy from every permu-
tation of cluster and class value is taken. The cluster
that achieved the highest accuracy for each label is
then assigned that ground truth label (this is done us-
ing the Hungarian algorithm).
The Rand Index (RI) works by measuring the
similarity between two sets of labels such as the pre-
dicted and actual class values. The RI is the number
of pairs that agree on a label divided by the total num-
ber of pairs. One of the limiting factors of RI is that
the score is inflated, especially when the number of
clusters is high. The Adjusted Rand Index (ARI)
compensates for this by adjusting the RI based on the
expected scores on a purely random model.
The Mutual Information (MI) score, is a func-
tion that measures the agreement of the two cluster-
ings or a clustering and a true labelling, based on
entropy. Normalised Mutual Information (NMI)
rescales MI onto [0,1].
We compare the performance of the following 10
clustering algorithms:
1. k-means-DBA and k-means-MBA: k-means
clustering with DTW/MSM barycentre averaging
(i.e. DBA/MBA respectively), and DTW/MSM
distance assignment.
2. k-means-ED, k-means-DTW and k-means-
MSM: k-means clustering with arithmetic mean
for centres, and ED, DTW and MSM distance
assignment, respectively.
3. k-shapes (Paparrizos and Gravano, 2015).
4. TTC: Two-step Time series Clustering
(TTC) (Aghabozorgi and Wah, 2014)
5. k-medoids-ED, k-medoids-DTW and k-
medoids-MSM: k-medoids clustering with
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure
57

ED, DTW and MSM distance assignment,
respectively.
k-means, k-medoids and k-shapes at their core are
variations of Lloyds algorithm (Lloyd, 1982) and so
share many of the same parameters. As such this
means by keeping many of the same parameters con-
stant better insight can be gained when comparing
metric performance. The values used in our experi-
ment for Lloyds based models are given in table 1.
Max iters is a maximum number of iterations the
algorithm can run before forceful termination. This
limit will only ever be reached if the algorithm does
not converge (i.e. an iterations cluster values do not
change compared to the previous iteration). Our ex-
periments found many of the algorithm converged
(the cluster values did not change between iterations)
in under 20 iterations and so the limit of 300 is set
for redundancy. The init algo is the initialisation al-
gorithm used to select the initial centres. Our experi-
ment uses the most common initialisation technique:
random initialisation (MacQueen et al., 1967). This
technique consists of choosing the initial centres ran-
domly from the dataset. The rationale behind this is
that random selection is likely to pick points from
dense regions. Rerunning the model multiple times
with random initialisation and taking the best clus-
tering (as measured by the sum of distances to their
closest cluster centres) is the most common way of
initialising k-means (Bradley and Fayyad, 1998). The
number of reruns is defined by n init which for our
experiment is set to 10 as this is the most common
value we could find from other similar experiments
and is the default value for scikit-learn
3
k-means
clusterer.
The metric parameter is our first independent vari-
able. These different metrics have been outlined in
Sections 2.1 and 2.3. Finally the centroid compu-
tation defines the technique used to compute a new
cluster centre from a collection of time series. MBA,
DBA and arithmetic mean have been defined in Sec-
tion 2.3. MSM medoids is given in Eq 5 and DTW
medoids is similar but instead of using the MSM dis-
tance in Eq 5 the DTW distance is employed. Finally
shape extraction is a shape based averaging technique
using SBD.
Results are expressed using an adaptation of the
critical difference diagram (Dem
ˇ
sar, 2006), replacing
the post-hoc Nemenyi test with a comparison of all
classifiers using pairwise Wilcoxon signed-rank tests,
and cliques formed using the Holm correction (Garc
´
ıa
and Herrera, 2008; Benavoli et al., 2016).
3
https://scikit-learn.org/stable/
Table 1: Lloyds based algorithm variation parame-
ters. For all models max iters = 300, n init=10 and
init algo=“random”.
metric centroid computation
k-means-MBA MSM MBA
k-means-MSM MSM arithmetic mean
k-means-DBA DTW DBA
k-means-DTW DTW arithmetic mean
k-means-ED ED arithmetic mean
k-shapes SBD SE
k-medoids-MSM MSM MSM medoid
k-medoids-DTW DTW DTW medoid
4.2 k-means Variants
Figures 3, 4 and 5 show the average ranks of the
five k-means clusters against our two benchmark al-
gorithms, k-shapes and TTC, for CL-ACC, ARI and
NMI. We had to exclude the HandOutlines dataset and
hence reduce the number of datasets in our study to
111, given the computational time required by this
dataset. The pattern of performance is the same for
the three measures: k-means-MBA is significantly
better than the other six algorithms. This is our
primary support for using k-means-MBA. There is
some consistency in the clique membership. k-means-
MSM and k-means-DBA are always in the same
clique and TTC is significantly better than k-shapes.
k-means-MSM (k-means with MSM using arithmetic
averaging to find centroids) is not significantly differ-
ent to k-means-DBA (k-means with DTW and DTW
barycentre averaging). Moreover, k-means-DTW (k-
means with DTW and arithmetic averaging) is the
worst performing algorithm and is significantly worse
than k-means-ED (k-means with ED and arithmetic
averaging) (confirming results presented in (Holder
et al., 2022)).
Figure 6 shows the scatter plot in terms of CL-
ACC of k-means-MBA against k-means-MSM. It
demonstrates the improvement provided by using
MBA.
Table 2 quantifies the summary performance
statistics of these clusterers. Using MBA increase
MSM based k-means by approximately 2% for all
three metrics. The improvement does come at a cost:
both DBA and MBA take much longer than arithmetic
averaging. We run our experiments on a shared com-
puting cluster in parallel, but we can say that whilst
k-means-MSM and k-means-DTW take on average a
few minutes per problem, k-means-DBA and MBA
average over an hour.
Deep learning results presented in (Lafabregue
et al., 2022) are available from the associated web-
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
58
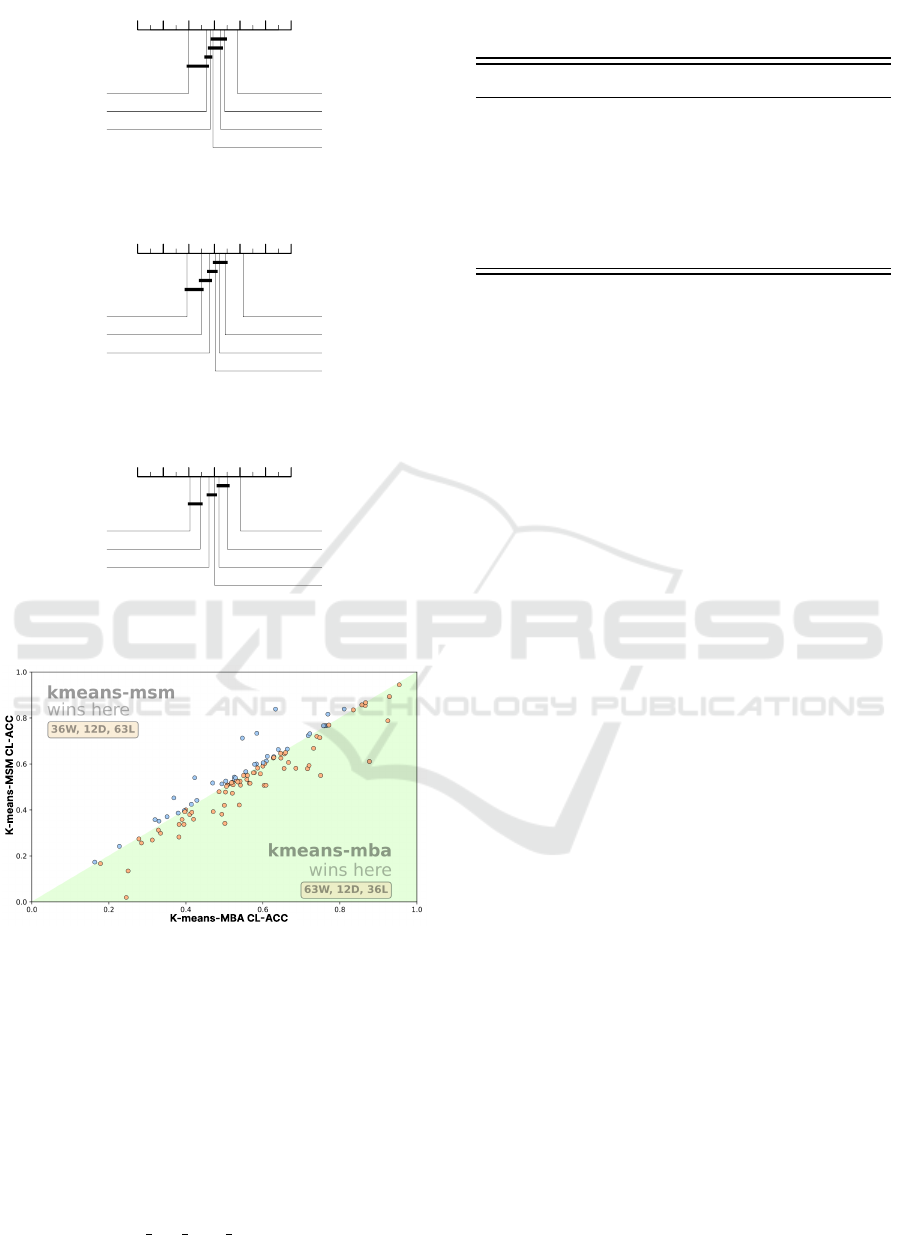
7 6 5 4 3 2 1
3.1081
k-means-MBA
3.6036
k-means-MSM
3.7568
TTC
4.0586
k-means-DBA
4.1667
k-means-ED
4.3063
k-shapes
5
k-means-DTW
Figure 3: Average ranks and cliques for CL-ACC for seven
clustering algorithms on 111 UCR datasets.
7 6 5 4 3 2 1
2.8739
k-means-MBA
3.5766
k-means-MSM
3.8018
k-means-DBA
3.9685
TTC
4.1937
k-means-ED
4.5135
k-shapes
5.0721
k-means-DTW
Figure 4: Average ranks and cliques for NMI for seven clus-
tering algorithms on 111 UCR datasets.
7 6 5 4 3 2 1
2.9775
k-means-MBA
3.4955
k-means-MSM
3.8243
k-means-DBA
3.991
TTC
4.2117
k-means-ED
4.5495
k-shapes
4.9505
k-means-DTW
Figure 5: Average ranks and cliques for ARI for seven clus-
tering algorithms on 111 UCR datasets.
Figure 6: CL-ACC scatter plot of k-means-MBA against k-
means-MSM. The yellow points show k-means-MBA was
better and the blue points show where k-means-MSM was
better.
site. They provide NMI results for over 300 different
clustering algorithms on the same UCR datasets we
use. These are not directly comparable, since they are
averaged over five runs and there may be other exper-
imental differences. However, they can give some in-
dication of relative performance. The best deep learn-
ing approach of the hundreds assessed, a cnn with
joint pretext loss and without clustering loss (key in
their results is res cnn joint None) achieved an aver-
age NMI of 0.3292. k-means-MBA obtained a com-
Table 2: Summary performance measures for k-means
based clustering.
CL-ACC ARI NMI
k-means-MBA 55.97% 23.91% 32.36%
k-means-MSM 54.07% 21.75% 29.87%
TTC 53.70% 22.38% 30.87%
k-means-DBA 53.96% 20.63% 29.86%
k-means-ED 51.59% 18.64% 27.31%
k-shapes 47.81% 11.24% 21.57%
k-means-DTW 48.95% 16.22% 23.30%
parable NMI of 0.3236.
4.3 k-means vs k-medoids
The alternative to using an elastic averaging technique
with k-means is to use k-medoids, which trade extra
memory to require fewer averaging operations. This
is because they require a precomputed pairwise dis-
tance matrix. This is not needed in k-means as only
distances to the generated centres each iteration are
needed. Holder et al. (2022) found that k-medoids
based on Lloyds algorithm was significantly better
than k-means for seven elastic distance measures. If
we use k-means-MBA, then the performance differ-
ence is removed and on NMI and ARI measures,
k-means-MBA significantly outperforms k-medoids-
MSM. Figures 7, 8 and 9 show the ranks by NMI, ac-
curacy and ARI of five k-means variants against three
k-medoids clusterers. The scatter plot of k-means-
MBA against k-medoids-MSM in terms of ARI is
shown in Figure 10.
As illustrated in Figures 7, 8, and 9, the choice
of elastic distance measure plays a critical role in the
performance of both k-means and k-medoids algo-
rithms. However, when we hold the distance met-
ric constant and solely vary the method for comput-
ing the cluster center - comparing k-medoids-MSM
against k-means-MBA we find that MBA significantly
enhances the quality of clustering across all evalua-
tion metrics. In addition this is true against the arith-
metic mean used in k-means-msm.
5 CONCLUSION
Distance functions play a crucial role in time series
machine learning, particularly with clustering where
it is commonly still used. A recent study found k-
medoids more effective than standard k-means on the
UCR data (Holder et al., 2022). However, k-medoids
has the problem of always requiring a complete dis-
tance matrix, and is usually slower than k-means. The
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure
59
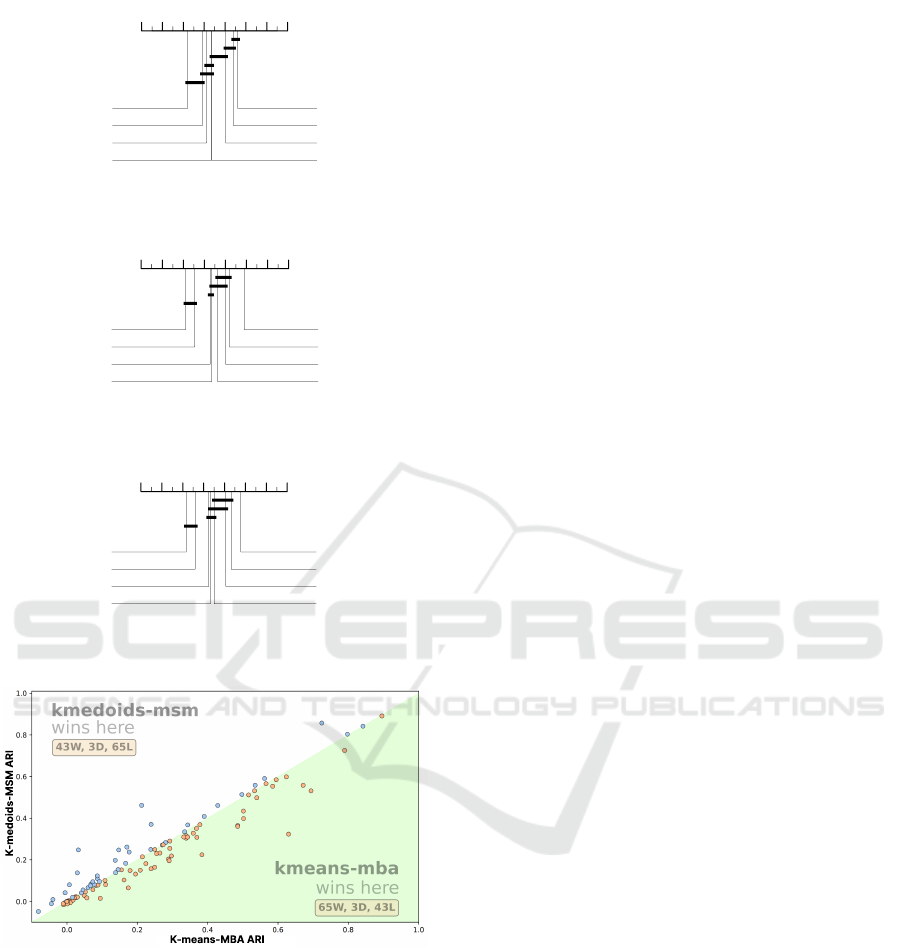
8 7 6 5 4 3 2 1
3.3919
k-means-MBA
3.5901
k-medoids-MSM
3.9685
k-means-MSM
4.6351
k-means-DBA
4.6396
k-means-ED
4.8829
k-medoids-DTW
5.0901
k-medoids-ED
5.8018
k-means-DTW
Figure 7: Average ranks and cliques for CL-ACC for five
k-means and three k-medoids methods.
8 7 6 5 4 3 2 1
3.0991
k-means-MBA
3.8063
k-medoids-MSM
4
k-means-MSM
4.3694
k-means-DBA
4.6577
k-medoids-DTW
4.7252
k-means-ED
5.4595
k-medoids-ED
5.8829
k-means-DTW
Figure 8: Average ranks and cliques for NMI for five k-
means and three k-medoids methods.
8 7 6 5 4 3 2 1
3.2207
k-means-MBA
3.6802
k-medoids-MSM
3.9369
k-means-MSM
4.4955
k-means-DBA
4.6757
k-medoids-DTW
4.7613
k-means-ED
5.3964
k-medoids-ED
5.8333
k-means-DTW
Figure 9: Average ranks and cliques for ARI for five k-
means and three k-medoids methods.
Figure 10: ARI scatter plot of k-means-MBA against k-
medoids-MSM. The yellow points show k-means-MBA was
better and the blue points show where k-medoids-MSM was
better.
problem with standard k-means is that arithmetic av-
eraging in the step to find centroids loses the elastic
information. A solution for k-means-DTW based on
barycentre averaging was proposed in (Petitjean et al.,
2011), known as DTW Barycentre Averaging (DBA).
However, it was also shown in (Holder et al., 2022)
that DTW is less effective at finding good clusterings
than alternative elastic distances. We have adapted
the Move-Split-Merge (MSM) distance (Stefan et al.,
2013) to be used with k-means barycentre averaging.
We have shown (see Table 2, Figures 7, 9, 8) that
using MSM with Barycentre Averaging (MBA) sig-
nificantly improves the results of k-means. k-means-
MBA is also significantly better than the popular k-
shapes and Two-step Time series Clustering (TTC)
algorithms, and similar to the best deep learning ap-
proach found through experimenting with over 300
models (see end of Section 4.2). We believe k-means-
MBA offers a good alternative to k-medoids-MSM for
Time Series Clustering (TSCL).
In future work we would seek to improve the time
complexity of MBA by employing a techniques such
as a bounding windows for MBA. Additionally we
have set out a framework to adapt other elastic dis-
tances for barycentre averaging. This could lead to
experimentation using other distances such as Time
Warp Edit (TWE) (Marteau, 2009) which achieved
similar performance to MSM for k-means (Holder
et al., 2022). Finally we would like to investigate
the possibility of using different elastic distances with
our framework to create an ensemble elastic distance
k-means clusterer similar to the elastic ensemble clas-
sifier proposed in (Lines and Bagnall, 2015).
ACKNOWLEDGEMENTS
This work has been supported by EPSRC (grant
reference EP/W030756/1) and partially sub-
sidised by “Agencia Espa
˜
nola de Investigaci
´
on
(Espa
˜
na)” (grant reference: PID2020-115454GB-
C22/AEI/10.13039/501100011033). David Guijo-
Rubio’s research has been subsidised by the
University of C
´
ordoba through grants to Public
Universities for the requalification of the Spanish
university system of the Ministry of Universities,
financed by the European Union - NextGenerationEU
(grant reference: UCOR01MS). Some of the exper-
iments were carried out on the High Performance
Computing Cluster supported by the Research and
Specialist Computing Support service at the Uni-
versity of East Anglia. We would like to thank all
those responsible for helping maintain the time series
classification archives and those contributing to open
source implementations of the algorithms.
REFERENCES
Aghabozorgi, S., Shirkhorshidi, A., and Wah, T. (2015).
Time-series clustering – a decade review. Information
Systems, 53:606–660.
Aghabozorgi, S. and Wah, T. Y. (2014). Clustering of
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
60

large time series datasets. Intelligent Data Analysis,
18:793–817.
Aghabozorgi, S., Ying Wah, T., Herawan, T., Jalab, H. A.,
Shaygan, M. A., and Jalali, A. (2014). A hybrid algo-
rithm for clustering of time series data based on affin-
ity search technique. The Scientific World Journal,
2014.
Bagnall, A., Lines, J., Bostrom, A., Large, J., and Keogh,
E. (2017). The great time series classification bake
off: a review and experimental evaluation of recent
algorithmic advances. Data Mining and Knowledge
Discovery, 31(3):606–660.
Benavoli, A., Corani, G., and Mangili, F. (2016). Should we
really use post-hoc tests based on mean-ranks? Jour-
nal of Machine Learning Research, 17:1–10.
Bradley, P. S. and Fayyad, U. M. (1998). Refining ini-
tial points for k-means clustering. In Proceedings
of the Fifteenth International Conference on Machine
Learning, ICML ’98, page 91–99, San Francisco, CA,
USA. Morgan Kaufmann Publishers Inc.
Brill, M., Fluschnik, T., Froese, V., Jain, B., Niedermeier,
R., and Schultz, D. (2019). Exact mean computation
in dynamic time warping spaces. Data Mining and
Knowledge Discovery, 33:252–291.
Chen, L. and Ng, R. (2004). On the marriage of Lp-norms
and edit distance. In proceedings of the 30th Interna-
tional Conference on Very Large Data Bases.
Dau, H., Bagnall, A., Kamgar, K., Yeh, M., Zhu, Y.,
Gharghabi, S., Ratanamahatana, C., Chotirat, A., and
Keogh, E. (2019). The UCR time series archive.
IEEE/CAA Journal of Automatica Sinica, 6(6):1293–
1305.
Dem
ˇ
sar, J. (2006). Statistical comparisons of classifiers
over multiple data sets. Journal of Machine Learning
Research, 7:1–30.
Ding, H., Trajcevski, G., Scheuermann, P., Wang, X., and
Keogh, E. (2008). Querying and mining of time series
data: Experimental comparison of representations and
distance measures. In proceedings of the 34th Inter-
national Conference on Very Large Data Bases.
Garc
´
ıa, S. and Herrera, F. (2008). An extension on “sta-
tistical comparisons of classifiers over multiple data
sets” for all pairwise comparisons. Journal of Ma-
chine Learning Research, 9:2677–2694.
Guijo-Rubio, D., Middlehurst, M., Arcencio, G., Silva,
D. F., and Bagnall, A. (2023). Unsupervised feature
based algorithms for time series extrinsic regression.
arXiv preprint arXiv:2305.01429.
Gupta, L., Molfese, D., Tammana, R., and Simos, P. (1996).
Nonlinear alignment and averaging for estimating the
evoked potential. IEEE Transactions on Biomedical
Engineering, 43(4):348–356.
Gusfield, D. (1997). Algorithms on Strings, Trees, and Se-
quences: Computer Science and Computational Biol-
ogy. Cambridge University Press.
Hills, J., Lines, J., Baranauskas, E., Mapp, J., and Bagnall,
A. (2014). Classification of time series by shapelet
transformation. Data Mining and Knowledge Discov-
ery, 28(4):851–881.
Holder, C., Middlehurst, M., and Bagnall, A. (2022).
A review and evaluation of elastic distance func-
tions for time series clustering. arXiv preprint
arXiv:2205.15181.
Holznigenkemper, J. and Seeger, C. K. B. (2023). On com-
puting exact means of time series using the move-
split-merge metric. Data Mining and Knowledge Dis-
covery, 37(2):595–626.
Lafabregue, B., Weber, J., Gancarski, P., and Forestier, G.
(2022). End-to-end deep representation learning for
time series clustering: a comparative study. Data Min-
ing and Knowledge Discovery, 36:29—-81.
Li, J., Izakian, H., Pedrycz, W., and Jamal, I. (2021).
Clustering-based anomaly detection in multivari-
ate time series data. Applied Soft Computing,
100:106919.
Liao, T. W. (2005). Clustering of time series data—a survey.
Pattern recognition, 38(11):1857–1874.
Lines, J. and Bagnall, A. (2014). Ensembles of elastic dis-
tance measures for time series classification. In pro-
ceedings of the 14th SIAM International Conference
on Data Mining.
Lines, J. and Bagnall, A. (2015). Time series classification
with ensembles of elastic distance measures. Data
Mining and Knowledge Discovery, 29:565–592.
Lines, J., Taylor, S., and Bagnall, A. (2018). Time se-
ries classification with HIVE-COTE: The hierarchi-
cal vote collective of transformation-based ensembles.
ACM Transactions Knowledge Discovery from Data,
12(5):1–36.
Lloyd, S. P. (1982). Least squares quantization in pcm.
IEEE Trans. Inf. Theory, 28:129–136.
MacQueen, J. et al. (1967). Some methods for classification
and analysis of multivariate observations. In Proceed-
ings of the fifth Berkeley symposium on mathematical
statistics and probability, volume 1, pages 281–297.
Marteau, P. (2009). Time warp edit distance with stiffness
adjustment for time series matching. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
31(2):306–318.
McDowell, I. C., Manandhar, D., Vockley, C. M., Schmid,
A. K., Reddy, T. E., and Engelhardt, B. E. (2018).
Clustering gene expression time series data using an
infinite gaussian process mixture model. PLoS com-
putational biology, 14(1):e1005896.
Middlehurst, M., Sch
¨
afer, P., and Bagnall, A. (2023). Bake
off redux: a review and experimental evaluation of
recent time series classification algorithms. arXiv
preprint arXiv:2304.13029.
Nikolaou, A., Guti
´
errez, P. A., Dur
´
an, A., Dicaire,
I., Fern
´
andez-Navarro, F., and Herv
´
as-Mart
´
ınez, C.
(2015). Detection of early warning signals in paleo-
climate data using a genetic time series segmentation
algorithm. Climate Dynamics, 44:1919–1933.
Paparrizos, J. and Gravano, L. (2015). k-shape: Efficient
and accurate clustering of time series. In Proceedings
of the 2015 ACM SIGMOD International Conference
on Management of Data, pages 1855–1870.
Paparrizos, J., Liu, C., Elmore, A., and Franklin, M.
(2020). Debunking four long-standing misconcep-
Barycentre Averaging for the Move-Split-Merge Time Series Distance Measure
61

tions of time-series distance measures. In proceed-
ings of the ACM SIGMOD international conference
on management of data.
Petitjean, F., Forestier, G., Webb, G. I., Nicholson, A. E.,
Chen, Y., and Keogh, E. (2016). Faster and more accu-
rate classification of time series by exploiting a novel
dynamic time warping averaging algorithm. Knowl-
edge and Information Systems, 47:1–26.
Petitjean, F., Ketterlin, A., and Gancarski, P. (2011). A
global averaging method for dynamic time warping,
with applications to clustering. Pattern Recognition,
44:678–.
Ratanamahatana, C. and Keogh, E. (2005). Three myths
about dynamic time warping data mining. In pro-
ceedings of the 5th SIAM International Conference on
Data Mining.
Shifaz, A., Pelletier, C., Petitjean, F., and Webb, G. (2023).
Elastic similarity and distance measures for multivari-
ate time series. Knowledge and Information Systems,
65(6).
Stefan, A., Athitsos, V., and Das, G. (2013). The Move-
Split-Merge metric for time series. IEEE Transactions
on Knowledge and Data Engineering, 25(6):1425–
1438.
Tan, C. W., Bergmeir, C., Petitjean, F., and Webb, G.
(2021). Time series extrinsic regression. Data Min-
ing and Knowledge Discovery, 35:1032––1060.
Zakaria, J., Mueen, A., and Keogh, E. (2012). Cluster-
ing time series using unsupervised-shapelets. In 2012
IEEE 12th International Conference on Data Mining,
pages 785–794. IEEE.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
62
