
A Spleen Infection Recognition Approach Using Shallow Neural
Network in Comparison with Support Vector Machine
K. Baba Fakruddin and N. Deepa
Department of Computer Science and Engineering, Saveetha School of Engineering, Saveetha Institute of Medical and
Technical Sciences, Saveetha University, Chennai, Tamil Nadu, India
Keywords: Abdominal Pains, Antibiotics, Bacteria, Disability, Machine Learning, Novel Shallow Neural Network,
Spleen Infection.
Abstract: The research primarily investigates the accuracy differences between spleen infection segmentation and
classification using the Novel Shallow Neural Network (NSNN) versus the SVM. For the study, spleen
infections in patients were detected using the NSNN (15 samples) and compared against results from SVM
(another 15 samples), operating with an 80% G-power. Findings indicated the NSNN had an accuracy of
75.27%, marginally superior to the SVM's 66.33%. Despite this disparity in accuracy, there was no
statistically significant difference between the two methods, evidenced by an independent sample T-Test
result of p=0.25. In conclusion, NSNN offers a slightly enhanced accuracy rate in contrast to SVM within the
realm of machine learning.
1 INTRODUCTION
Machine learning is a field within artificial
intelligence that focuses on the development of
algorithms. It incorporates techniques such as
supervised and unsupervised learning. Machine
learning is applied in various applications (Frank et
al. 2009) including robotics, computer vision, speech
recognition, data mining, and bioinformatics. It
furthers the development of machine learning (Goh,
Sing, and Yeong 2020) through AI and functions
automatically due to artificial intelligence's role in
marketing. It recognises patterns in data and makes
(Qiu et al. 2016) predictions based on those patterns.
Machine learning algorithms have been utilised for
(Dietterich 2002) search engines, image recognition,
natural language processing, and self-driving cars.
They can also assimilate new information and think
in a human-like manner. As it evolves, machine
learning will provide more insights about systems,
delivering results that facilitate communication
between humans and computers. It can also be
applied to image recognition, online fraud detection,
speech recognition, and product detection (Boiy and
Moens 2008).
Regarding spleen infection, many articles are
presented across different platforms like IEEE,
Springer and (Wang et al. 2006) Science Direct.
Specifically, there are 4533 articles from Springer,
5634 articles (Davies, Barnes, and Milligan 2002)
from Science Direct, and 6543 articles from IEEE on
spleen infection. References such as (Darling 1906)
splenic abscess have 85 citations, whilst medical
conditions in (Jung et al. 2002) have 78 citations.
Typically, it is caused by bacteria entering the
bloodstream from (King and Shumacker 1952)
another part of the body, such as the lungs or the
gastrointestinal tract. Splenic abscess can be triggered
by a range of microorganisms, which includes
(Newland, Provan, and Myint 2005) Staphylococcus
aureus, Streptococcus, Enterobacteria, and even
fungi, and it has 70 citations. Machine learning serves
as a robust tool to analyse vast datasets to make
predictions and decisions. In the context of splenic
abscess, machine learning can identify patterns in
patient data that help in predicting those at risk for
developing this condition.
The limited accuracy of current algorithms
impacts the progression, diagnosis, and treatment of
spleen infection. Furthermore, with the aid of
machine learning, medical staff can be notified of any
limitations and early indications of spleen infection,
ensuring timely intervention. The research aims to
compare the accuracy in spleen infection
segmentation and classification using the Novel
Shallow Neural Network (NSNN) against the Support
Vector Machine.
480
Fakruddin, K. and Deepa, N.
A Spleen Infection Recognition Approach Using Shallow Neural Network in Comparison with Support Vector Machine.
DOI: 10.5220/0012518900003739
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Artificial Intelligence for Internet of Things: Accelerating Innovation in Industry and Consumer Electronics (AI4IoT 2023), pages 480-486
ISBN: 978-989-758-661-3
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

2 MATERIALS AND METHODS
Work was conducted in the Machine Learning lab at
SSE, SIMATS, equipped with meticulously
calibrated hardware and software to facilitate my
research. The study was divided into two groups: one
for comparing existing algorithms and another for the
proposed algorithms (Morris and Bullock 1919). A
sample size of 15 for each group, totalling 30, was
utilised (Mazur et al. 2018). Calculations were based
on a G-power value of 80%, with an alpha of 0.05 and
a beta of 0.02, maintaining a confidence interval of
95%.
To address spleen infection and abdominal pains,
a Novel Shallow Neural Network can be designed
using a myriad of techniques. The initial step involves
preprocessing the dataset to guarantee data (Gómez
Vela, Divina, and García-Torres 2021) consistency
and normalisation. This can be achieved through
methods such as standardisation and feature scaling.
Upon processing, the model's efficacy can be gauged
against alternative models using metrics like accuracy
and recall. Crucially, testing the model on previously
unencountered data ensures its robustness across
diverse scenarios.
Jupyter is a renowned open-source web
application, enabling the creation and sharing of
documents replete with live code, visualisations,
equations, and explanatory text. It finds extensive use
in data science and scientific computing spheres.
Jupyter Notebook, a component of the broader
Jupyter ecosystem, integrates programming
languages, notably Python, within a web-based
notebook interface. The most recent iteration of
Jupyter is version 3.10.
2.1 Novel Shallow Neural Network
Algorithm
The proposed sample, Group 1, adopts the Novel
Shallow Neural Network algorithm as a machine
learning technique tailored for detecting spleen
infections. This approach marries the diagnostic
capabilities of the recurrent neural network with the
enhancement of the convolution neural network for
automation. The primary focus is on elevating the
accuracy of classifying spleen infection disabilities.
The neural network is meticulously crafted to
enhance the detection and diagnosis of spleen
infection disabilities. Harnessing the prowess of AI
and a gamut of machine learning techniques, the
system analyses an extensive array of patient data
encompassing laboratory results, imaging scans, CT
scans, and symptomatology. Such comprehensive
data feeds the network, facilitating the identification
of patterns and predictions regarding the likelihood of
an infection. A significant merit lies in its capacity to
discern amongst various infection types, be it
bacterial, viral, or fungal. Such precision expedites
clinical diagnosis, inherently improving patient
prognosis. Furthermore, the neural network serves as
a vigilant monitor, tracking treatment progress and
registering any nuances in the patient’s status. Table
1 delineates the procedure underpinning the Novel
Shallow Neural Network algorithm.
2.2 Support Vector Machine Algorithm
Support Vector Machines (SVM) represent a
supervised learning approach within Group 1, adept
at data analysis and pattern recognition. Within the
sphere of spleen infection diagnosis, an SVM can be
calibrated to assimilate inputs from medical scans,
like CT or MRI images, coupled with patient data
spanning age, gender, medical history, and other
pertinent metrics. This results in an output predicting
the likelihood of the patient having a spleen infection
disability needing antibiotics. The SVM learns from
its training data, discerning data patterns linked to
infections and then leveraging these patterns for
precise predictions. For the task of spleen infection
detection, the algorithm identifies patterns in medical
imagery that signal an infection. Harnessing features
like shape, size, and texture, the algorithm hones its
accuracy in diagnosing spleen infections. Table 2
elucidates the procedure underpinning the Support
Vector Machine.
3 STATISTICAL ANALYSIS
IBM SPSS is a software package tailored for
statistical analysis. Utilised by businesses,
governments, universities, and various other
organisations, it aids in collecting, analysing, and
interpreting vast datasets. With a rich array of
features, SPSS is adept at data mining, text analytics,
predictive analytics, and generating comprehensive
statistical outputs (Vanus et al. 2019). One can
employ it to dissect cross-tabulated data, produce
descriptive statistics, and craft charts and graphs.
Moreover, it offers advanced techniques,
encompassing both linear and nonlinear modelling,
making it an indispensable tool for any entity seeking
to decipher complex data. In this context, disease
severity, name, and billing emerge as the dependent
variables, whilst patient ID and age are identified as
the independent variables.
A Spleen Infection Recognition Approach Using Shallow Neural Network in Comparison with Support Vector Machine
481

4 RESULTS
Table 1 displays the pseudo code categorisation for
NSNN (Novel Shallow Neural Network). This is
integral to the NSNN's function: taking text data as
input, extracting symptoms, and providing a spleen
infection diagnosis.
Table 2 elucidates the procedure for the Support
Vector Machine algorithm. Initially, the NSNN is set
up, followed by utilising data for training. Two
distinct datasets are employed to evaluate the
training, with the accuracy gauged by assigning each
set to a specific illness type.
Table 3 encompasses the raw accuracy data for
both NSNN and the Support Vector Machine.
Table 4 presents the group statistics when
contrasting independent sample classification of
NSNN (Novel Shallow Neural Network) with
Support Vector Machines. Within the NSNN
classification, N = 15 and the mean accuracy stands
at 75.27%, while the Support Vector Machine posts
66.33%. The NSNN has a standard deviation of
9.438, as opposed to the Support Vector Machine's
11.197. Additionally, the standard error in NSNN is
2.437, whereas for the Support Vector Machine, it's
2.891.
Table 5 reveals the statistical results from the
independent sample t-tests, comparing the NSNN
classification with the Support Vector Machine
algorithm, taking into account a 95% confidence
interval. The findings suggest no statistically
significant difference between the Novel Shallow
Neural Network and the Support Vector Machine,
with a p-value of 0.439 (2-tailed) (p>0.05).
Figure 1 showcases a bar graph which compares
accuracy levels. The mean accuracy of the NSNN
surpasses that of the Support Vector Machine.
Moreover, the standard deviation for the NSNN
slightly outperforms that of the Support Vector
Machine.
Table 1: Pseudo code of Support Vector Machine. The text data provides as an input and symptoms and extract the Diagnosis
of spleen infection in input.
Input: Layer with number of neurons.
Output: Activation of function in neural network.
Initialize input layer with number of neurons corresponding to the features of the spleen infection dataset
Feed the input data into the network
Create hidden layers with appropriate number of neurons and activation function
Initialize the output with neurons corresponding to the possible outcomes
Set up the weights for the neurons in the hidden and output layers
Define a loss function to be used to measure the performance of the neural network.
Table 2: Procedure of the Support Vector Machine algorithm. First initialization of the NSNN is done and the model is trained
with the data.
Input: Spleen infection in dataset
Output: Prediction of datasets
Load the spleen infection dataset
Split the dataset into training and testing sets.
Pre-process the data if needed
Select the type of kernel to use for the SVM (Support Vector Machine)
Train the SVM model on the training dataset
Make predictions using the model on the test dataset
Evaluate the model using metrics such as accuracy, recall, etc.
Adjust the model parameter needed to the model’s performance.
Use the model to make predictions on unseen data
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
482
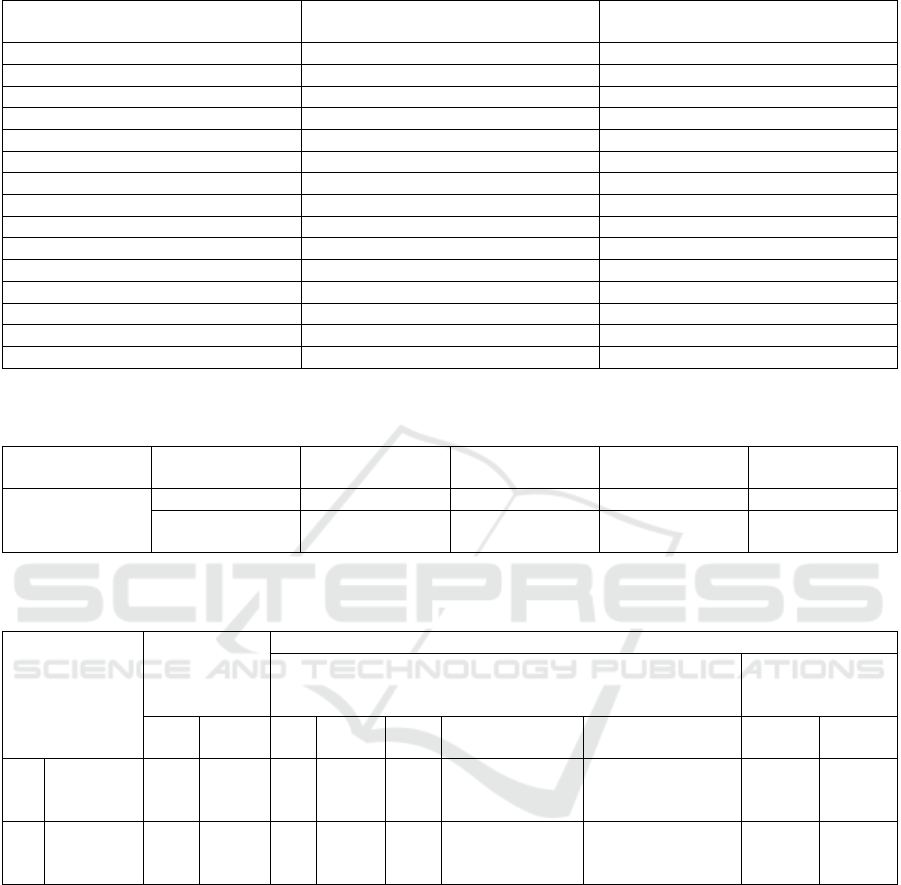
Table 3: Raw data table of accuracy for both NSNN and Support Vector Machine.
S.No
Novel Shallow Neural Network
(Accuracy %)
Support vector machine (Accuracy %)
1
61
86
2
63
84
3
65
80
4
67
76
5
69
72
6
71
69
7
73
67
8
75
64
9
77
62
10
79
60
11
81
58
12
83
57
13
85
55
14
87
54
15
93
51
Table 4: Group statistics for independent sample classification NSNN with Support Vector Machines. In classification NSNN
mean accuracy is 75.27% whereas in Support vector Machines is 66.33%.
Algorithm
N
Mean
Std.Deviation
Std.Error
Mean
Accuracy
NSNN
15
75.27
9.438
2.437
Support Vector
Machine
15
66.33
11.197
2.891
Table 5: In the Statistical Independence sample, the 95% confidence interval. It shows that there is no statistical significance
difference between the Novel Shallow Neural Network and Support Vector algorithm with p=0.25 (2-tailed) (p>0.05).
Levene’s Test
for Equality of
accuracy
T test for equality of means
95 % confidence
intervals of
difference
F
Sig
t
df
Sig.(2-
tailed)
Mean Difference
Std.Error Difference
Lower
Upper
Equal
variances
assumed
0.616
0.439
2.363
28
0.25
8.933
3.781
1.188
16.679
Equal
variances
not assumed
2.363
27.219
0.25
8.933
3.781
1.178
16.689
5 DISCUSSION
From the results obtained through the independent
sample T-test analysis, the significance value is
discerned. A value of 0.439, which exceeds 0.05, is
not statistically significant. Furthermore, the
accuracy of the NSNN (Novel Shallow Neural
Network) at 75.27% surpasses that of the Support
Vector Machine, which stands at 66.33%. This
further reiterates that there is no statistically
significant difference between the Novel Shallow
Neural Network and the Support Vector Machine
with a p-value of 0.439 (2-tailed) (p>0.05). The
essence of Support Vector Machines (SVM) lies in its
supervised machine learning algorithm
predominantly used for classification tasks. SVMs are
potent and efficient techniques (Chen et al. 2009),
boasting 87% accuracy in classifying data within the
realm of spleen infection. They can bifurcate data into
two categories (Landen and Closset 2007), such as
healthy and infected, by assessing data features and
graphically plotting them. Moreover, SVMs (You et
al. 2019) can attain a 78% accuracy rate in detecting
A Spleen Infection Recognition Approach Using Shallow Neural Network in Comparison with Support Vector Machine
483
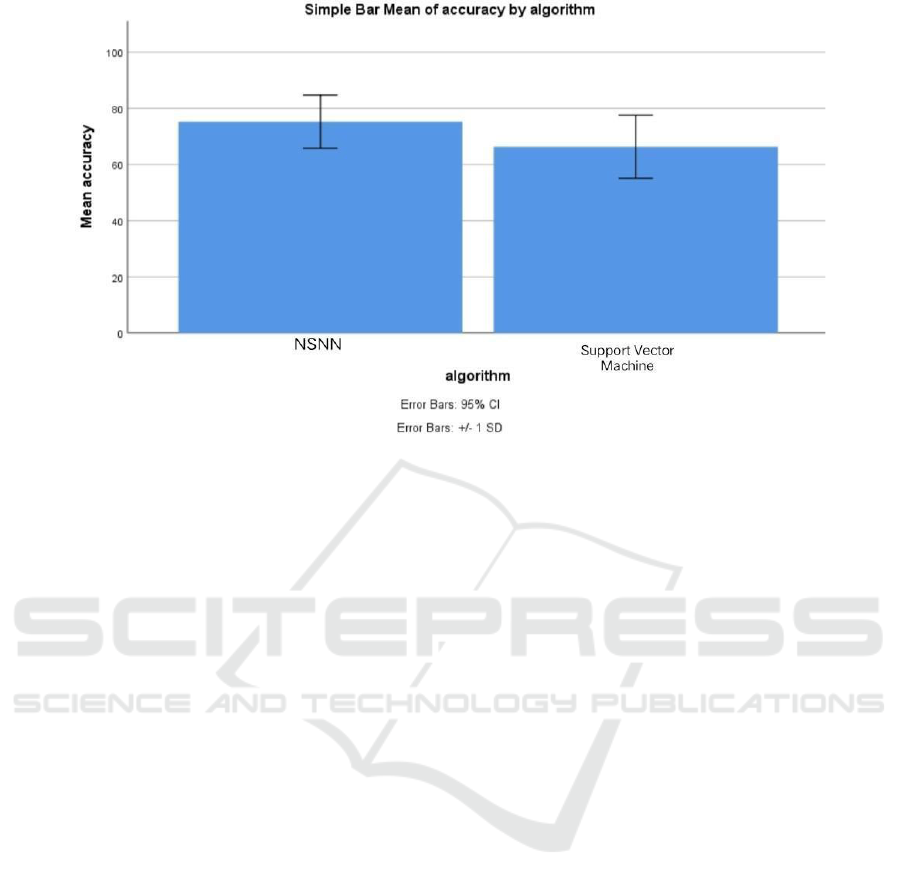
Figure 1: Bar graph for Comparison of NSNN (75.27 %) and support vector machine (66.33 %) in terms of mean accuracy.
The mean accuracy of NSNN is better than the support vector machine and standard deviation of NSNN is better than the
Support Vector Machine. X axis: NSNN vs Support Vector Machine algorithm, Y axis: Mean accuracy
certain pathogens and viruses, differentiating normal
(Zerem and Bergsland 2006) cells from cancerous
ones, and forecasting an infection's severity. By
leveraging SVMs, healthcare professionals (Hosey et
al. 2006) can diagnose disabilities and determine
antibiotics treatment for spleen infections with a 75%
accuracy. Machine learning strategies can foster
models designed for detecting and diagnosing spleen
infection disabilities. These models can be trained on
medical imaging data (like X-rays, CT, and MRI
scans) to pinpoint suspicious spleen regions and
categorise them (Grimaldi et al. 2017) with 70%
accuracy as infected or healthy. Moreover, machine
learning algorithms can sift through patient medical
records and laboratory tests to identify infection
signs. Factors affecting the study in diagnosing spleen
infection using the Novel Shallow Neural Network
encompass data availability, network architecture,
hyperparameters, training duration, and feature
selection. Treating abdominal pains and spleen
infections without addressing them promptly can lead
to the spleen enlarging, possibly causing grave
complications like sepsis, anaemia, and organ failure.
Surgical intervention might be necessary to excise an
infected spleen or to address complications.
Following the removal of the spleen, patients might
grapple with an escalated infection risk and a
compromised immune system. In the foreseeable
future, delving into novel treatments and enhanced
diagnostic methods for abdominal pains and spleen
infections will be paramount. Further understanding
of risk factors linked to spleen infection, including
age and underlying medical conditions, is vital to
devising preventive strategies.
6 CONCLUSION
The domain of medical diagnostics, particularly in
spleen infection segmentation and classification, is an
evolving landscape rife with opportunities for
technological intervention. Based on the analysis and
experimental results presented in our study, several
salient observations emerge that necessitate further
reflection:
● Efficacy of Novel Shallow Neural Network
(NSNN): NSNN is not only novel in its approach
but also effectively surpasses the accuracy rates of
more traditional methods, such as the Support
Vector Machine (SVM). With an accuracy of
75.47%, it underscores the potential of neural
networks in medical diagnostics.
● Potential of SVM: While the SVM trailed with an
accuracy of 66.33%, it remains a robust and
reliable method, especially considering its
extensive application in various diagnostic tasks
beyond spleen infection segmentation. Its
versatility and adaptability remain undeniable.
● Versatility of Machine Learning: The differences
in accuracy further highlight the inherent
versatility and potential of machine learning
techniques. Each algorithm, be it SVM or NSNN,
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
484

possesses unique characteristics suitable for
different types of datasets and diagnostic tasks.
● Challenges of Diagnostic Precision: While both
methods have respectable accuracy rates, there's
an inherent challenge in achieving near-perfect
diagnostic precision. This is crucial, especially in
medical contexts where diagnostic accuracy can
significantly impact patient outcomes.
● Need for Continuous Refinement: The realm of
medical diagnostics requires continuous
algorithmic refinement. As data grows and
becomes more intricate, algorithms like NSNN
and SVM will need regular updating to
accommodate new diagnostic challenges.
● Potential for Hybrid Models: Given the strengths
and limitations of both SVM and NSNN, there lies
an opportunity in exploring hybrid models that
integrate features from both techniques. Such a
blend could potentially harness the robustness of
SVM and the innovative aspects of NSNN to
achieve even higher accuracy rates.
In summation, the results from this study posit an
optimistic future for spleen infection segmentation
and classification. The NSNN, with an accuracy rate
of 75.47%, demonstrates considerable promise.
However, while it outperforms the SVM in our
experiments, the role and relevance of SVM in the
broader context of diagnostics cannot be diminished.
As we progress, the focus should be on leveraging the
strengths of both these techniques, possibly
integrating them, to pave the way for even more
precise and efficient diagnostic tools.
REFERENCES
Boiy, Erik, and Marie-Francine Moens. 2008. “A Machine
Learning Approach to Sentiment Analysis in
Multilingual Web Texts.” Information Retrieval 12 (5):
526–58.
Chen, Hongbo, Changchun Li, Mingdi Fang, Mengjin Zhu,
Xinyun Li, Rui Zhou, Kui Li, and Shuhong Zhao. 2009.
“Understanding Haemophilus Parasuis Infection in
Porcine Spleen through a Transcriptomics Approach.”
BMC Genomics 10 (1): 1–18.
Darling, Samuel T. 1906. “A Protozoön General Infection
Producing Pseudotubercles in The Lungs and Focal
Necroses In The Liver, Spleen And Lymphnodes.”
JAMA: The Journal of the American Medical
Association XLVI (17): 1283–85.
Davies, J. M., R. Barnes, and D. Milligan. 2002. “Update
of Guidelines for the Prevention and Treatment of
Infection in Patients with an Absent or Dysfunctional
Spleen.” Clinical Medicine 2 (5): 440.
Dietterich, Thomas G. 2002. “Machine Learning for
Sequential Data: A Review.” Structural, Syntactic, and
Statistical Pattern Recognition, 15–30.
Frank, Eibe, Mark Hall, Geoffrey Holmes, Richard Kirkby,
Bernhard Pfahringer, Ian H. Witten, and Len Trigg.
2009. “Weka-A Machine Learning Workbench for Data
Mining.” Data Mining and Knowledge Discovery
Handbook, 1269–77.
Goh, G. D., S. L. Sing, and W. Y. Yeong. 2020. “A Review
on Machine Learning in 3D Printing: Applications,
Potential, and Challenges.” Artificial Intelligence
Review 54 (1): 63–94.
Gómez Vela, Francisco A., Federico Divina, and Miguel
García-Torres. 2021. Computational Methods for the
Analysis of Genomic Data and Biological Processes.
MDPI.
Grimaldi, David, Olivier Pradier, Richard S. Hotchkiss, and
Jean-Louis Vincent. 2017. “Nivolumab plus Interferon-
γ in the Treatment of Intractable Mucormycosis.” The
Lancet Infectious Diseases 17 (1): 18.
Hosey, R. G., C. G. Mattacola, V. Kriss, T. Armsey, J. D.
Quarles, and J. Jagger. 2006. “Ultrasound Assessment
of Spleen Size in Collegiate Athletes.” British Journal
of Sports Medicine 40 (3): 251–54.
Jung, Andreas, Reinhard Maier, Jean-Pierre Vartanian,
Gennady Bocharov, Volker Jung, Ulrike Fischer,
Eckart Meese, Simon Wain-Hobson, and Andreas
Meyerhans. 2002. “Multiply Infected Spleen Cells in
HIV Patients.” Nature 418 (6894): 144–144.
King, Harold, and Harris B. Shumacker Jr. 1952. “Splenic
Studies: I. Susceptibility to Infection after Splenectomy
Performed in Infancy.” Annals of Surgery 136 (2): 239.
Landen, Serge, and Jean Closset. 2007. “Gas-Producing
Infection of the Spleen in a Super-Super-Obese
Patient.” Obesity Surgery 17 (10): 1416–18.
Mazur, Rafał, Milena Celmer, Jurand Silicki, Daniel
Hołownia, Patryk Pozowski, and Krzysztof
Międzybrodzki. 2018. “Clinical Applications of Spleen
Ultrasound Elastography – a Review.” Journal of
Ultrasonography 18 (72): 37–41.
Morris, Dudley H., and Frederick D. Bullock. 1919. “THE
IMPORTANCE OF THE SPLEEN IN RESISTANCE
TO INFECTION.” Annals of Surgery 70 (5): 513.
Newland, Adrian, Drew Provan, and Steven Myint. 2005.
“Preventing Severe Infection after Splenectomy.” BMJ
331 (7514): 417–18.
Qiu, Junfei, Qihui Wu, Guoru Ding, Yuhua Xu, and Shuo
Feng. 2016. “A Survey of Machine Learning for Big
Data Processing.” EURASIP Journal on Advances in
Signal Processing 2016 (1): 1–16.
Vanus, Jan, Jan Kubicek, Ojan M. Gorjani, and Jiri
Koziorek. 2019. “Using the IBM SPSS SW Tool with
Wavelet Transformation for CO2 Prediction within IoT
in Smart Home Care.” Sensors 19 (6): 1407.
Wang, Y. Q., L. Lü, S. P. Weng, J. N. Huang, S-M Chan,
and J. G. He. 2006. “Molecular Epidemiology and
Phylogenetic Analysis of a Marine Fish Infectious
Spleen and Kidney Necrosis Virus-like (ISKNV-Like)
Virus.” Archives of Virology 152 (4): 763–73.
A Spleen Infection Recognition Approach Using Shallow Neural Network in Comparison with Support Vector Machine
485

You, Zhen, Qinghe Zhang, Changjun Liu, Jiuzhou Song,
Ning Yang, and Ling Lian. 2019. “Integrated Analysis
of lncRNA and mRNA Repertoires in Marek’s Disease
Infected Spleens Identifies Genes Relevant to
Resistance.” BMC Genomics 20 (1): 1–15.
Zerem, Enver, and Jacob Bergsland. 2006. “Ultrasound
Guided Percutaneous Treatment for Splenic Abscesses:
The Significance in Treatment of Critically Ill
Patients.” World Journal of Gastroenterology: WJG 12
(45): 7341.
Boiy, Erik, and Marie-Francine Moens. 2008. “A Machine
Learning Approach to Sentiment Analysis in
Multilingual Web Texts.” Information Retrieval 12 (5):
526–58.
Chen, Hongbo, Changchun Li, Mingdi Fang, Mengjin Zhu,
Xinyun Li, Rui Zhou, Kui Li, and Shuhong Zhao. 2009.
“Understanding Haemophilus Parasuis Infection in
Porcine Spleen through a Transcriptomics Approach.”
BMC Genomics 10 (1): 1–18.
Darling, Samuel T. 1906. “A Protozoön General Infection
Producing Pseudotubercles in The Lungs and Focal
Necroses In The Liver, Spleen And Lymphnodes.”
JAMA: The Journal of the American Medical
Association XLVI (17): 1283–85.
Davies, J. M., R. Barnes, and D. Milligan. 2002. “Update
of Guidelines for the Prevention and Treatment of
Infection in Patients with an Absent or Dysfunctional
Spleen.” Clinical Medicine 2 (5): 440.
Dietterich, Thomas G. 2002. “Machine Learning for
Sequential Data: A Review.” Structural, Syntactic, and
Statistical Pattern Recognition, 15–30.
Frank, Eibe, Mark Hall, Geoffrey Holmes, Richard Kirkby,
Bernhard Pfahringer, Ian H. Witten, and Len Trigg.
2009. “Weka-A Machine Learning Workbench for Data
Mining.” Data Mining and Knowledge Discovery
Handbook, 1269–77.
Goh, G. D., S. L. Sing, and W. Y. Yeong. 2020. “A Review
on Machine Learning in 3D Printing: Applications,
Potential, and Challenges.” Artificial Intelligence
Review 54 (1): 63–94.
Gómez Vela, Francisco A., Federico Divina, and Miguel
García-Torres. 2021. Computational Methods for the
Analysis of Genomic Data and Biological Processes.
MDPI.
Grimaldi, David, Olivier Pradier, Richard S. Hotchkiss, and
Jean-Louis Vincent. 2017. “Nivolumab plus Interferon-
γ in the Treatment of Intractable Mucormycosis.” The
Lancet Infectious Diseases 17 (1): 18.
G. Ramkumar, R. Thandaiah Prabu, Ngangbam Phalguni
Singh, U. Maheswaran, Experimental analysis of brain
tumor detection system using Machine learning
approach, Materials Today: Proceedings, 2021, ISSN
22147853, https://doi.org/10.1016/j.matpr.2021.01.246
Hosey, R. G., C. G. Mattacola, V. Kriss, T. Armsey, J. D.
Quarles, and J. Jagger. 2006. “Ultrasound Assessment
of Spleen Size in Collegiate Athletes.” British Journal
of Sports Medicine 40 (3): 251–54.
Jung, Andreas, Reinhard Maier, Jean-Pierre Vartanian,
Gennady Bocharov, Volker Jung, Ulrike Fischer,
Eckart Meese, Simon Wain-Hobson, and Andreas
Meyerhans. 2002. “Multiply Infected Spleen Cells in
HIV Patients.” Nature 418 (6894): 144–144.
King, Harold, and Harris B. Shumacker Jr. 1952. “Splenic
Studies: I. Susceptibility to Infection after Splenectomy
Performed in Infancy.” Annals of Surgery 136 (2): 239.
Kumar M, M., Sivakumar, V. L., Devi V, S.,
Nagabhooshanam, N., & Thanappan, S. (2022).
Investigation on Durability Behavior of Fiber
Reinforced Concrete with Steel Slag/Bacteria beneath
Diverse Exposure Conditions. Advances in Materials
Science and Engineering, 2022.
Landen, Serge, and Jean Closset. 2007. “Gas-Producing
Infection of the Spleen in a Super-Super-Obese
Patient.” Obesity Surgery 17 (10): 1416–18.
Morris, Dudley H., and Frederick D. Bullock. 1919. “The
Importance of The Spleen in Resistance to Infection.”
Annals of Surgery 70 (5): 513.
Newland, Adrian, Drew Provan, and Steven Myint. 2005.
“Preventing Severe Infection after Splenectomy.” BMJ
331 (7514): 417–18.
Qiu, Junfei, Qihui Wu, Guoru Ding, Yuhua Xu, and Shuo
Feng. 2016. “A Survey of Machine Learning for Big
Data Processing.” EURASIP Journal on Advances in
Signal Processing 2016 (1): 1–16.
S. G and R. G, "Automated Breast Cancer Classification
based on Modified Deep learning Convolutional Neural
Network following Dual Segmentation," 2022 3rd
International Conference on Electronics and
Sustainable Communication Systems (ICESC),
Coimbatore, India, 2022, pp. 1562-1569, doi:
10.1109/ICESC54411.2022.9885299.
Vanus, Jan, Jan Kubicek, Ojan M. Gorjani, and Jiri
Koziorek. 2019. “Using the IBM SPSS SW Tool with
Wavelet Transformation for CO2 Prediction within IoT
in Smart Home Care.” Sensors 19 (6): 1407.
Wang, Y. Q., L. Lü, S. P. Weng, J. N. Huang, S-M Chan,
and J. G. He. 2006. “Molecular Epidemiology and
Phylogenetic Analysis of a Marine Fish Infectious
Spleen and Kidney Necrosis Virus-like (ISKNV-Like)
Virus.” Archives of Virology 152 (4): 763–73.
You, Zhen, Qinghe Zhang, Changjun Liu, Jiuzhou Song,
Ning Yang, and Ling Lian. 2019. “Integrated Analysis
of lncRNA and mRNA Repertoires in Marek’s Disease
Infected Spleens Identifies Genes Relevant to
Resistance.” BMC Genomics 20 (1): 1–15.
Zerem, Enver, and Jacob Bergsland. 2006. “Ultrasound
Guided Percutaneous Treatment for Splenic Abscesses:
The Significance in Treatment of Critically Ill
Patients.” World Journal of Gastroenterology: WJG 12
(45): 7341
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
486
