
Comparison of Superpixel and Correlation Based Segmentation for
Improved WBC Segmentation in Microscopic Images
Rayala Ramanjaneyulu and V. Rajmohan
Saveetha University Chennai, India
Keywords: Segmentation, Novel Superpixel Algorithm, Correlation Based Segmentation, White Blood Cells,
Microscopicimage, Accuracy, Medical.
Abstract: The objective of this investigation is to refine the precision of segmentation for white blood cells (WBCs) by
employing an innovative Superpixel algorithm. Additionally, the efficacy of this innovative approach will be
evaluated by comparing it to the correlation-based segmentation technique. Methods and Materials: The
specimens were examined using the new Superpixel Algorithm as group 1, encompassing N=10 samples,
while adopting the Correlation-based method as group 2 segmentation, also comprising N=10 samples. A
preliminary test power of 80%, alongside alpha and beta values of 0.05 and 0.2, respectively, were assumed,
along with a 95% confidence interval. The analysis was conducted using the MATLAB software. Results:
The novel Superpixel Algorithm demonstrated an accuracy of 87% in segmenting white blood cells, whereas
the Correlation-based segmentation achieved 74% accuracy. A statistically significant p-value of 0.001 (p <
0.05, two-tailed), determined through SPSS statistical analysis, indicates a substantial disparity in the
segmentation data. This difference affirms the absence of errors in the acquired data. Conclusion: The
Superpixel method surpasses the correlation-based segmentation approach in terms of enhancing the accuracy
of white blood cell segmentation.
1 INTRODUCTION
Accurate segmentation of white blood cells (WBCs)
within microscopic images holds paramount
importance in the domain of medical image analysis.
This significance arises from the fact that the
accuracy of the segmentation outcomes directly
impacts the overall precision of subsequent analyses
and diagnostic interpretations. Segmentation
algorithms play a pivotal role in this procedure by
delineating the target entities from the background
(Alharbi et al. 2022). Various methodologies for
WBC segmentation in microscopic images have been
explored, including thresholding, edge detection, and
region-based techniques. Among these approaches,
two of the most extensively utilized methods
encompass the innovative superpixel algorithm and
the correlation-based segmentation (Khan and Mir
2022). The novel superpixel algorithm partitions the
image into small, homogenous regions and
aggregates them into larger segments, whereas the
correlation-based segmentation employs the intensity
correlations among pixels to effectuate image
segmentation. Nonetheless, owing to the diverse
dissimilarities in shape, size, edges, and positions of
WBC cells, data collection can prove intricate in
medical devices. Furthermore, the lighting conditions
during image capture influence the extent of contrast
between the background and cell boundaries (AS et
al. 2013). The analysis of WBCs involves both cell
segmentation and feature extraction. During the white
blood cell segmentation process, leukocytes are
extracted from blood smear images. This extraction
endeavors to unveil distinctive attributes that
facilitate their differentiation from other categories of
blood cells (Mask R-CNN 2022).
A comprehensive compilation of research
materials encompasses 80 papers on GitHub, 40
papers on Google Scholar, and 30 papers within the
MathWorks articles repository. These publications
have surfaced within the last five years and pertain to
the specific research domain at hand. The
compositional analysis of white blood cells imparts
valuable medical insights into the patients' condition
(Vickram, A. S et al 2020). The findings derived from
the estimation provide crucial understanding of the
patient's health status, thereby streamlining the
diagnostic process. As a result, the accuracy of the
algorithmic differential white blood cell count
assumes a pivotal role in this context. The
Ramanjaneyulu, R. and Rajmohan, V.
Comparison of Superpixel and Correlation Based Segmentation for Improved WBC Segmentation in Microscopic Images.
DOI: 10.5220/0012543100003739
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Artificial Intelligence for Internet of Things: Accelerating Innovation in Industry and Consumer Electronics (AI4IoT 2023), pages 31-36
ISBN: 978-989-758-661-3
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
31

methodology frequently entails prevalent techniques
like image segmentation, feature extraction, and
classification to accomplish its objectives. Previous
studies indicate that this leads to diminished
accuracy, subsequently raising the accuracy level. To
address this limitation, a comparison is made between
the novel superpixel algorithm for WBC
segmentation and the correlation-based segmentation
(Lagerberg and Korte 2020) (Ramkumar, G. et al.
2021).
The microscopic examination of medical blood
smears, encompassing the quantification of diverse
types of white blood cells (WBCs), constitutes a
widely employed blood test in the realm of medical
hematology. Consequently, the enhancement of
precision for white blood cell images within the
microscopic view during the segmentation process
emerges as a crucial objective. This enhancement is
pursued through the integration of the innovative
superpixel algorithm with correlation-based
segmentation methodologies (Faried Effendy. 2022)
(Nanmaran et al 2022). As observed, some of the
drawbacks associated with existing techniques, such
as Correlation-based segmentation, involve
susceptibility to noise and artifacts in the image,
potentially leading to false positives or false
negatives. This approach necessitates a reference
image or template, which may not be available or
suitable for WBCs within microscopic images.
Additionally, Correlation-based segmentation might
struggle to capture the intricate shapes and structures
of WBCs, resulting in inaccurate segmentation
outcomes. The process is computationally demanding
and may necessitate substantial time and resources for
the analysis of extensive images or datasets.
The objective of this study is to surmount the
limitations associated with correlation-based
segmentation by implementing the super pixel
algorithm. The integration of the super pixel
algorithm renders the segmentation process more
resilient to noise and image artifacts, thereby
augmenting segmentation accuracy and conferring
greater adaptability compared to correlation-based
segmentation. This adaptability stems from the ability
to tailor super pixel algorithms to match various
image types and objects. For instance, distinct super
pixel algorithms can be employed based on the size
and shape of the segmented WBCs (Junaid Mir,
2022). From a broader perspective, the application of
a super pixel algorithm exhibits the potential to refine
the precision of white blood cell (WBC) segmentation
in microscopic images, surpassing the capabilities of
the correlation-based segmentation technique.
2 MATERIALS AND METHODS
This study was conducted within the Digital Image
Processing Laboratory, located in the Department of
Electronics and Communication Engineering at
Saveetha School of Engineering, which is a part of
SIMATS (Saveetha Institute of Medical and
Technical Sciences), situated in Tamil Nadu, India.
The central focus of this investigation was to enhance
the precision of white blood cell segmentation within
microscopic images. This improvement was
accomplished through the incorporation of an
innovative super pixel algorithm, and its performance
was assessed in comparison to correlation-based
segmentation. The determination of the sample size
was based on findings from a prior study (Nithyaa et
al. 2021). The current analysis was executed using the
clinical.com platform, maintaining a statistical power
of 80% with a confidence interval set at 0.86% and a
threshold set at 86%. Each group consisted of a
sample size of 10, resulting in an overall sample size
of 20. Furthermore, all images were uniformly resized
to a standard dimension of 512x512.
Super pixel Algorithm
Steps to super pixel algorithm :
• Input image
• SLIC segmentation
• DBSCAN clustering.
• Combining the diminutive and noisy super
pixels.
• Segmentation followed by amalgamation
of sizable and noisy super pixels.
For the purpose of obtaining merged regions, the
initial clusters are combined by considering factors
such as colour similarity and spatial adjacency.
Noisy super pixels with mixed colours are
segmented and subsequently integrated within each
of the merged regions.
Segmented image
The algorithmic steps and calculations are
performed according to the equation provided below
(Equation 1). The calculation of the number of pixels
in the input image (N) is derived from the input
image, and the number of super pixels employed for
segmenting the input image (K). The segmentation
output is obtained by dividing N by K.
S = √N/K (1)
N/K gives Approximate size of each super pixel.
Correlation based segmentation
Steps to correlation-based segmentation
1. Both sequences should be loaded.
2. Codification of images.
3. The correlation procedure.
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
32
4. Determine the peak location and intensity
5. peak>threshold? If yes go to step 6, if no
follow step 7.
6. The item is in the scene
7. The item is not the scene
8. What was the last object? If yes go to follow
step 9, if no go back to step 3
9. Evaluation.
Table 1: The process of collecting data for the Superpixel
Algorithm.
Iterations
Accuracy (%)
1
84.67
2
84.21
3
85.14
4
85.95
5
86.32
6
86.84
7
88.54
8
89.36
9
89.34
10
90.24
The correlation coefficient (ρ) acts as an indicator
of both the strength and direction of the linear
correlation existing between two variables,
designated as x and y. The covariance (cov(x,y))
between x and y measures the degree to which these
variables display concurrent fluctuations. Equation 2
is expressed as the mean value of the products of the
deviations of x and y from their individual means.
ρx,y = cov/(x,y)/σx σy (2)
Where, (cov) means covariance between x & y
σx, σy are variance of x & y
Data Collection: A dataset of microscopic images
featuring white blood cells (WBCs) was assembled
from publicly accessible sources such as medical
image repositories. The selection of images aimed to
encompass a diverse spectrum of WBC types and
imaging conditions.
Image Pre-processing: The amassed medical
microscope images underwent pre-processing
procedures designed to eliminate noise, artifacts, and
background details. Techniques like image denoising
and image thresholding were employed for this
purpose.
Segmentation Algorithms: The MATLAB
environment was employed to implement both the
Superpixel algorithm and the correlation-based
segmentation algorithm. The Superpixel algorithm
was utilized to generate Superpixels, which in turn
were applied for segmenting WBCs (R. Aarthi,
2019). On the other hand, the correlation-based
segmentation algorithm adopted a correlation-centric
approach for the WBC segmentation process.
Evaluation Metrics: The appraisal of the
effectiveness of the two algorithms encompassed the
utilization of diverse metrics, encompassing
accuracy, sensitivity, and specificity. These metrics
were computed for each image within the dataset and
were averaged across the entire dataset, furnishing a
comprehensive comparative assessment of the two
algorithms.
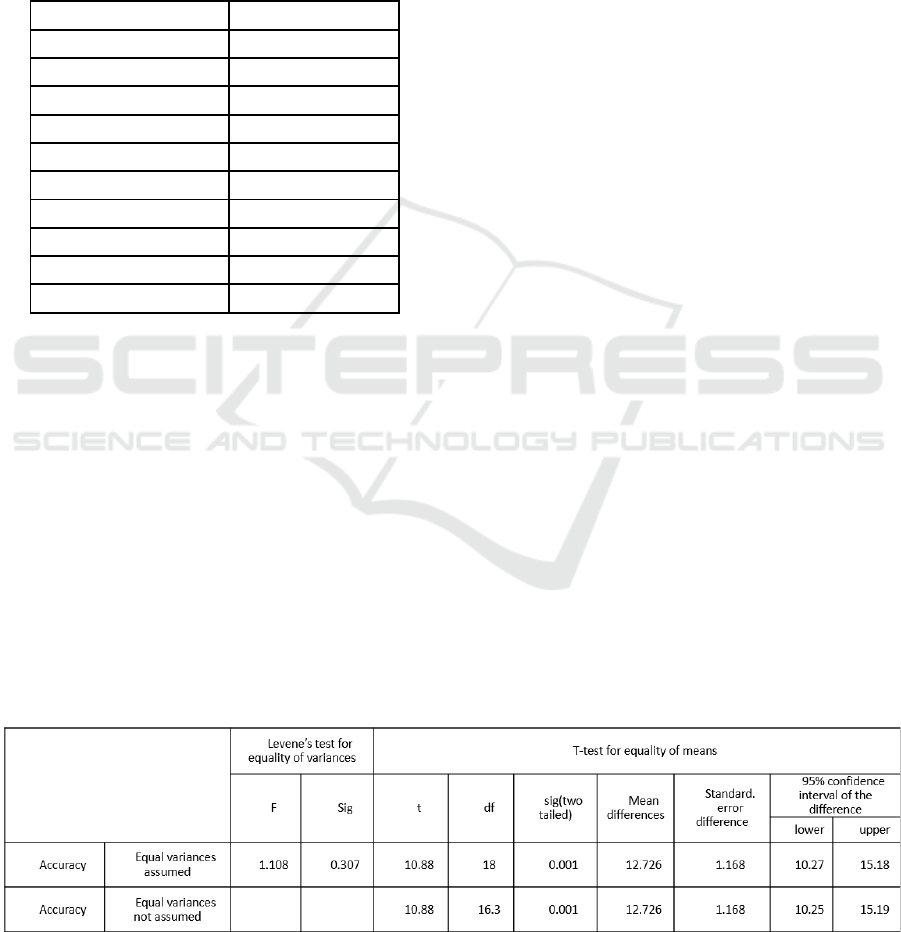
3 STATISTICAL ANALYSIS
Statistical analysis of the collected data for the
segmentation of white blood cells was carried out
using SPSS version 21 (Kelly Small, 2022). Within
the SPSS software, the Independent Sample t-test and
group statistics were calculated. In this context,
sensitivity, specificity, and noise were considered as
independent variables, whereas accuracy was treated
as the dependent variable.
Table 2: A comparison between the Superpixel algorithm and correlation-based segmentation.
Comparison of Superpixel and Correlation Based Segmentation for Improved WBC Segmentation in Microscopic Images
33
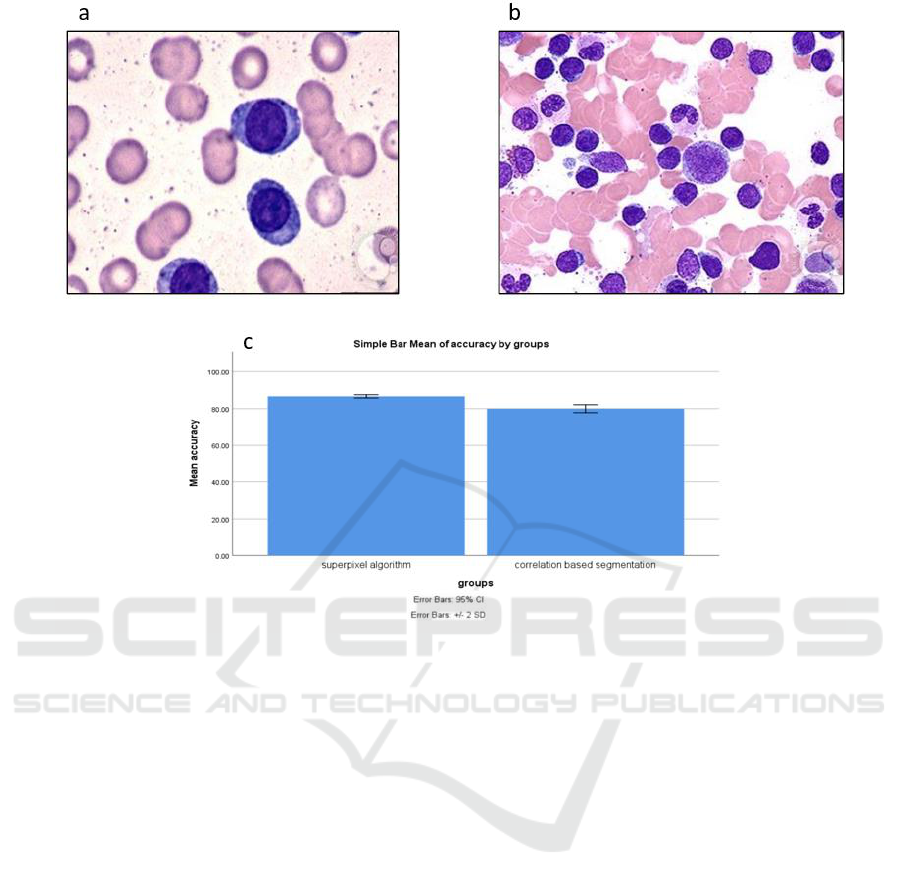
Figure 1: (a)&(b) White blood cells extracted from microscopic images (c) Mean accuracy Superpixel algorithm and the
Correlation-based segmentation algorithm.
4 RESULTS
Table 1 presents the compiled data for the Superpixel
Algorithm, utilizing a sample size of 10 (N = 10),
pertaining to the segmentation of white blood cells
through microscopic images. The results highlight
notable improvements in accuracy. Table 2 exhibits
the collected data concerning the Correlation-Based
Segmentation Algorithm, employing a sample size of
10 (N = 10), focusing on the segmentation of white
blood cells within microscopic images. Remarkably,
the findings suggest comparatively diminished
accuracy.
Figure 1a: White blood cells extracted from
microscopic images (datasets) were obtained from
blood samples for analysis. These cells were
employed as input for data collection via image
extraction. The resultant data encompassed counts of
both Red Blood Cells (RBCs) and White Blood Cells
(WBCs), along with corresponding accuracy
measurements.
Figure 1b: White blood cells extracted from
microscopic images (datasets) were sourced from
blood samples for examination. These cells served as
input for data collection through image extraction,
facilitating the determination of counts for both Red
Blood Cells (RBCs) and White Blood Cells (WBCs),
alongside accuracy measurements.
With an impressive overall accuracy of 87.06%,
this approach underscores its effectiveness in
successfully segmenting white blood cells within
microscopic images. It has been observed that a
comparison between the Superpixel and correlation-
based segmentation yields a plotted mean accuracy.
The highest accuracy is attained by the Superpixel
algorithm for image segmentation at 87.06%
5 DISCUSSIONS
To enhance the precision of white blood cell (WBC)
segmentation in microscopic images, two
methodologies were employed. In a comparative
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
34

assessment with correlation-based segmentation, the
Superpixel technique exhibited superior performance
in enhancing segmentation accuracy, yielding an
average accuracy score of 88% (Sera Tort. 2020). In
the SPSS statistical analysis, the resultant data
showcased a significantly lower p-value of 0.001%
(p<0.05), indicative of its statistical significance.
Employing the Superpixel method yielded a
segmentation accuracy of 87.06% for white blood
cells in images, whereas the utilization of the
correlation-based segmentation approach resulted in
an accuracy of 74%. These outcomes distinctly
underscore the superior performance of the
Superpixel algorithm (Shahzad et al. 2020). With the
application of the superpixel technique, the accuracy
was determined to be 87.06%, signifying a 13%
enhancement. Comparatively, the accuracy obtained
from a different algorithm (clustering) was measured
at 85% (Deshpande et al. 2022). The segmentation
process, implemented through the Superpixel
algorithm, exhibited a heightened accuracy of 87%,
consequently bolstering precision. Regarding this
research, no contradictory findings were encountered
(Al-Dulaimi et al. 2021).
A limitation of this study lies in the fact that
correlation-based segmentation might necessitate a
substantial volume of training data to achieve precise
WBC segmentation. This requirement could
potentially lead to a time-consuming and costly
process. Furthermore, correlation-based
segmentation could encounter challenges when
dealing with intricate objects, including overlapping
cells or cells exhibiting irregular shapes. In contrast,
super pixel algorithms might demand less training
data and prove to be more time-efficient. Thus, the
super pixel algorithm exhibits superiority over
correlation-based segmentation (Fatichah et al.
2022).
For future endeavours, the focus is on developing
algorithms that can attain high-accuracy
segmentation within a reduced timeframe. The goal is
to produce algorithms capable of achieving optimal
accuracy levels while streamlining the time required
for the segmentation process.
6 CONCLUSION
Through an evaluation of performance metrics and a
comparison of two algorithms, two different
techniques for calculating the accuracy of WBC
segmentation were scrutinized. The results revealed
that the super pixel algorithm achieved a notably
higher mean accuracy of 87% in comparison to the
correlation-based segmentation, which yielded a
mean accuracy of 74%. Consequently, the super pixel
algorithm demonstrated enhanced efficacy in
enhancing image accuracy. Additionally, the standard
deviation for the super pixel algorithm was
determined to be 2.16385, while the correlation-based
segmentation exhibited a higher standard deviation of
2.99625.
REFERENCES
Al-Dulaimi, Khamael, Jasmine Banks, Kien Nguyen,
Aiman Al-Sabaawi, Inmaculada Tomeo-Reyes, and
Vinod Chandran. (2021). “Segmentation of White
Blood Cell, Nucleus and Cytoplasm in Digital
Haematology Microscope Images: A Review-
Challenges, Current and Future Potential Techniques.”
IEEE Reviews in Biomedical Engineering 14
(January): 290–306.
Alharbi, Amal H., C. V. Aravinda, Meng Lin, P. S.
Venugopala, Phalgunendra Reddicherla, and Mohd
Asif Shah. (2022). “Segmentation and Classification of
White Blood Cells Using the UNet.” Contrast Media &
Molecular Imaging 2022 (July): 5913905.
Almassri, Hasan, Tim Dackermann, and Norbert Haala.
(2019). “mDBSCAN: Real Time Superpixel
Segmentation by DBSCAN Clustering Based on
Boundary Term.” Proceedings of the 8th International
Conference on Pattern Recognition Applications and
Methods. https://doi.org/10.5220/0007249302830291.
AS, Vickram, Raja Das, Srinivas MS, Kamini A. Rao, and
Sridharan TB. (2013)"Prediction of Zn concentration in
human seminal plasma of Normospermia samples by
Artificial Neural Networks (ANN)." Journal of assisted
reproduction and genetics 30 (2013): 453-459.
Axel, Leon, Puneet Bhatla, Dan Halpern, Silvia Magnani,
Jadranka Stojanovska, and Chirag Barbhaiya. (2023).
“Correlation of MRI Premature Ventricular Contraction
Activation Pattern in Bigeminy with Electrophysiology
Study-Confirmed Site of Origin.” The International
Journal of Cardiovascular Imaging 39 (1): 145–52.
Burch, Jane, and Sera Tort. (2020). “What Is the Accuracy
of Changes in White Blood Cell (WBC) Count for
Diagnosing COVID-19?” Cochrane Clinical Answers.
https://doi.org/10.1002/cca.3398.
Casler, Kelly Small. (2022). “White Blood Cells and the
Differential Count.” Laboratory Screening and
Diagnostic Evaluation.
https://doi.org/10.1891/9780826188434.0165.
“Classification of Acute Lymphoblastic Leukemia on
White Blood Cell Microscopy Images Based on
Instance Segmentation Using Mask R-CNN.” (2022).
International Journal of Intelligent Engineering and
Systems. https://doi.org/10.22266/ijies2022.1031.54.
Deshpande, Nilkanth Mukund, Shilpa Gite, Biswajeet
Pradhan, Ketan Kotecha, and Abdullah Alamri. (2022).
“Improved Otsu and Kapur Approach for White Blood
Comparison of Superpixel and Correlation Based Segmentation for Improved WBC Segmentation in Microscopic Images
35

Cell Segmentation Based on LebTLBO Optimization
for the Detection of Leukemia.” Mathematical
Biosciences and Engineering: MBE 19 (2): 1970–2001.
Fatichah, Chastine, Diana Purwitasari, Victor Hariadi, and
Faried Effendy. (2022). “Overlapping White Blood
Cell Segmentation And Counting On Microscopic
Blood Cell Images.” International Journal on Smart
Sensing and Intelligent Systems.
https://doi.org/10.21307/ijssis-2017-705.
Khan, Rehan, and Junaid Mir. (2022). “White Blood Cells
Segmentation and Classification Using U-Net CNN and
Hand-Crafted Features.” 2022 International
Conference on IT and Industrial Technologies (ICIT).
https://doi.org/10.1109/icit56493.2022.9988955.
Nanmaran, R., Srimathi, S., Yamuna, G., Thanigaivel, S.,
Vickram, A. S., Priya, A. K., ... & Muhibbullah, M.
(2022). Investigating the role of image fusion in brain
tumor classification models based on machine learning
algorithm for personalized medicine. Computational
and Mathematical Methods in Medicine, 2022.
Nithyaa1, A. N. Nithyaa A. N., A. N. Nithyaa A. N.
Nithyaa, Kumar R. Prem, Gokul M Gokul, and Geetha
Aananthi C. (2021). “Matlab Based Potent Algorithm
for WBc Cancer Detection and Classification.”
Biomedical and Pharmacology Journal.
https://doi.org/10.13005/bpj/2328.
Ramalakshmi, M., & Vidhyalakshmi, S. (2021). GRS
bridge abutments under cyclic lateral push. Materials
Today: Proceedings, 43, 1089-1092.
Ramkumar, G. et al. (2021). “Energetic Glaucoma
Segmentation and Classification Strategies Using
Depth Optimized Machine Learning Strategies”
Contrast Media & Molecular Imaging Volume 2021,
Article ID 5709257, 11 Pages, 2021
https://doi.org/10.1155/2021/5709257
S. K. Sarangi, Pallamravi, N. R. Das, N. B. Madhavi, P.
Naveen, and A. T. A. K. Kumar, “Disease Prediction
Using Novel Deep Learning Mechanisms,” J. Pharm.
Negat. Results, vol. 13, no. 9, pp. 4267–4275, (2022),
doi: 10.47750/pnr.2022.13.S09.530
Shahzad, Muhammad, Arif Iqbal Umar, Muazzam A. Khan,
Syed Hamad Shirazi, Zakir Khan, and Waqas Yousaf.
(2020). “Robust Method for Semantic Segmentation of
Whole-Slide Blood Cell Microscopic Images.”
Computational and Mathematical Methods in Medicine
2020 (January): 4015323.
V, Arsha P., P. V. Arsha, and Pillai Praveen Thulasidharan.
(2018). “A Survey Of White Blood Cell Segmentation
In Medical Image Analysis.” International Journal of
Computer Sciences and Engineering.
https://doi.org/10.26438/ijcse/v6si6.9194.
Verma, J. P. (2013). “Data Analysis in Management with
SPSS Software.” https://doi.org/10.1007/978-81-322-
0786-3.
Vickram, A. S., Samad, H. A., Latheef, S. K., Chakraborty,
S., Dhama, K., Sridharan, T. B., ... & Gulothungan, G.
(2020). Human prostasomes an extracellular vesicle–
Biomarkers for male infertility and prostrate cancer:
The journey from identification to current knowledge.
International journal of biological macromolecules,
146, 946-958.
Khan, Rehan, and Junaid Mir. (2022). “White Blood Cells
Segmentation and Classification Using”
V. P. Parandhaman, "A Secured Mobile Payment
Transaction Handling System using Internet of Things
with Novel Cipher Policies," (2023) International
Conference on Advances in Computing,
Communication and Applied Informatics (ACCAI),
Chennai, India, 2023, pp. 1-8, doi:
10.1109/ACCAI58221.2023.10200255.
U-Net CNN and Hand-Crafted Features.”
(2022)International Conference on IT and Industrial
Technologies (ICIT).
https://doi.org/10.1109/icit56493.2022.9988955.
Ware, Alisha D. (2020). “The Complete Blood Count and
White Blood Cell Differential.”
Contemporary Practice in Clinical Chemistry.
https://doi.org/10.1016/b978-0-12-815499-1.00025-9.
AI4IoT 2023 - First International Conference on Artificial Intelligence for Internet of things (AI4IOT): Accelerating Innovation in Industry
and Consumer Electronics
36
