
Towards Understanding Crossover for Cartesian Genetic Programming
Henning Cui
1 a
, Andreas Margraf
2 b
, Michael Heider
1 c
and J
¨
org H
¨
ahner
1 d
1
Institute for Computer Science, University of Augsburg, Am Technologiezentrum 8, 86159 Augsburg, Germany
2
Fraunhofer IGCV, Am Technologiezentrum 2, 86159 Augsburg, Germany
Keywords:
Cartesian Genetic Programming, CGP, Crossover, Reorder, Evolutionary Algorithm.
Abstract:
Unlike in traditional Genetic Programming, Cartesian Genetic Programming (CGP) does not commonly fea-
ture a recombination/crossover operator, although recombination plays an important role in other evolutionary
techniques, including Genetic Programming from which CGP originates. Instead, CGP mainly depends on
mutation and selection operators in their evolutionary search. To this day, it is still unclear as to why CGP’s
performance does not generally improve with the addition of crossover. In this work, we argue that CGP’s
positional bias might be a reason for this phenomenon. This bias describes a skewed distribution of active and
inactive nodes, which might lead to destructive behaviour of standard recombination operators. We provide
a first assessment with preliminary results. No final conclusion to this hypothesis can be drawn yet, as more
thorough evaluations must be done first. However, our first results show promising trends and may lay the
foundationf or future work.
1 INTRODUCTION
Cartesian Genetic Programming (CGP) is a form
of Genetic Programming (GP), based on directed
acyclic graphs whose nodes are arranged in a two-
dimensional grid (cf. Section 2 for a detailed descrip-
tion).
Since its inception in 1999 by Miller (Miller,
1999), the crossover operator is an active research
topic, as no universal recombination operator has
been found yet—as contrasted with GP, for which
numerous different crossover algorithms exist (Lang-
don et al., 2008). As for CGP, an effective crossover
operator relies on extensions to the CGP formula
and highly depends on the specific use case (Husa
and Kalkreuth, 2018). Moreover, it is still unclear
as to why crossover raises issues in the context of
CGP (Miller, 1999; Clegg et al., 2007; Miller, 2020;
Kalkreuth, 2020).
Regarding possible explanations for CGP’s issues
with crossover, we hypothesize that the positional
bias might be influential. In CGP, the positional bias
is an effect which potentially limits CGP to fully ex-
plore its search space (Goldman and Punch, 2013a).
a
https://orcid.org/0000-0001-5483-5079
b
https://orcid.org/0000-0002-2144-0262
c
https://orcid.org/0000-0003-3140-1993
d
https://orcid.org/0000-0003-0107-264X
The basic problem is a non-uniform distribution of
nodes contributing to an output. This, in turn, might
lead to problems for standard recombination opera-
tors like the point-, multi-n-, or uniform-crossover. To
counteract positional bias, Goldman and Punch intro-
duced Reorder, an operator to shuffle CGPs genome
ordering without changing its phenotype (Goldman
and Punch, 2013a). Thereby, the effect of the posi-
tional bias is supposedly lessened. To test our hypoth-
esis, we take advantage of Reorder. If the positional
bias plays a role in CGP’s issues with crossover, this
extensions might benefit from the usage of crossover
and provide a better understanding of this issue.
We provide a quick overview of the core princi-
ples of CGP and reintroduce the Reorder operator,
as well as the positional bias in the following Sec-
tion 2. Afterwards, Section 3 gives an overview of
related work. In Section 4, we discuss our hypothesis
in-depth and discuss preliminary results in Section 5.
At last, Section 6 summarizes our findings and dis-
cusses future research directions.
2 CARTESIAN GENETIC
PROGRAMMING
The core principles of the supervised learning algo-
rithm called Cartesian Genetic Programming (CGP)
308
Cui, H., Margraf, A., Heider, M. and Hähner, J.
Towards Understanding Crossover for Cartesian Genetic Programming.
DOI: 10.5220/0012231400003595
In Proceedings of the 15th International Joint Conference on Computational Intelligence (IJCCI 2023), pages 308-314
ISBN: 978-989-758-674-3; ISSN: 2184-3236
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

are reintroduced in the following section—with the
first description given by Miller (Miller, 1999). In ad-
dition, we present Reorder, which is introduced by
Goldman and Punch (Goldman and Punch, 2013a)
and extends the classic CGP formula.
2.1 Representation
Originally, CGP is represented as a directed, acyclic
and feed-forward graph. The graph contains nodes ar-
ranged in a c × r grid, with c ∈ N
+
and r ∈ N
+
defin-
ing the number of columns and rows in the grid re-
spectively
1
.
The set of nodes can be divided into three groups:
input-, computational- and output-nodes. Input nodes
directly receive a program’s inputs. Computational
nodes are represented by a function gene and a con-
nection genes, with a ∈ N
+
being the maximum ar-
ity of all functions in the predefined function set. At
last, output nodes redirect the calculated output of a
previous input- or computational node and define the
models output. They are represented by a single con-
nection gene.
Additionally, computational nodes can be clas-
sified into active and inactive nodes. The former
nodes are part of a path to an output node and—as
a consequence—contribute to a program output. On
the contrary, inactive nodes do not contain a path to
an output node and, thereby, do not contribute to the
models output. This is also why their operations are
not computed for the evaluation of a fitness value and
during inference time. However, inactive nodes are
essential to CGP as they probably aid the evolution-
ary process through genetic drift (Turner and Miller,
2015).
As is standard in most CGP variants, an elitist
(µ + λ)–evolution strategy with µ = 1 and λ = 4 is
used to select the best graph. This is combined with
neutral search to improve performance and conver-
gence time (Turner and Miller, 2015; Yu and Miller,
2001; Vassilev and Miller, 2000).
Regarding its mutation operator, a point mutation
is oftentimes found in literature (Miller, 2011; Miller,
2020). The caveat of the point mutation, however,
is its possibility that that only inactive genes are mu-
tated. Hence, it is not possible to evaluate the quality
of the mutation, as the phenotype did not change. This
could potentially lead to fitness plateaus and longer
training times. In order to prevent such plateaus,
Goldman and Punch propose an alternative mutation
operator called Single (Goldman and Punch, 2013b).
1
Please note that, with today’s standards, a CGP model
typically consists of only one row (Miller, 2011). That
means, r = 1.
n
0
:
INPUT
n
1
:
INPUT
n
2
:
ADD
n
3
:
SUB
n
4
:
ADD
n
5
:
OUTPUT
Figure 1: Example graph defined by a CGP genotype. The
dashed node and connections are inactive due to not con-
tributing to the output.
With Single, the point mutation is applied to randomly
selected genes until a gene corresponding to an active
node is mutated. This forces changes in the phenotype
with no wasted evaluations being performed. Thus,
when the optimal mutation rate is not known, the au-
thors Goldman and Punch claim that Single should be
preferred (Goldman and Punch, 2013b).
An illustrative example of a graph defined by a
CGP genotype can be seen in Figure 1. It shows a
graph with c = 6 and r = 1, which is supposed to solve
a task consisting of two inputs and one output. Here,
the first two nodes (n
0
and n
1
) are input nodes and
provide two different program inputs. The follow-
ing nodes (n
2
–n
4
) are computational nodes. At last,
the output is provided by one output node n
5
. In this
example, both inputs are subtracted, with this inter-
mediate result being added to itself and taken as the
program output. Thus, every node drawn with a solid
line is an active node. The node n
2
is an inactive node
(marked by dashed lines) since it does not contribute
to the program output nor contains a path to an output
node.
2.2 Positional Bias and Reorder
CGP has many advantages, such as the absence of
bloat (Miller, 1999) and its small solutions (Miller,
2011). Nonetheless, it is far from perfect, with one of
its shortcomings being the positional bias.
2.2.1 Positional Bias
Goldman and Punch found a negative impact in
CGP’s search space by enforcing a feed forward
grid (Goldman and Punch, 2013a). Namely, a posi-
tional bias: A non-uniformity of the probability that
a node is active. Computation nodes near input nodes
have a higher probability of becoming active, while
nodes closer to the output nodes have the lowest prob-
ability of becoming active. In turn, the nodes near the
output nodes have a higher probability of becoming
inactive.
During the mutation of a connection gene, each
previous node has the same probability of being cho-
sen as the new starting node for the connection. How-
ever, nodes near input nodes have more options for
successor nodes, as directed edges are only allowed
Towards Understanding Crossover for Cartesian Genetic Programming
309
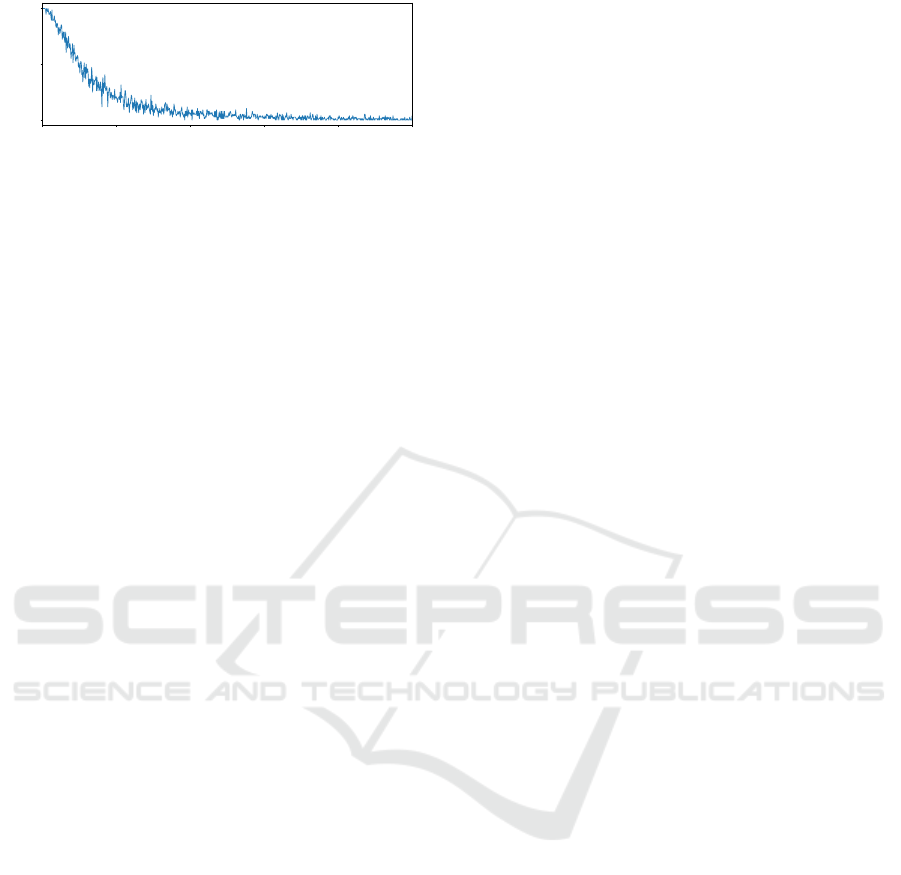
0 20 40 60 80 100
Relative node index in % to the graph's total length
0.0
0.5
1.0
% active
Figure 2: Example distribution of active nodes in CGP
graph solutions. The x-axis represents a node’s position in
the graph, the y-axis its percentage of being active, gener-
ated from 50 independent runs on the same dataset.
from left to right. This is why they have a higher
chance of becoming active, as a node is only active
when it is part of a path to an output node. Nodes
closer to output nodes have less options of being cho-
sen by other nodes, as they have less nodes in front.
With the help of Figure 1, the positional bias can
be exemplified. In this example, n
1
’s possible suc-
cessors are n
2
–n
5
whereas n
4
’s possible successor is
just n
5
. Hence, four nodes can potentially mutate their
connection to n
1
and if one of those nodes is active, n
1
becomes active, too. Contrary, n
4
has only one node
in front: n
5
. This is also the only node which could
mutate its connection to n
4
.
An illustrative example of the active node distribu-
tion can be seen in Figure 2. We show the distribution
of active nodes, obtained from 50 independent runs
on the same dataset. Additionally, by modeling the
distribution of active nodes, the positional bias could
be quantitatively measured and described.
2.2.2 Reorder
The positional bias makes it difficult or impossible to
solve certain problems (Goldman and Punch, 2013a;
Payne and Stepney, 2009). To weaken the effects of
this bias, Goldman and Punch proposed two new op-
erators which are executed before mutation: DAG and
Reorder (Goldman and Punch, 2013a).
The first extension, DAG, allows each node to mu-
tate connections to every other node in the genome as
long as the new connection does not create a cycle.
However, as we do not build upon it, we will not dis-
cuss DAG in further detail.
Reorder shuffles the ordering of the genotype but
respects the sequence of operation of active nodes.
Hence, the phenotype does not change but, as a re-
sult, active nodes are dispersed through the grid.
This Reorder operator begins by creating a de-
pendency set D. It contains each computational node
and its information from which node it gets its input
from. Additionally, a new genome is initialized con-
taining all input and output nodes from the original
genome, as they do not get assigned a new position.
Afterwards, the set of addable nodes Q is created.
Initially, this set contains only computational nodes
whose dependencies are satisfied. In this context, sat-
isfied means the following: Let b be a computational
node, and a be a computational or input node from
which b gets one of its input from, with (b, a) ∈ D.
This means, there is a directed edge connecting a to
b via b’s connection gene. Then, b is satisfied when
all nodes from which b gets its input from are mapped
into the new genome.
With both sets initialized, the shuffling of the
genome is done by repeating the following steps:
1. Select and remove a random node a ∈ Q.
2. Map a sequentially to the next, free location in the
new genome.
3. For each node pair (b, a) ∈ D with b depending on
a, do:
If all dependencies of b are satisfied, add b to
Q.
This operator ends when all computational nodes
have been assigned a new location in the genotype.
For better readability, we will refer to a CGP vari-
ant with the Reorder-extension as REORDER.
3 RELATED WORK
Various previous works investigated different
crossover operators for CGP and their workings.
However, to the best of our knowledge, we are the
first to investigate the influence of the positional bias
on CGP’s crossover operator.
Cai et al. argued that CGP is not positional in-
dependent (Cai et al., 2006). This means, CGP’s
components and their workings depend on their po-
sition in the graph. As crossover does not consider
these dependencies, useful structures are destroyed.
Hence, the authors introduced a new crossover oper-
ator, which considers such dependencies. While sim-
ilar notions are made for this article, their argument
differs as they did not regard the distribution of active
nodes.
Other similar arguments to this works’ were pre-
sented by Kalkreuth et al. (Kalkreuth et al., 2017).
They argued that swapping arbitrary genes is not
preferable and introduced a subgraph-crossover oper-
ator which only recombines active nodes. However,
contrary to this work, they also did not consider the
positional bias.
Based on the just described work of Kalkreuth et
al. (Kalkreuth et al., 2017) and embedded CGP (Kauf-
mann and Platzner, 2008), Husa and Kalkreuth in-
troduced the block-crossover, which swap blocks of
ECTA 2023 - 15th International Conference on Evolutionary Computation Theory and Applications
310

consecutive active nodes (Husa and Kalkreuth, 2018;
Kalkreuth, 2020).
It is also possible to use a floating-point represen-
tation for CGP, as was done by Clegg et al. (Clegg
et al., 2007) and Wilson et al. (Wilson et al., 2018).
They tested different crossover operators and found
that their method benefits from it.
In the context of image processing, Karel et al.
used the original grid-structure of CGP to evolve im-
age filters (Slan
´
y and Sekanina, 2007). They also
included the single-point and multi-point crossover
without modifications to CGP in their studies. By
observing the fitness landscape of their solutions, the
authors concluded that the single-point crossover im-
proved the evolutionary search.
Another domain specific approach was done by
Torabi et al.. They used CGP in the context of
neural architecture search and adapted a specialized
crossover mechanism to design convolutional neural
networks (Torabi et al., ).
Some other more recent methods, such as from
Kalkreuth, developed an operator which recombines
only active nodes (Kalkreuth, 2022).
4 POSITIONAL BIAS AND
CROSSOVER
We hypothesize that the positional bias might play
a role in CGP’s issue with recombination opera-
tors. Active nodes accumulate near input nodes,
while inactive nodes concentrate near output nodes.
This means, important nodes—which generate the
output—are clustered together. However, traditional
crossover operators like the n−point or uniform
crossover do not consider such clusters. It is possible
that, by applying crossover, important semantic struc-
tures get destroyed. Other methods from Husa and
Kalkreuth (Husa and Kalkreuth, 2018), Cai et al. (Cai
et al., 2006) or Kalkreuth et al. (Kalkreuth et al., 2017)
do however consider such structures, which may be
one explanation of their performance gain.
In addition, nodes near the output nodes may also
lead to problems. They are mostly inactive, and re-
combining them should not harm the performance.
On the contrary, it should diversify the genome and
lead to more neutral genetic drift, which is a needed
trait in CGP (Turner and Miller, 2015; Yu and Miller,
2001). However, a problem might occur when active
nodes are recombined. This could completely change
the recombined phenotype, as these nodes might con-
nect to inactive nodes. These inactive nodes become
active, and connections to various other nodes might
be established. New nodes become active or inactive,
which can drastically change the phenotype.
Hence, traditional recombination operators may
lead to too much destructive behaviour for CGP. Im-
portant structures of active nodes near the input nodes
might get destroyed, while stray active nodes near the
output nodes might drastically change the phenotype.
With REORDER, the positional bias can be par-
tially circumvented. Hence, the problems stated
above could be dampened, too. As a result, REORDER
should improve with an additional crossover operator.
5 PRELIMINARY RESULTS AND
DISCUSSION
In order to get a first grasp of REORDER’s influence
on popular, elemental crossover operators, we con-
ducted an empirical study on Boolean benchmarks. In
the following sections, we describe our experimental
design and preliminary results.
5.1 Experimental Design
We present first results in Table 1, in which we report
three different values for each CGP variant: Its mean
number of iterations until a solution is found (I2S), its
respective standard deviation and the favourable num-
ber of computational nodes.
For our results, we tested six different CGP vari-
ants. The standard implementation described in Sec-
tion 2.1 serves as a baseline, as it is the most com-
monly found CGP implementation. In addition, a
pure REORDER variant is included as another base-
line, as four crossover operators are used with RE-
ORDER: Single-, two-, three-point- as well as a uni-
form crossover.
Each variant was trained with the Single muta-
tion. This avoids the need to optimize a mutation
rate and each variant can be compared fairer. All
variants using the crossover operator where trained
with an elitist (4 + 10)-ES, as prior work showed
that smaller population sizes are favoured (Husa and
Kalkreuth, 2018). Nonetheless, we did a hyperparam-
eter search for a good number of nodes n and tested
n ∈
{
50, 100, 150, ··· , 2000
}
. Each configuration
was tested for 50 times with independent repetitions
and random seeds.
For a first grasp, four Boolean benchmark datasets
were used: 3-bit Parity (Wegener, 2005), 16-4-bit
Encode, 4-16-bit Decode (Kaufmann and Kalkreuth,
2020) and 3-bit Multiply (White et al., 2013). For the
remainder of this work, we will refer to them as Par-
ity, Encode, Decode and Multiply respectively.
Towards Understanding Crossover for Cartesian Genetic Programming
311

Table 1: Preliminary results, showing the mean number of iterations until a solution is found (I2S) and its standard deviation,
as well as the best number of nodes found. Results for each Dataset are ordered according to their mean value.
Variant M ean(I2S) Std(I2S) # Computational Nodes
Parity
Standard (1 + 4) 342 330 600
Uniform + REORDER 437 430 50
Three-point + REORDER 678 833 50
REORDER 739 702 200
Two-point + REORDER 800 1,349 100
Single-point + REORDER 1,096 1,702 50
Encode
Three-point + REORDER 3,629 661 550
Two-point + REORDER 4,568 687 550
Uniform + REORDER 4,905 2,884 300
Standard (1 + 4) 6,213 3,368 200
Single-point + REORDER 11,927 10,500 200
REORDER 14,733 9,974 600
Decode
Uniform + REORDER 9,812 4,477 400
REORDER 15,017 10,397 700
Standard (1 + 4) 23,132 5,539 400
Two-point + REORDER 25,294 11,509 200
Three-point + REORDER 25,520 12,196 300
Single-point + REORDER 26,255 11,684 250
Multiply
Uniform + REORDER 76,117 65,594 350
REORDER 90,370 131,042 750
Standard (1 + 4) 129,739 59,127 700
Three-point + REORDER 133,506 109,385 300
Two-point + REORDER 140,705 73,147 450
Single-point + REORDER 148,384 97,937 350
All benchmarks used the same, standard fitness
function for Boolean benchmarks: The ratio of cor-
rectly mapped inputs. Furthermore, we used the stan-
dard Boolean function set, containing the Boolean op-
erators AND, OR, NAND and NOR.
5.2 Discussion
As for the results shown in Table 1, some trends can
be seen.
For Parity, a standard elitist (1 + 4)–evolution
strategy performs best. However, as is argued by
White et al. (White et al., 2013), Parity is deemed
as too easy by the evolutionary algorithm community.
Hence, results from this dataset should be taken with
reservation.
As for the other three benchmarks, REORDER
with a crossover variation performs best according
to the mean I2S. Both Decode and Multiply favour
the uniform recombination, which leads to better re-
sults than both baselines. Interestingly, for these two
benchmarks, the n−point crossover performs worst.
Furthermore, single-point crossover almost always
leads to the worst results across all benchmarks. Con-
sidering the two- and three-point crossover, Encode is
the only benchmark benefiting from them.
We also report the best number of computational
nodes found by our hyperparameter search. However,
there is only one trend which can be read from Ta-
ble 1. REORDER in combination with a crossover op-
erator generally needs less computational nodes than
a pure REORDER variant. Considering Multiply, the
number of nodes needed is more than halved when
uniform crossover + REORDER variant is compared
to its baseline—while its mean I2S is also reduced by
about 14,000 iterations.
ECTA 2023 - 15th International Conference on Evolutionary Computation Theory and Applications
312

However, it is still unclear why the uniform
crossover is preferable for Decode and Multiply,
while Encode shows different trends. Both former
benchmarks are harder benchmarks compared to En-
code and Parity, though. It is possible that the
n−point crossovers are too destructive for harder
problems, as too many useful structures are dissi-
pated. This may indicate that the positional bias
plays only a minor role in CGP’s performance with
its crossover operators. Nevertheless, we can val-
idate the results of Husa and Kalkreuth (Husa and
Kalkreuth, 2018), that different recombination oper-
ators are needed for different settings.
6 CONCLUSION
The crossover operator has been an active research
topic for Cartesian Genetic Programming (CGP)
since its introduction. To this day, it is unclear why
CGP does not generally benefit from a recombination
algorithm and finding universally well-performing
operators might improve CGP’s performance greatly.
In the process to find answers to this question,
we argued that the influence of CGP’s positional bias
might be a possible issue. The uneven distribution of
active and inactive nodes might lead to problems for
untailored crossover operators, as simply swapping
genes might destroy useful structures. To mitigate the
effects of the positional bias, the reorder extension to
CGP was reintroduced.
Our hypothesis was tested empirically and prelim-
inary results were presented. By comparing differ-
ent CGP variants with different crossover operators, a
first conclusions can be given: The uniform crossover
generally benefits CGP with reorder.
However, no clear conclusion to our hypothesis
can be drawn yet. Further evaluations and configura-
tions must be tested before the influence of the posi-
tional bias can be truly assessed. For future work, a
more diverse set of benchmarks must be tested. Test-
ing only Boolean benchmarks are not sufficient and
evaluations must be extended to regression and real-
world benchmarks. Furthermore, we did not com-
pare our results to the standard CGP formula with
crossover operators, which is an important step to
assess REORDERs influence on the crossover opera-
tors. In addition, statistical analysis methods should
be added to the evaluations as well. By only compar-
ing mean values and their standard deviations, a first
assessment can be made. However, without proper
statistical analysis, it is not possible to truly judge the
results.
Along with these evaluations mentioned, the ideas
of Kalkreuth et al. should be included (Kalkreuth
et al., 2017). By only recombining active nodes in
CGP with reorder, some effects might be observed
which could help to accept or reject our hypothesis.
Additionally, new crossover operators should be
tested to evaluate our hypothesis. New algorithms
similar to those introduced by Cai et al. could recom-
bine genes differently or with different probabilities,
based on the genes location in the graph (Cai et al.,
2006). Other operators could be influenced by exist-
ing works and be based on the density of active nodes.
Furthermore, the counts of successful crossovers,
the fitness changes per crossover, distributions of ac-
tive and inactive nodes, etc. should be also accounted
for.
REFERENCES
Cai, X., Smith, S. L., and Tyrrell, A. M. (2006). Posi-
tional independence and recombination in cartesian
genetic programming. In Collet, P., Tomassini, M.,
Ebner, M., Gustafson, S., and Ek
´
art, A., editors, Ge-
netic Programming, pages 351–360, Berlin, Heidel-
berg. Springer Berlin Heidelberg.
Clegg, J., Walker, J. A., and Miller, J. F. (2007). A new
crossover technique for cartesian genetic program-
ming. In Annual Conference on Genetic and Evolu-
tionary Computation.
Goldman, B. W. and Punch, W. F. (2013a). Length bias
and search limitations in cartesian genetic program-
ming. In Proceedings of the 15th Annual Conference
on Genetic and Evolutionary Computation, GECCO
’13, page 933–940, New York, NY, USA. Association
for Computing Machinery.
Goldman, B. W. and Punch, W. F. (2013b). Reducing
wasted evaluations in cartesian genetic programming.
In Genetic Programming, pages 61–72, Berlin, Hei-
delberg. Springer Berlin Heidelberg.
Husa, J. and Kalkreuth, R. (2018). A comparative study
on crossover in cartesian genetic programming. In
Castelli, M., Sekanina, L., Zhang, M., Cagnoni, S.,
and Garc
´
ıa-S
´
anchez, P., editors, Genetic Program-
ming, pages 203–219, Cham. Springer International
Publishing.
Kalkreuth, R. (2020). A comprehensive study on subgraph
crossover in cartesian genetic programming. In IJCCI,
pages 59–70.
Kalkreuth, R. (2022). Towards discrete phenotypic re-
combination in cartesian genetic programming. In
Rudolph, G., Kononova, A. V., Aguirre, H., Kerschke,
P., Ochoa, G., and Tu
ˇ
sar, T., editors, Parallel Prob-
lem Solving from Nature – PPSN XVII, pages 63–77,
Cham. Springer International Publishing.
Kalkreuth, R., Rudolph, G., and Droschinsky, A. (2017).
A new subgraph crossover for cartesian genetic pro-
gramming. In McDermott, J., Castelli, M., Sekanina,
Towards Understanding Crossover for Cartesian Genetic Programming
313

L., Haasdijk, E., and Garc
´
ıa-S
´
anchez, P., editors, Ge-
netic Programming, pages 294–310, Cham. Springer
International Publishing.
Kaufmann, P. and Kalkreuth, R. (2020). On the parame-
terization of cartesian genetic programming. In 2020
IEEE Congress on Evolutionary Computation (CEC),
pages 1–8.
Kaufmann, P. and Platzner, M. (2008). Advanced tech-
niques for the creation and propagation of modules
in cartesian genetic programming. In Proceedings of
the 10th Annual Conference on Genetic and Evolu-
tionary Computation, GECCO ’08, page 1219–1226,
New York, NY, USA. Association for Computing Ma-
chinery.
Langdon, W. B., Poli, R., McPhee, N. F., and Koza, J. R.
(2008). Genetic Programming: An Introduction and
Tutorial, with a Survey of Techniques and Applica-
tions, pages 927–1028. Springer Berlin Heidelberg,
Berlin, Heidelberg.
Miller, J. F. (1999). An empirical study of the efficiency of
learning boolean functions using a cartesian genetic
programming approach. In Proceedings of the 1st An-
nual Conference on Genetic and Evolutionary Com-
putation - Volume 2, GECCO’99, page 1135–1142,
San Francisco, CA, USA. Morgan Kaufmann Publish-
ers Inc.
Miller, J. F. (2011). Cartesian Genetic Programming.
Springer Berlin Heidelberg, Berlin, Heidelberg.
Miller, J. F. (2020). Cartesian genetic programming: its sta-
tus and future. Genetic Programming and Evolvable
Machines, 21(1):129–168.
Payne, A. J. and Stepney, S. (2009). Representation and
structural biases in cgp. In 2009 IEEE Congress on
Evolutionary Computation, pages 1064–1071.
Slan
´
y, K. and Sekanina, L. (2007). Fitness landscape anal-
ysis and image filter evolution using functional-level
cgp. In Ebner, M., O’Neill, M., Ek
´
art, A., Van-
neschi, L., and Esparcia-Alc
´
azar, A. I., editors, Ge-
netic Programming, pages 311–320, Berlin, Heidel-
berg. Springer Berlin Heidelberg.
Torabi, A., Sharifi, A., and Teshnehlab, M. Using cartesian
genetic programming approach with new crossover
technique to design convolutional neural networks.
Turner, A. J. and Miller, J. F. (2015). Neutral genetic
drift: an investigation using cartesian genetic pro-
gramming. Genetic Programming and Evolvable Ma-
chines, 16(4):531–558.
Vassilev, V. K. and Miller, J. F. (2000). The advantages
of landscape neutrality in digital circuit evolution. In
Evolvable Systems: From Biology to Hardware, pages
252–263, Berlin, Heidelberg. Springer Berlin Heidel-
berg.
Wegener, I. (2005). Complexity theory: exploring the limits
of efficient algorithms. Springer Science & Business
Media.
White, D., Mcdermott, J., Castelli, M., Manzoni, L., Gold-
man, B., Kronberger, G., Ja
´
skowski, W., O’Reilly, U.-
M., and Luke, S. (2013). Better gp benchmarks: Com-
munity survey results and proposals. Genetic Pro-
gramming and Evolvable Machines, 14:3–29.
Wilson, D. G., Miller, J. F., Cussat-Blanc, S., and Luga,
H. (2018). Positional cartesian genetic programming.
CoRR, abs/1810.04119.
Yu, T. and Miller, J. (2001). Neutrality and the evolvability
of boolean function landscape. In Genetic Program-
ming, pages 204–217, Berlin, Heidelberg. Springer
Berlin Heidelberg.
ECTA 2023 - 15th International Conference on Evolutionary Computation Theory and Applications
314
