
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2
Variants’ Effects
Ruba Al Khalaf
1 a
, Anna Bernasconi
1 b
and Alberto Garc
´
ıa S.
2 c
1
Politecnico di Milano, Milan, Italy
2
Universitat Polit
`
ecnica de Val
`
encia, Valencia, Spain
Keywords:
Ontological Analysis, OntoUML, SARS-CoV-2, Variants Impacts.
Abstract:
The SARS-CoV-2 virus continuously accumulates genetic variation through mutations; mutations are the
virus’ way to achieve viral adaptation. Although the huge amount of information accumulated on the virus
during the COVID-19 pandemic, the knowledge that contributes to explaining and supporting the research
related to SARS-CoV-2 characteristics and evolution is not currently organized, nor systematized. Here, we
present OntoEffect, an ontology that captures and represents such information systematically. Specifically, we
aim to represent the dimensions of the virus and its mutations, discussing their impacts on the virus itself, as
well as on public health, prevention, and treatment protocols. Aiming to obtain ontological clarity in such a
complex domain, OntoEffect was built using OntoUML, an ontology-driven conceptual modeling language,
grounded on the Unified Foundational Ontology (UFO). In the highly specialized context of virology, we show
the powerful ability of ontological models to provide clear and precise explanations of a domain and allow its
shared understanding among stakeholders.
1 INTRODUCTION
The severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2) was initially detected in China in De-
cember 2019, causing the coronavirus disease 2019
(COVID-19). Consequently, after the continuous
spreading of the virus across the globe, the World
Health Organization (WHO) declared a Public Health
Emergency of International Concern (PHEIC) on 30
January 2020. Subsequently, on 11 March 2020, the
WHO officially classified the outbreak as a pandemic.
After more than three years into the pandemic,
specifically on 5 May 2023, the WHO announced that
the disease no longer met the criteria for a PHEIC due
to its established and ongoing nature. According to
the same announcement, this decision does not imply
that the pandemic itself has ended, but rather that the
global emergency it caused has subsided, at least for
the time being (World Health Organization, 2023).
The impact of the global COVID-19 pandemic has
affected all human lives worldwide on multiple levels.
Since its emergence, the SARS-CoV-2 virus has con-
a
https://orcid.org/0000-0002-5645-5886
b
https://orcid.org/0000-0001-8016-5750
c
https://orcid.org/0000-0001-5910-4363
tinued to evolve genetically with an expanding range
of variants. As it happens in other RNA viruses,
achieving genetic diversity is an essential aspect for
the survival and continuation of SARS-CoV-2; in-
deed, it brings viral survival, fitness, and pathogenesis
(Lauring et al., 2013; Mattenberger et al., 2021). The
observed impacts are continuously present, and they
vary depending on the specific variant that spreads
at given times of the outbreak. For example, it was
widely known that the viral transmission increased
with the Alpha variant of SARS-CoV-2 virus (Tanaka
et al., 2021). Therefore, collecting information about
the impacts of genetic variations in SARS-CoV-2 in
a systematic way, structuring it, and drawing the re-
lations between these impacts is of great importance.
In this research, we aim to fill this gap; specifically,
we draw the relationships between the virus and the
impacts of its mutations and variants formation over
the progression of the pandemic.
In the context of COVID-19, it has been recog-
nized that many ontologies are being used to ad-
dress the challenges related to the pandemic analyt-
ics – with different intents like structural representa-
tion and semantics of data, visualizing the status of
infections, querying, and ontology editing and extrac-
tion (Ahmad et al., 2021).
62
Al Khalaf, R., Bernasconi, A. and S., A.
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects.
DOI: 10.5220/0012183900003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 2: KEOD, pages 62-72
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

Mapping knowledge into standardized models,
such as ontologies, allows for a precise understand-
ing of a particular domain (Romanenko et al., 2022;
Guizzardi and Guarino, 2023), which can be used to
support data representation, integration, sharing, and
analysis.
Along this direction, we present OntoEffect to for-
mally represent the set of relevant concepts in this
domain. More specifically, we capture the concepts
related to the impact of viral variation over the over-
all outcome of the virus behavior. OntoEffect also
captures the relationships between these concepts in
a structured format that can be used to integrate data
and facilitate knowledge sharing among researchers.
As methodological support for this process, we
employ OntoUML, a well-founded ontology-driven
conceptual modeling language whose meta-model
complies with the theoretically well-grounded Uni-
fied Foundational Ontology (UFO) (Guizzardi, 2005).
OntoUML has previously been employed for on-
tological analysis of specialized life sciences do-
mains (Guizzardi et al., 2021; Garc
´
ıa S et al., 2022b)
and proven effective in improving the explanation of
complex domains with respect to traditional concep-
tual models (Garc
´
ıa S et al., 2022a; Verdonck et al.,
2019).
The scope of this research is broad, including
concepts that encompass -but are not restricted to-
epidemiology, immunology, and structural biology.
For achieving the highest possible clarification, we
follow the principles of the ontological unpacking
method (Bernasconi et al., 2022), proposed to analyze
and break down a complex concept or phenomenon
into its constituent parts based on an ontology that de-
fines the relationships between different types of en-
tities, finally resulting in a more granular understand-
ing of that complex system. In other words, the on-
tological unpacking method is a procedure in which
Ontology – as a field of study – is utilized to apply
the theory of ontological analysis, thereby uncover-
ing the ontological conceptual model that depicts an
information structure.
The remainder of the paper is structured as fol-
lows. Section 2 reviews related works, including
well-known ontologies concerning the SARS-CoV-2
domain, and motivates our effort. Section 3 overviews
the methodology used to conduct the ontological anal-
ysis, listing the most relevant OntoUML class and re-
lationship stereotypes. Section 4 presents the Onto-
Effect ontology and its advantages. Finally, Section
5 briefly describes the validation and concludes the
work.
2 RELATED WORK
Ontologies have gained attention during the pandemic
of COVID-19, and several researchers have built on-
tologies to solve diverse problems they were facing.
Within the list of UFO and OntoUML ontol-
ogy models presented in the OntoUML/UFO-catalog
(Barcelos et al., 2022) – a catalog for ontology-driven
conceptual modeling research (https://github.com/
unibz-core/ontouml-models), sharing all the models
that employ OntoUML for advancing ontology-driven
research – a pair of works related to what we are pre-
senting can be found:
• (Bernasconi et al., 2020) presented the Viral Con-
ceptual Model (VCM), which is a conceptual
model that focuses on viral genomic data struc-
turing to characterize genomic sequences.
• The same model went through an ontological un-
packing process (Guizzardi et al., 2021) achieved
with the use of the OntoUML language; the ob-
tained ontology represents the semantic areas of
Virus Infection, Tissue Sampiling, Virus Sequenc-
ing, and Virus Sequence Annotation. While the
VCM only served as a base for building a genomic
data structure, its ontological unpacking provided
a detailed analysis of the complex notions hidden
within the original VCM model.
• A domain ontology for modeling lockdown inter-
ventions during the COVID-19 pandemic was pre-
sented in (Fabio et al., 2021). It aims to provide a
comprehensive understanding of what a lockdown
entails by proposing an ontology-based modeling
approach. The authors highlight the need for a
well-grounded domain ontology that incorporates
real-world semantics to capture the complexities
and variations of lockdown measures.
Other related ontologies were developed with a dif-
ferent upper-level ontology, the Basic Formal Ontol-
ogy (BFO) (Arp et al., 2015). For instance, He et al.
presented the Coronavirus Infectious Disease Ontol-
ogy (CIDO, (He et al., 2020)) at the beginning of the
COVID-19 pandemic. CIDO is a community-based
open-source biomedical ontology that supports coro-
navirus disease knowledge, along with data standard-
ization, integration, sharing, and analysis. In addition
to being based on BFO, CIDO follows the Open Bi-
ological and Biomedical Ontologies (OBO) Foundry
Principles (Smith et al., 2007).
To our knowledge and after studying the domain,
we believe OntoEffect is unique in its scope and de-
sign approach as it systematically and comprehen-
sively describes the impact of viral variations on dif-
ferent levels and aspects.
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects
63

3 METHODOLOGY
The workflow for building OntoEffect is shown in
Figure 1. To identify the effects of interest that are
suited to be considered as entities in OntoEffect, we
first built a taxonomy (i.e., controlled vocabulary) of
effects. In this article, we propose the transformation
of the taxonomy into a structured ontology.
We started from the beginning of the COVID-
19 pandemic, following a knowledge acquisition pro-
tocol to search the literature from different sources
considering both peer-reviewed articles and preprints,
with more focus on the peer-reviewed ones. Such
sources are Google Scholar, PubMed, GISAID/COG-
UK reports, MedRxiv, and BioRxiv. For keywords
search and to achieve relativeness to our field of inter-
est, we used keywords composed of a virus-related
term (e.g., SARS-CoV-2), a known clinical impact
(e.g., infectivity), and a known mutation and/or vari-
ant (e.g., Alpha). The criteria for inclusion of an ef-
fect in the taxonomy of effect are its high representa-
tiveness in the literature and connection with at least
one non-synonymous mutation (or variant) of SARS-
CoV-2.
Along with the evolution of the virus, we con-
tinued curating the newest related publications in the
field and adding new terms to the taxonomy. To struc-
ture the final version of the taxonomy, we classified
the selected terms into four categories according to
the field of impact.
They include Epidemiology (clustering the im-
pacts of a variant on epidemiological features); Im-
munology (impacts on the host immune system); Vi-
ral kinetics and dynamics (of viral proteins and the
viral cell cycle); and Diagnosis, prevention, and
treatments (impacts on these measures).
Note that different versions of the taxonomy were
proposed in past works: a preliminary idea is in
(Al Khalaf et al., 2021); consolidated in (Alfonsi
et al., 2022) (employed within a providing a complete
schema representing COVID-19 knowledge and data
interplay); finalized in (Serna Garc
´
ıa et al., 2023b),
used within the CoVEffect system, which mines the
effects of SARS-CoV-2 mutations and variants based
on deep learning (Serna Garc
´
ıa et al., 2023a).
Figure 1: Schematic workflow to build OntoEffect.
Lastly, after an informal exploration of the do-
main supported by experts in molecular biology and
bioinformatics, we started the ontological unpacking
process (Guizzardi et al., 2021) leading to identifying
units of our model (classes), starting from the previ-
ously developed terms’ definitions. Then, we also de-
fined the relations among them. The transformation
process from each term in the taxonomy into OntoEf-
fect entities was performed manually and very care-
fully following the previously listed theories of On-
toUML, finally obtaining an ontological representa-
tion that reveals the ontological semantics of the built
taxonomy.
Here, we explored an alternative way to cluster
the effects of the taxonomy during the application of
the ontological unpacking process. We considered
what actor is impacted by each effect; this concep-
tual classification allowed a clearer representation of
the relations between the different entities of the on-
tology. As a consequence, we obtained four clusters:
i) Viral-related, which contains the impacts related to
the virus characterization regardless of the host; ii)
Host-related, which contains the impacts related to
the host characterization regardless of the virus; iii)
Virus-host interaction, which contains the impacts re-
lated to the interactions between the virus and the host
of interest; and iv) Disease and disease management,
which contains the impacts involved after the disease
is established in the host and those related to the man-
agement of the disease (diagnosis, prevention, and
treatment protocols).
3.1 UFO and OntoUML Language
Foundational ontologies are comprehensive systems
of domain-independent categories and their connec-
tions based on solid philosophical principles. These
systems consist of axioms that define various el-
ements such as objects, events, causality, spatial-
temporal relationships, dependencies, and more.
They play a crucial role in representing phenom-
ena across different material domains. Some exam-
ples of foundational ontologies include Descriptive
Ontology for Linguistic and Cognitive Engineering
(DOLCE) (Borgo and Masolo, 2009), General Formal
Ontology (GFO) (Poli et al., 2010), Suggested Upper
Merged Ontology (SUMO) (Niles and Pease, 2001),
and Unified Foundational Ontology (UFO) (Guiz-
zardi, 2005). By providing a conceptual framework,
they enable us to grasp the fundamental nature of spe-
cific domains and make it easier to identify connec-
tions and interesting associations and/or relationships
among data that align with their ontological counter-
parts (Amaral et al., 2021).
According to (Verdonck and Gailly, 2016), UFO
and UFO-based conceptual modeling language (On-
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
64

toUML) have emerged among the most used ap-
proaches in the field, and since the choice of an
upper-level ontology depends on the specific needs
and requirements of the application or domain be-
ing modeled. OntoUML is an ontology-driven con-
ceptual modeling language whose meta-model com-
plies with the ontological distinctions and axiomati-
zation of UFO (Guizzardi and Wagner, 2004). We
believe that UFO is the most suitable top-level ontol-
ogy to fulfill our needs of explanation of a complex
domain – as UFO informs OntoUML, which has been
used recently for this purpose in the life sciences do-
mains (Guizzardi et al., 2021; Bernasconi et al., 2022;
Garc
´
ıa S et al., 2022b). Therefore, we developed On-
toEffect using OntoUML language (Guizzardi, 2005).
OntoUML is an extension of the Unified Mod-
eling Language (UML) (Booch et al., 1997) based
on UFO. It incorporates ontological concepts and fo-
cuses on modeling not only software systems but also
the underlying conceptual structure and semantics of
the domains. OntoUML aims to represent entities, re-
lationships, and categories in a way that aligns with
formal ontology principles, enabling a more compre-
hensive understanding of complex systems by model-
ing them into ontologies. There are three theories in
OntoUML specification listed as follows:
• Types and Individuals, OntoUML is founded
upon the essential distinction between types and
individuals. Types are abstract things that are
formed to assist in the perception and categoriza-
tion of the world around us. These things serve as
bundles of characteristics that can be anticipated
to be encountered in other specific entities - indi-
viduals which it can be endurants (e.g., objects)
or perdurants (e.g., events).
• Identity Principle, is a sort of function that is be-
ing used to differentiate two individuals, and ev-
ery individual must have exactly one identity prin-
ciple (e.g., a fingerprint is an identity principle of
a person).
• Rigidity Principle, endurants can be either rigid
when an individual instantiates the same type in
all possible scenarios; or anti-rigid when it instan-
tiates a type only in a specific scenario.
Table 1 and Table 2 schematically represent the onto-
logical distinctions used in OntoEffect, both as class
stereotypes and relationships.
4 OntoEffect ARCHITECTURE
Here, we present the four views of OntoEffect,
corresponding to the listed clusters. The mas-
ter view that connects all these views (along with
the complete views) is available at https://tinyurl.
com/5n8c7k8t–not shown in the paper due to space
limitations. The schemata were drawn with Vi-
sual Paradigm Community Edition (v17.0) and the
OntoUML plugin, release 0.5.3 (https://github.com/
OntoUML/ontouml-vp-plugin). In the following sub-
sections, we wrote in bold-type the classes (i.e., enti-
ties) of the ontology and italic-type the relations be-
tween the entities.
4.1 Viral-Related View
We present a partition of the viral-related view in Fig-
ure 2. OntoEffect aims to represent the impact of viral
variation over the overall outcome of the virus behav-
ior; thus, the SARSCOV2 entity represents the virus
of our interest, which is already a mutated version
of the virus. We note that OntoEffect in its current
state does not include entities related to the formation
of a SARS-CoV-2 variant nor the composition of its
sequence. This is already modeled in the Virus Se-
quence Annotation view of the ontological unpacking
of the Viral Conceptual Model (VCM) (Bernasconi
et al., 2022; Guizzardi et al., 2021), which can be eas-
ily integrated with OntoEffect.
SARSCOV2 is an instance of VirusSpecies,
which itself is an instance of the Species.
Several SARSCOV2 can group forming
SARSCOV2Collective. A virus has a viral
membrane, a genome, and a proteome represented
as ViralMembrane, SARSCOV2Genome, and
SARSCOV2Proteome respectively.
A SARSCOV2Gene is a member of
SARSCOV2Genome, and it is linked through a
material relation with SARSCOV2Protein which
is a member of SARSCOV2Proteome. As an
example of SARSCOV2Protein, we present Spike
protein that is composed of two subunits S1 (which
contains the receptor binding domain, RBD) and
S2. Moreover, A viral protein could have in its
structure a region called Epitope that is a region
responsible for provoking an immune response in
the host. Each protein has its own characteristics,
such as Flexibility, Stability, Functioning, and
StructuralOptimization. Moreover, a protein might
include multiple NonSynonymousMutation.
Different types of Interactions could occur either
inside the same molecule (known as Intramolecular-
Interactions) or between molecules (known as In-
termolecularInteraction). Instead, IntraviralPro-
teinProteinInteractions is a bundle of relational dis-
positions connecting SARSCOV2Protein with itself
through an association.
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects
65

Table 1: The types of OntoUML class stereotypes used in OntoEffect.
Stereotype Definition
Type are types whose instances are themselves types.
Category is a rigid mixin that does not require a dependency to be specified. It is used to aggregate
essential properties of individuals who follow different identity principles.
Collective is used to represent rigid concepts that provide an identity principle for their instances.
The main characteristic of a hCollectivei is that it has a homogenous internal structure.
Kind is used to represent rigid concepts that provide an identity principle for their instances
and do not require a relational dependency. A hKindi represents a Functional Complex;
its parts contribute in different ways to its functionality.
Subkind is a construct used to represent rigid specializations of identity providers. By default, its
usage does not require a relational dependency.
Phase is used to represent anti-rigid subtypes of identity providers that are instantiated by
changes in intrinsic properties (e.g., the age of a person).
Role is a construct used to represent anti-rigid specializations of identity providers that are
instantiated in relational contexts. Like hPhasei , all instances of a hRolei must follow
the same identity principle.
Relator is used to represent truth-makers of material relations, i.e., the “things” that must exist
in order for two or more individuals to be connected by material relations.
Mode is a particular type of intrinsic property that has no structured value. Modes are individ-
uals that existentially depend on their bearers.
Quality is a particular type of intrinsic property that has a structured value. Qualities are things
that are existentially dependent on the things they characterize.
Other characteristics of the virus are the Incuba-
tionPeriod, which is the interval between the moment
a host is infected and the onset of the disease, and the
ViralVirulence which is the competence of the infec-
tious agent to produce pathologic effects.
Finally, a virus could incubate in a specific Cul-
ture, which can be specified through CultureCon-
ditions (such as Temperature), CultureMediums,
and the employed CellType and CellLine. We rep-
resent this incubation through a ViralCulture relator
in which we can assess the ViralFitness of the virus
in a specific environment.
4.2 Host-Related View
In Figure 3, a partition of the host-related view is pre-
sented. A Human is an instance of the HostSpecies,
which itself is an instance of the Species. This schema
comprises two sections; the right one describes the
different possible phases a human can go through,
while the left describes the human’s molecules, cells,
and systems.
A Sample is taken from a Human and is mediated
through a Diagnosis relator that results in a TestRe-
sult which could be a Positive or Negative which
characterize if the Human is in Infected phase or
Notinfected phase, respectively. A Human is in the
Infected phase and plays a HumanHost role. This
phase could have different generalizations:
• Livinig or Deceased host.
• Treated (i.e., playing a treatedIndividual role)
or Untreated host.
• According to the host’s history, the infected host
could be FirstTimeInfected or ReInfected with
the virus.
The NotInfected host plays a HumanReceiver role,
that is the human who has the ability to get the viral
infection from another host. Therefore, a material re-
lation links the HumanReceiver to the HumanHost,
and both are mediated through a ViralTransmission
relator. The NotInfected host is either NeverInfected
with the virus or got the infection earlier and already
Recovered from it.
Also, a Human could receive a vaccine and enter
the Vaccinated phase playing a VaccinatedIndivid-
ual role, or could stay UnVaccinated.
Human is more complex species than a Virus;
hence, it is composed of different systems, e.g. Im-
muneSystem which involves WhiteBloodCells. A
subkind of the WhiteBloodCells is Lymphocytes
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
66
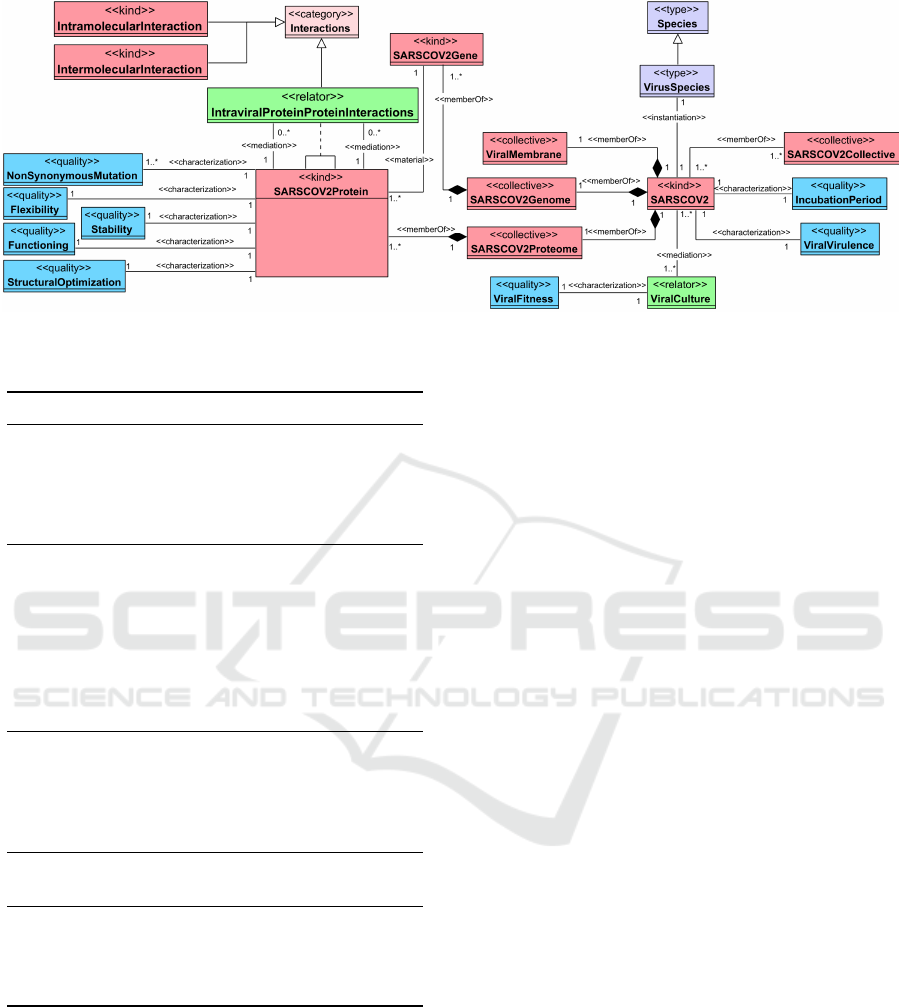
Figure 2: A partition of the viral related view.
Table 2: The types of relations used in OntoEffect.
Relation Definition
Material hMateriali relations have a mate-
rial structure on their own. The
relata of a material relation are
mediated by individuals that are
called relators.
Mediation A relation between a hRelatori
and the entities it connects. It is
a type of existential dependence
relation. It can be derived from
the relation between the relata
and the individuals that compose
the relator and that inhere in the
relata.
Characterization A relation between a bearer type
and its feature. A feature is the
intrinsic (inherent) moment of
its bearer type, and thus existen-
tially dependent on the bearer.
ComponentOf Is a parthood relation between
two complexes.
MemberOf Is a parthood relation between
a functional complex or a
hCollectivei (as a part) and a
hCollectivei (as a whole).
which play a role in the immune response for fight-
ing infectious agent (Wheelock and Toy, 1973). The
two main types of lymphocytes are Tcells and Bcells.
A HumanGene is a member of HumanGenome,
which is found exclusively in the HumanCells. The
HumanGene is linked through a material relation
with HumanProtein which is a member of Human-
Proteome. Also, HumanGene and HumanProtein
are participants of the HUMANProteinCodingPro-
cess.
The HostInfectedCells are specifically the group
of HumanCells that are infected with the virus (or
have the ability to be infected). Each of these cells has
a membrane (represented as HostCellMembrane)
that encloses different organelles, apparatus, and pro-
teins (represented as InfectedCellProtein).
Since the HumanProtein is a big class of
molecules, we only represent:
• InfectedCellProteins.
• ImmuneSystemProteins, which are the proteins
of the immune system, e.g. Immunoglobulin and
Antibody.
• HumanEnzyme like the Angiotensin-converting
enzyme 2 (ACE2) which plays a role as a Recep-
tor of the virus. Other enzymes are the Human-
Protease, like TMPRSS2 (i.e., Transmembrane
protease serine 2, playing a role as Transmem-
braneProtease), Furin (playing a role as Pro-
ProteinConvertase), and Cathepsin L.
Finally, we have the Neutralization category, which
involves different types of NeutralizingAgents that
can neutralize the virus and prevent it from enter-
ing the host cell. Such agents (besides the Anti-
bodies that are naturally part of the ImmuneSystem)
are TherapeuticAntibody as the laboratory manu-
factured antibodies, ConvalescentSera as the blood
serum that is obtained from an individual who has Re-
covered from a previous SARSCOV2 infection, and
VaccinatedSera as the blood serum that is obtained
from an individual who has been Vaccinated with one
of the SARS-CoV-2 vaccines. Finally, measurements
of the sensitivity of the virus to each of these agents
can be characterized as qualities of SensitivityToAn-
tibodies, SensitivityToConvalescentSera, and Sen-
sitivityToVaccinatedSera.
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects
67
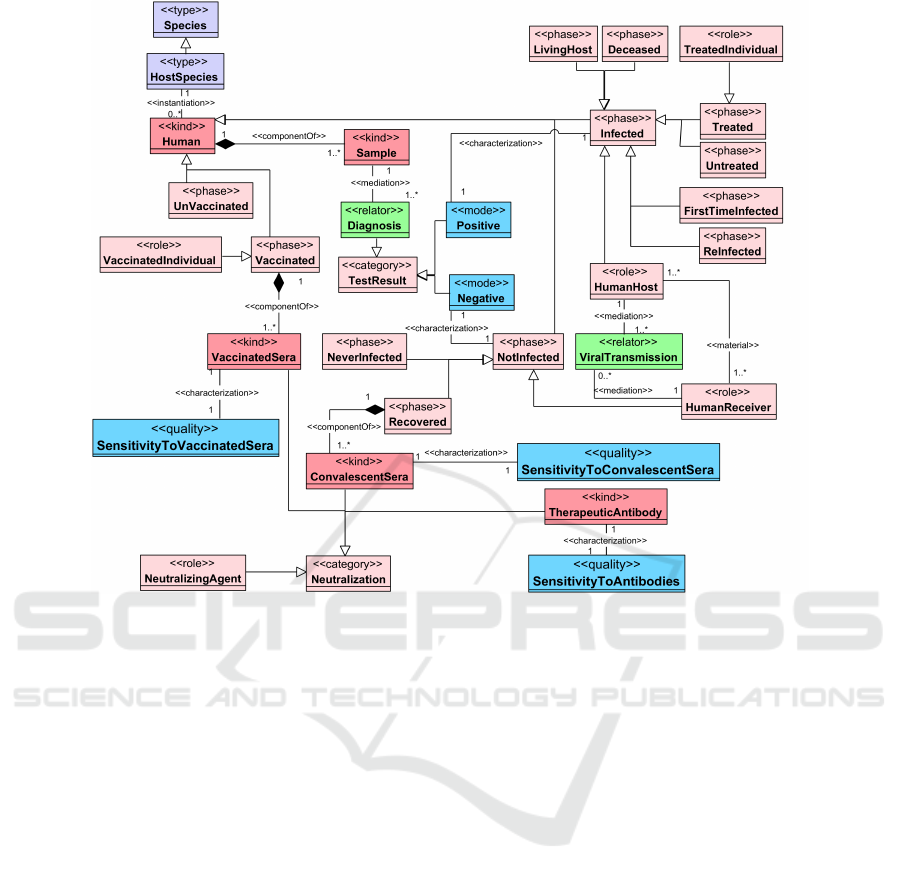
Figure 3: A partition of the host-related view.
4.3 Virus-Host Interaction View
In Figure 4, a partition of the virus-host interac-
tion view is presented. To establish a viral infec-
tion, there are plenty of interactions between the virus
and its host that are needed. We collected these in-
teractions in one category that is called HostVirus-
Interaction, which is an instance of Interacrions.
ViralInfectionFormation, is a relator that mediates
SARSCOV2Collective and Human. To say that
a cell is infected with a virus, three main steps
must have happened (represented as instances of the
HostVirusInteraction):
1. ViralEntry, a category that represents the pro-
cess of a virus entering a host cell through one of
two entry pathways; CellSurfaceEntry and En-
dosomalEntry. The efficiency of this process can
be measured, and it is represented with a quality
called EntryEfficiency.
2. ViralReplication, after that the virus enters the
cell or its genome enters the host cell (through
EndosomalEntry or CellSurfaceEntry, respec-
tively). The SARSCOV2genome will be repli-
cated and translated into SARSCOV2Protein
through SARSCOV2ProteinCodingProcess
with the help of InfectedCellProtein.
3. ViralRelease, after that a host cell becomes in-
fected with the virus and starts to replicate the
virus and biosynthesis it, this cell becomes a
virus-producer cell which releases the mature
virus to infect other host cells in a process called
ViralRelease.
We are focusing on the viral entry process, as it is the
initial step of the ViralInfectionFormation. Prevent-
ing this process, either by a vaccine or a treatment,
means preventing the disease. Both entry pathways
start with the virus binding to the host cell receptor
in which the RBD of the Spike protein binds to the
ACE2 receptor. This process is presented through
the BindingToHostReceptorInteraction relator (has
BindingAffinityToHostReceptor as a quality). This
binding induces conformational changes in the Spike,
which is followed by the S2 cleavage through Cleav-
ingInteraction. Depending on the entry route, the S2
is cleaved by different HumanProtease; using TM-
PRSS2 in the CellSurfaceEntry or using Cathepsin
L in the other pathway. Regardless of which entry
pathway the virus follows, a membrane fusion must
occur that is represented through MembraneFusion-
Interaction relator that mediates ViralMembrane
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
68
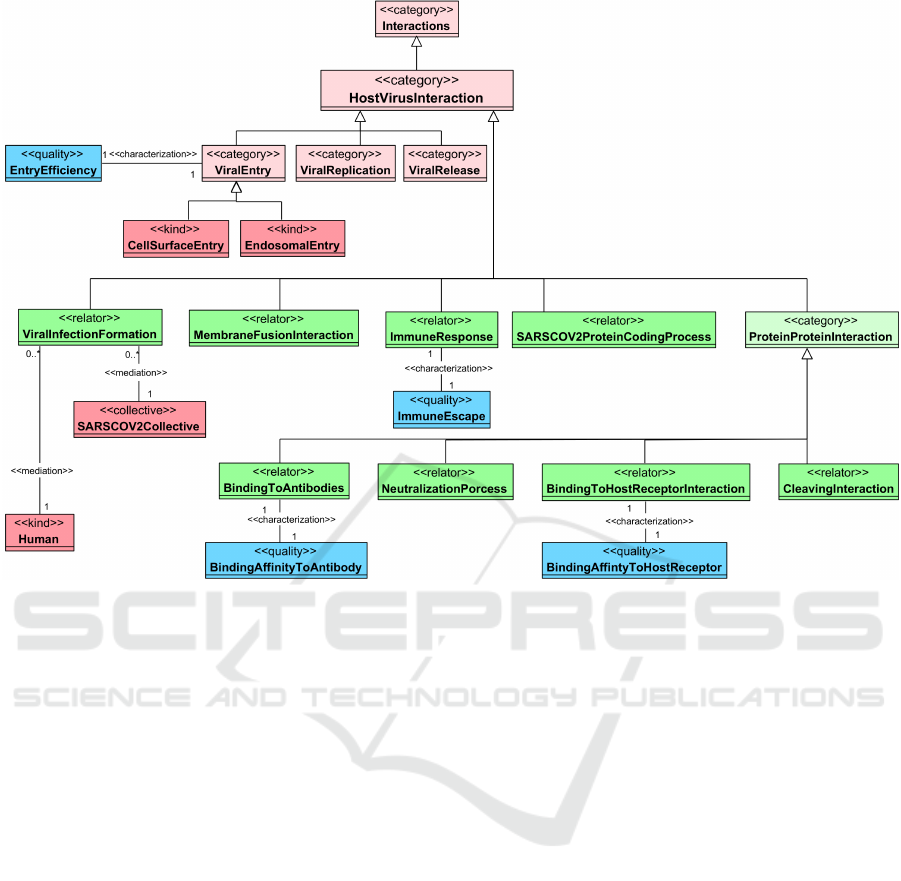
Figure 4: A partition of the Host-virus interaction view.
and HostCellMembrane since the fusion of these
two membranes is crucial to uncoating the genome
of SARS-CoV-2 to start the ViralReplication.
Just like MERS-CoV, the spike protein of SARS-
CoV-2 virus is cleaved by Furin into S1 and S2 sub-
units during their biosynthesis; therefore, this cleav-
age is also mediated through the CleavingInterac-
tion.
Another class of HostVirusInteraction repre-
sents the ImmuneReponse of the host toward the
presence of the virus inside it. To have such a re-
sponse, Epitope from a viral protein must be rec-
ognized by an Antibody or Immunoglobulin. The
immune response is a complex cascade of immune
mechanisms recruiting various immune cells and
chemical mediators. Therefore, we simplified it into
one relator called ImmuneResponse. A virus could
obtain the power to escape the host immune system
and minimize or completely vanish the ImmuneRe-
sponse; this phenomenon can be measured and is rep-
resented as ImmuneEscape. The binding between
Epitope and Antibody is mediated through Bind-
ingToAntibody relator (has BindingAffinityToAnti-
body as a quality). Moreover, there is Neutraliza-
tionProcess relator that mediates the neutralization of
SARSCOV2 inside a HumanHost by the previously
mentioned NeutralizingAgent.
Since BindingToHostReceptorInteraction,
CleavingInteraction, NeutralizationProcess, and
BindingToAntibody are interactions between a
HumanProtein and a SARSCOV2Protein, we
generalized them in one category; ProteinProteinIn-
teraction.
4.4 Disease and Disease Management
View
In Figure 5, we present a partition of the view of On-
toEffect that characterizes the disease and its manage-
ment through diagnosis, treatment, and prevention.
We started this view form Human that could admit
into a Hospital (for many reasons, e.g., suspicion of
infection, receiving treatment or vaccine, or having
severe symptoms that require hospitalization) through
HospitalAdmission relator; also, many factors could
increase or decrease the RiskOfHospitalization of a
Human in a Hospital.
COVID19 is the Disease caused by SARSCOV2
infection and beside it an Infected host could suf-
fer from (or have) OtherDisease. Such a scenario
might lead to complications between COVID19 and
that OtherDisease and worsen the overall health sta-
tus of the Infected host; this was mediated through a
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects
69

Complications relator. The FatalityRate is a quality
that is calculated from the InfectedCollective and is
the proportion of persons who Deceased after the vi-
ral infection over the number of confirmed Infected
people.
COVID19, as any other viral disease, is charac-
terized by InfectionDuration, which is the duration
of the host being infected with the virus. This disease
has different ranges of symptoms, which can be rep-
resented as phases which are: WithoutSymptoms,
MildSymptoms, ModerateSymptoms, and Sever-
Symptoms. The symptoms of the disease determine
its severity; therefore, the different phases of symp-
toms are associated with the DiseaseSeverity rela-
tor. Furthermore, an InfectedSample can be obtained
from the HumanHost to measure the ViralLoad in-
side it; ViralLoad is the number of copies of RNA of
a given virus per milliliter.
Lastly, there is a risk of a second infection, repre-
sented as Reinfection relator, by the same virus after
being Recovered from (or during the course of) a pri-
mary infection; such risk can be measured, and it is
represented with the RiskOfReinfection quality.
For disease diagnosis, as previously mentioned,
a Sample is taken from a Human and mediated
with Diagnostics collective through a Diagnosis re-
lator. A sample is used in a diagnostic test to de-
tect either the presence of SARSCOV2Protein (with
AntigenicTest, or MolecularTestPCR which has a
CtValue) or the presence of ImmuneSystemPro-
tein (with SerologicalTest) through DetectionPro-
cess. Such Diagnostics have measurements for their
effectiveness in detecting the presence of its target
molecules; we characterized it with a quality called
EffectivnessOfDiagnostics.
Considering the treatment of the disease, a cate-
gory of AntiviralDrug is mediated with the Treat-
ment relator and associated with different kinds
of drugs such as EntryInhibitors, AttachmentIn-
hibitors, and PolymeraseInhibitors. Another me-
diation relation with the Treatment relator is with
TreatedIndividual that is the role played by an
Treated host, and it represents an Infected host who
is Treated with any kind of AntiviralDrugs. The ef-
ficacy of a drug to treat the infected host can be mea-
sured, and it is represented as EffectivnessOfDrug
quality.
Finally, considering the prevention protocols of
the disease, we are addressing the Vaccination rather
than other protocols (like self-isolation) since we are
focusing on the effects of the viral variation on the
outcome of the virus and its resulting disease. A Hu-
man enters the Vaccinated phase and plays a Vacci-
natedIndividual role if a Vaccine was received, and
the Vaccination relator mediates this process. A Vac-
cine is a NeutralizingAgent, and there are multiple
kinds of vaccines that were developed to prevent the
disease, e.g., Pfizer, Moderna, and AstraZeneca.
The efficacy of a vaccine to neutralize the virus and
prevent viral infection formation can be measured,
and it is represented as EffectivnessOfVaccine qual-
ity.
5 VALIDATION AND
CONCLUSIONS
This paper presents OntoEffect, an OntoUML-based
ontology that explains SARS-CoV-2 variants’ effects.
As a core part of the research, we presented the On-
toUML model that represents classes and relation-
ships describing the domain. OntoEffect aims to pro-
vide a detailed analysis of the complex notions of the
impacts of SARS-CoV-2 variations. The main chal-
lenges of this effort are related to the complexity of
describing the biological domain with knowledge rep-
resentation artifacts. Many concepts are entangled or
nested, and their semantics are non-homogenous; thus
a deep understanding is needed and can be achieved
through an ontological representation.
Our current version of the ontology is undergo-
ing continuous updating of information. We designed
semi-supervised methods for extracting content from
the CORD-19 (Wang et al., 2020) literature cor-
pus to continuously collect instances of knowledge-
related entities (Serna Garc
´
ıa et al., 2023a). Un-
derstanding and mastering the complex domain of
SARS-CoV-2 mutation effects has proven extremely
useful, in our data-driven analysis of SARS-CoV-2
phenomena, where data science methods were in-
tegrated with knowledge-driven hypothesis making
(Bernasconi et al., 2021b; Bernasconi et al., 2021a).
As a preliminary assessment of this effort, we
presented the ontology to three domain experts, who
have a background in molecular biology and previ-
ously worked on several projects with viral genomic
sequences; they were asked to evaluate the ontology
for accuracy, relevance, and completeness. We had
two focus groups with the experts, resulting in a se-
ries of observations that will inspire our next refine-
ment of the views. Generally, our modeling choices
were confirmed (after we properly guided their un-
derstanding of OntoUML stereotypes). This work re-
quires broader validation by means of empirical stud-
ies, along the lines of (Verdonck et al., 2019; Garc
´
ıa S
et al., 2022a; Garc
´
ıa S. et al., 2023)–showing the
benefits of this representation when the explanation
of such a complex domain is required to non-expert
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
70
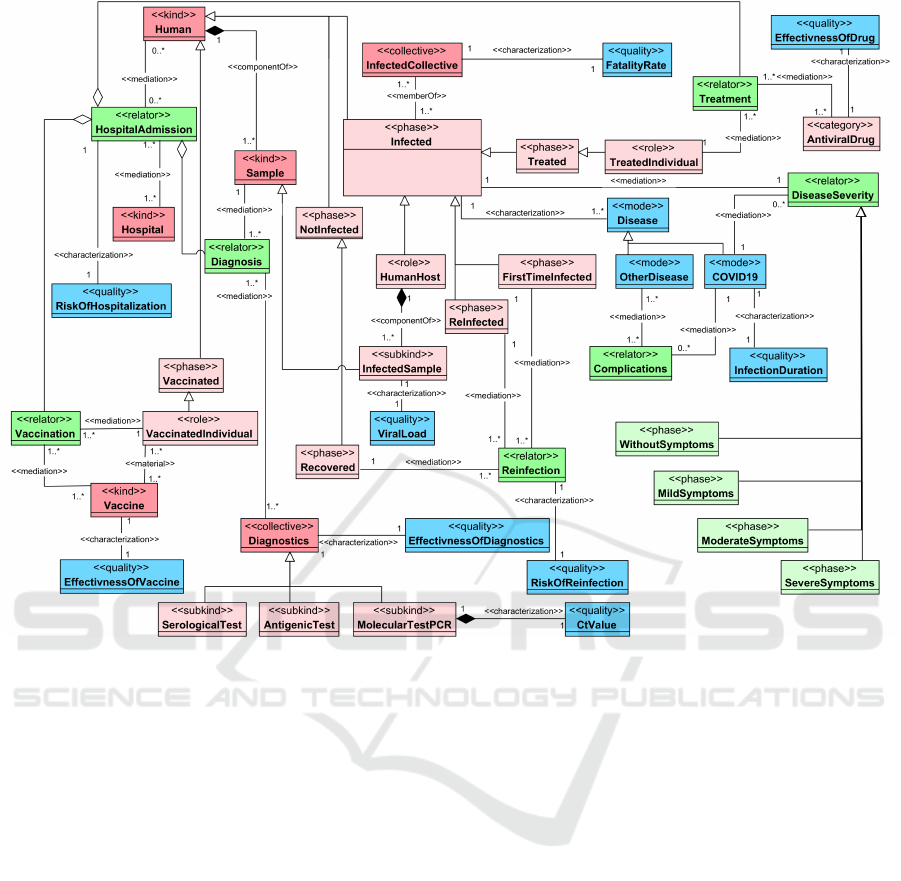
Figure 5: A partition of the disease and disease management view.
stakeholders. All in all, we here defend that OntoEf-
fect presents a first step towards the full understand-
ing of SARS-CoV-2 impacts to non-domain experts,
powering intra-domain knowledge exchange and in-
formation interoperability. This schema will further
enable knowledge-based applications for annotation
and reasoning.
REFERENCES
Ahmad, A., Bandara, M., Fahmideh, M., Proper, H. A.,
Guizzardi, G., and Soar, J. (2021). An overview of on-
tologies and tool support for COVID-19 analytics. In
2021 IEEE 25th International Enterprise Distributed
Object Computing Workshop (EDOCW), pages 1–8.
IEEE.
Al Khalaf, R., Alfonsi, T., Ceri, S., and Bernasconi, A.
(2021). Cov2k: a knowledge base of sars-cov-2 vari-
ant impacts. In Research Challenges in Information
Science: 15th International Conference, RCIS 2021,
Limassol, Cyprus, May 11–14, 2021, Proceedings,
pages 274–282. Springer.
Alfonsi, T., Al Khalaf, R., Ceri, S., and Bernasconi, A.
(2022). Cov2k model, a comprehensive representation
of sars-cov-2 knowledge and data interplay. Scientific
Data, 9(1):260.
Amaral, G., Baiao, F., and Guizzardi, G. (2021). Founda-
tional ontologies, ontology-driven conceptual model-
ing, and their multiple benefits to data mining. Wiley
Interdisciplinary Reviews: Data Mining and Knowl-
edge Discovery, 11(4):e1408.
Arp, R., Smith, B., and Spear, A. D. (2015). Building on-
tologies with basic formal ontology. Mit Press.
Barcelos, P. P. F., Sales, T. P., Fumagalli, M., Fonseca,
C. M., Sousa, I. V., Romanenko, E., Kritz, J., and
Guizzardi, G. (2022). A fair model catalog for
ontology-driven conceptual modeling research. In
Conceptual Modeling: 41st International Conference,
ER 2022, Hyderabad, India, October 17–20, 2022,
Proceedings, pages 3–17. Springer.
Bernasconi, A., Canakoglu, A., Pinoli, P., and Ceri, S.
(2020). Empowering virus sequence research through
conceptual modeling. In Conceptual Modeling: 39th
International Conference, ER 2020, Vienna, Austria,
November 3–6, 2020, Proceedings 39, pages 388–
402. Springer.
Bernasconi, A., Guizzardi, G., Pastor, O., and Storey, V. C.
(2022). Semantic interoperability: ontological un-
packing of a viral conceptual model. BMC bioinfor-
matics, 23(Suppl 11):491.
Bernasconi, A., Gulino, A., Alfonsi, T., Canakoglu, A.,
Pinoli, P., Sandionigi, A., and Ceri, S. (2021a).
OntoEffect: An OntoUML-Based Ontology to Explain SARS-CoV-2 Variants’ Effects
71

VirusViz: comparative analysis and effective visual-
ization of viral nucleotide and amino acid variants.
Nucleic Acids Research, 49(15):e90–e90.
Bernasconi, A., Mari, L., Casagrandi, R., and Ceri, S.
(2021b). Data-driven analysis of amino acid change
dynamics timely reveals SARS-CoV-2 variant emer-
gence. Scientific Reports, 11(1):21068.
Booch, G., Rumbaugh, J., and Jackobson, I. (1997). The
Unified Modeling Language User Guide. Addison-
Wesley Object Technology Series – Pearson Educa-
tion (US).
Borgo, S. and Masolo, C. (2009). Handbook on Ontologies.
Springer Verlag.
Fabio, I., Amaral, G. C., Griffo, C., Bai
˜
ao, F., and Guiz-
zardi, G. (2021). “What exactly is a lockdown?”: to-
wards an ontology-based modeling of lockdown inter-
ventions during the COVID-19 pandemic. In ONTO-
BRAS, pages 151–165.
Garc
´
ıa S, A., Bernasconi, A., Guizzardi, G., Pastor, O.,
Storey, V. C., and Costa, M. (2022a). An Initial Empir-
ical Assessment of an Ontological Model of the Hu-
man Genome. In Advances in Conceptual Modeling,
pages 55–65, Cham. Springer International Publish-
ing.
Garc
´
ıa S., A., Bernasconi, A., Guizzardi, G., Pastor, O.,
Storey, V. C., and Panach, I. (2023). Assessing the
value of ontologically unpacking a conceptual model
for human genomics. Information Systems, page
102242.
Garc
´
ıa S, A., Guizzardi, G., Pastor, O., Storey, V. C., and
Bernasconi, A. (2022b). An ontological characteri-
zation of a conceptual model of the human genome.
In Intelligent Information Systems: CAiSE Forum
2022, Leuven, Belgium, June 6–10, 2022, Proceed-
ings, pages 27–35. Springer.
Guizzardi, G. (2005). Ontological foundations for struc-
tural conceptual models. CTIT, Centre for Telematics
and Information Technology, Twente, Netherlands.
Guizzardi, G., Bernasconi, A., Pastor, O., and Storey, V. C.
(2021). Ontological unpacking as explanation: the
case of the viral conceptual model. In Conceptual
Modeling: 40th International Conference, ER 2021,
Virtual Event, October 18–21, 2021, Proceedings 40,
pages 356–366. Springer.
Guizzardi, G. and Guarino, N. (2023). Semantics, ontology
and explanation. arXiv preprint arXiv:2304.11124.
Guizzardi, G. and Wagner, G. (2004). A unified founda-
tional ontology and some applications of it in business
modeling. In Proceedings of the Open InterOp Work-
shop on Enterprise Modelling and Ontologies for In-
teroperability, co-located with CAiSE’04 Conference.
He, Y., Yu, H., Ong, E., Wang, Y., Liu, Y., Huffman, A.,
Huang, H.-h., Beverley, J., Hur, J., Yang, X., et al.
(2020). CIDO, a community-based ontology for coro-
navirus disease knowledge and data integration, shar-
ing, and analysis. Scientific data, 7:181.
Lauring, A. S., Frydman, J., and Andino, R. (2013). The
role of mutational robustness in RNA virus evolution.
Nature Reviews Microbiology, 11(5):327–336.
Mattenberger, F., Vila-Nistal, M., and Geller, R. (2021). In-
creased rna virus population diversity improves adapt-
ability. Scientific Reports, 11:6824.
Niles, I. and Pease, A. (2001). Towards a standard upper on-
tology. In Proceedings of the international conference
on Formal Ontology in Information Systems-Volume
2001, pages 2–9.
Poli, R., Healy, M., and Kameas, A. (2010). Theory
and applications of ontology: Computer applications.
Springer.
Romanenko, E., Calvanese, D., and Guizzardi, G. (2022).
Towards pragmatic explanations for domain ontolo-
gies. In Knowledge Engineering and Knowledge
Management: 23rd International Conference, EKAW
2022, Bolzano, Italy, September 26–29, 2022, Pro-
ceedings, pages 201–208. Springer.
Serna Garc
´
ıa, G., Al Khalaf, R., Invernici, F., Ceri, S., and
Bernasconi, A. (2023a). Coveffect: interactive sys-
tem for mining the effects of sars-cov-2 mutations
and variants based on deep learning. GigaScience,
12:giad036.
Serna Garc
´
ıa, G., Al Khalaf, R., Invernici, F., Ceri,
S., and Bernasconi, A. (2023b). Supporting
data for ”coveffect: Interactive system for min-
ing the effects of sars-cov-2 mutations and vari-
ants based on deep learning”, GigaScience Database.
http://dx.doi.org/10.5524/102386. Last accessed: Aug
2023.
Smith, B., Ashburner, M., Rosse, C., Bard, J., Bug, W.,
Ceusters, W., Goldberg, L. J., Eilbeck, K., Ireland, A.,
Mungall, C. J., et al. (2007). The OBO Foundry: coor-
dinated evolution of ontologies to support biomedical
data integration. Nature biotechnology, 25(11):1251–
1255.
Tanaka, H., Hirayama, A., Nagai, H., Shirai, C., Taka-
hashi, Y., Shinomiya, H., Taniguchi, C., and Ogata, T.
(2021). Increased transmissibility of the SARS-CoV-2
Alpha variant in a Japanese population. International
journal of environmental research and public health,
18(15):7752.
Verdonck, M. and Gailly, F. (2016). Insights on the use
and application of ontology and conceptual modeling
languages in ontology-driven conceptual modeling.
In Conceptual Modeling: 35th International Confer-
ence, ER 2016, Gifu, Japan, November 14-17, 2016,
Proceedings 35, pages 83–97. Springer.
Verdonck, M., Gailly, F., Pergl, R., Guizzardi, G., Mar-
tins, B., and Pastor, O. (2019). Comparing traditional
conceptual modeling with ontology-driven conceptual
modeling: An empirical study. Information Systems,
81:92–103.
Wang, L. L., Lo, K., Chandrasekhar, Y., Reas, R., Yang,
J., Eide, D., Funk, K., Kinney, R., Liu, Z., Merrill,
W., et al. (2020). CORD-19: The COVID-19 Open
Research Dataset. ArXiv.
Wheelock, E. F. and Toy, S. T. (1973). Participation of lym-
phocytes in viral infections. Advances in immunology,
16:123–184.
World Health Organization (2023). Statement on
the fifteenth meeting of the IHR (2005) Emer-
gency Committee on the COVID-19 pan-
demic. https://www.who.int/news/item/05-05-
2023-statement-on-the-fifteenth-meeting-of-the-
international-health-regulations. Last accessed: Aug
2023.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
72
