
Hyperparameter Optimization Using Genetic Algorithm for
Extracting Social Determinants of Health Text
Navya Martin Kollapally
1a
and James Geller
2b
1
Department of Computer Science, New Jersey Institute of Technology, Newark, U.S.A.
2
Department of Data Science, New Jersey Institute of Technology, Newark, U.S.A.
Keywords: Hyperparameter Optimization, Clinical BioBERT, Social Determinants of Health (SDoH), Ontology,
Electronic Health Record (EHR), Genetic Algorithm, Simulated Annealing.
Abstract: Clinical factors account only for a small portion, about 10-30%, of the controllable factors that affect an
individual's health outcomes. The remaining factors include where a person was born and raised, where he/she
pursued their education, what their work and family environment is like, etc. These factors are collectively
referred to as Social Determinants of Health (SDoH). Our research focuses on extracting sentences from
clinical notes, using an SDoH ontology (called SOHO) to provide appropriate concepts. We utilize recent
advancements in Deep Learning to optimize the hyperparameters of a Clinical BioBERT model for SDoH
text. A genetic algorithm-based hyperparameter tuning regimen improved with principles of simulated
annealing was implemented to identify optimal hyperparameter settings. To implement a complete classifier,
we pipelined Clinical BioBERT with two subsequent linear layers and two dropout layers. The output predicts
whether a text fragment describes an SDoH issue of the patient. The proposed model is compared with an
existing optimization framework for both accuracy of identifying optimal parameters and execution time.
1 INTRODUCTION
Social determinants of health (SDoH) are the non-
clinical factors such as where an individual was born,
lives, studies, works, plays, etc. that affect a wide
range of clinical outcomes (US Department of Health
and Human Services, 2023). Existing research has
indicated that most of the SDoH data in Electronic
Health Records (EHRs) are represented as
unstructured text (US Department of Health and
Human Services, 2023; EHRIntelligence, 2021).
Ontologies play an important role in clinical text
mining. Medical ontologies/terminologies are used to
identify and extract information from clinical
documents. The UMLS Metathesaurus (Bodenreider,
2004) is a large biomedical resource that includes
standard biomedical vocabularies such as SNOMED
CT (Cote & Robboy, 1980), ICD-10-CM (Janca &
Bedirhan, 1993), MeSH (Rogers, 1965), and over 180
other vocabularies. Many user-generated phrases
a
https://orcid.org/0000-0003-4004-6508
b
https://orcid.org/0000-0002-9120-525X
To avoid confusion between SDoH and SOHO we have
used Agency FB font for SOHO in this manuscript.
such as “verbally responsive,” “vitals stable on
admission” and “unresponsive patient with abnormal
vitals” that clinicians use daily may not be captured
at the granularity required using only concepts from
the UMLS. Hence, we are utilizing concepts from the
specialized SOHO
1
ontology (Kollapally, Chen, Xu, &
Geller, 2022), along with regular expression (regex)-
based programming techniques for identifying
relevant text.
We are utilizing a deep neural network, the Clinical
BioBERT model, for clinical note classification. The
performance of a machine learning model depends on
the quality of data it is trained with, but an equally
important factor is the correct choice of
hyperparameters. There are various methods to
identify optimal hyperparameters of a model. They
include Bayesian optimization (Klein, 2017), grid
search (Bergstra & Bengio, 2012), evolutionary
optimization (Hutter, 2018), meta learning
(Vanschoren, Soares, & Brazdil, 2014), and bandit-
300
Kollapally, N. and Geller, J.
Hyperparameter Optimization Using Genetic Algorithm for Extracting Social Determinants of Health Text.
DOI: 10.5220/0012310300003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 300-307
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

based methods (Li, Jamieson, & DeSalvo, 2017), etc.
All these techniques have a search space defined by the
choice and range of parameters under consideration.
The goal of this paper is to identify, from a large
database of clinical text (the MIMIC-III database)
(Johnson, 2016), text samples that express an SDoH
sentiment about the described patient (but not about
people related to the patient). This is achieved in a
two-step process. First, we extract text samples with
a regular expression that looks for concepts from the
SDoH ontology (SOHO) in the input text. However,
some text samples use a SOHO term in an incidental
way, not really referring to an SDoH issue of the
patient. To classify text input as being SDoH text or
not, we use a neural network pipeline. We combine a
Clinical BioBERT (Alsentzer, Murphy, & Boag,
2019) model with a neural network classifier
framework. To achieve a better performance, we
optimized the selected hyperparameters of the model
using a genetic algorithm (GA). We considered three
ML optimizers, namely AdamW (Zhang, 2018),
Adafactor (Noam Shazeer, 2018) and LAMB (You,
2020). Alongside the GA operations called n-bit
crossover and random bit flip mutations, we also used
roulette-wheel selection to obtain the optimal
candidate solution using the genetic algorithm.
We framed this problem as an entity recognition
task and used the latest advancements in large language
models (LLM), specifically Universal NER (Zhou &
Zhang, 2023). Universal NER uses a smaller model
with minimal parameters that it learned from its teacher
LLM model gpt-3.5-tubo-0301, by applying target
distillation. Additionally, we employed the state-of-
the-art hyperparameter optimization framework
Optuna (Akiba, Sano, & Yanase, 2019) to compare the
results with our model. The Optuna hyperparameter
optimization framework is among the latest
advancements in this field and is unique because of its
define-by-run and pruning strategies. The comparison
studies of our model with Universal NER and Optuna
will be presented in the Discussion Section.
2 METHODS
2.1 Model Architecture
The Clinical BioBERT model architecture is a multi-
layer bidirectional transformer encoder
implementation. The input data is converted into token
embeddings, each as a 768-dimensional vector
representation. This (768) is the standard size in the
BERT architecture. The input embeddings are first
passed through a multi-head self-attention
mechanism. The self-attention mechanism generates a
set of attention weights that are used to weigh the
importance of each token in the input sequence. The
context vector is passed through a position-wise feed-
forward neural network, which further transforms it.
The classification layer takes the CLS token of the last
layer and predicts the context of the text sample. This
layer is made up of two linear layers separated by two
drop out layers. Figure 1 shows the model architecture
of Clinical BioBERT for SDoH text classification.
2.2 Dataset
We utilized the SOHO ontology (Kollapally, Chen, Xu,
& Geller, 2022), available in BioPortal, as a reference
terminology for extracting concepts from MIMIC-III
v1.4. The concepts in the SOHO branch “Social
determinants of health” were used for concept
extraction from MIMIC-III clinical notes. MIMIC-III
contains data associated with 53,423 distinct hospital
admissions for patients 16 years and up, admitted to
critical care units between 2001 and 2012. We
specifically utilized clinical notes available in the
Note_events table, which is a 4GB data file.
The Stanford NLTK library was used for text pre-
processing. After stop word removal and converting
the text to all lower case, the clinical notes from
MIMIIC-III were fed to a regex-based Python script to
extract text fragments with SDoH concepts in them.
Using regular expressions, whenever we found a
concept in the Note_events file that matched a
concept in the SOHO ontology, we extracted the
preceding four sentences, and the succeeding four
sentences from Note_events. Preliminary
observations showed that this is typically sufficient to
capture the SDoH context. Not all rows of data
returned by the Python regex script expressed a strong
SDoH sentiment about the patient under
consideration. Hence, we performed a manual review
of a subset of approximately 1500 rows of extracted
text, and we annotated 1054 rows of them with the
label “1” for training the Clinical BioBERT
architecture. Those sentences described SDoH
statements about the patient. Negative training
samples (1130 rows) were extracted from admission
labs, discharge labs and discharge instructions and
labelled as “0.” These do not describe SDoH
statements about the patient. The resulting 2184 rows
of data were split into 80% training and 20% test data.
2.3 Choice of Optimizers
Adaptive optimization algorithms such as Adam tend
to have a better performance compared to Stochastic
Hyperparameter Optimization Using Genetic Algorithm for Extracting Social Determinants of Health Text
301
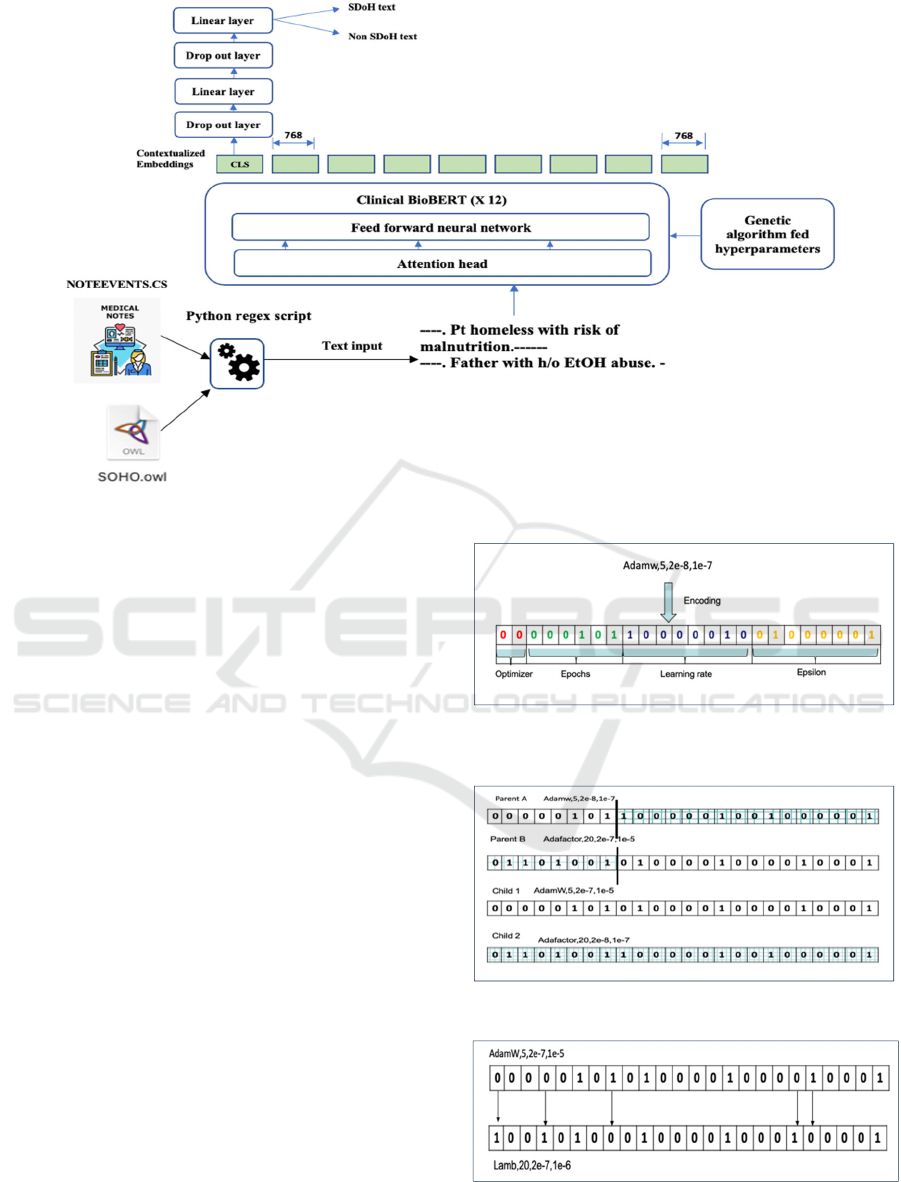
Figure 1: Model architecture of Clinical BioBERT for SDoH text classification, modified from (E. Alsentzer, June 2019).
Gradient Descent (SGD) optimization (Loshchilov &
Hutter, 2017). An improved version of Adam, called
AdamW, exhibits a better performance. Layer-wise
Adaptive Moments optimizer for Batch training
(LAMB) uses an accurate layer-wise trust ratio to
adjust the Adam optimizer’s learning rate. Thus, the
three optimizer types that we compared in this
research were AdamW, Adafactor and LAMB. The
hyperparameters chosen for this study are optimizer
type, epoch number, learning rate (, and epsilon (ε)
these were selected, based on benchmarks provided
by previous scholarly articles (You, 2020). Epoch
counts chosen were 5, 10, 15, 20, 25, 35, and 50. The
learning rates ranged from 2e-8 (i.e., 2*10
-8
) to 1e-1.
2.4 Evolutionary Strategies
Following the terminology of genetic programming,
each of the hyperparameters is encoded as a
“chromosome,” using binary encoding. Each
chromosome consists of four genes and is 24 bits
long. We used two bits to represent the optimizer, six
bits for the epoch number, eight bits for the learning
rate, and eight bits for ε (Figure 2). We started with a
random initial population of 20 chromosomes per
generation.
Roulette-wheel selection is a probabilistic
approach that ensures that the population does not just
consist of elite candidates; it also contains some weak
solutions. Roulette-wheel selection ensures diversity
Figure 2: The 24-bit chromosome representing a candidate
in the population.
Figure 3: Sample encoding of 1-point crossover encoded.
Figure 4: Sample encoding of bit flip mutation.
HEALTHINF 2024 - 17th International Conference on Health Informatics
302
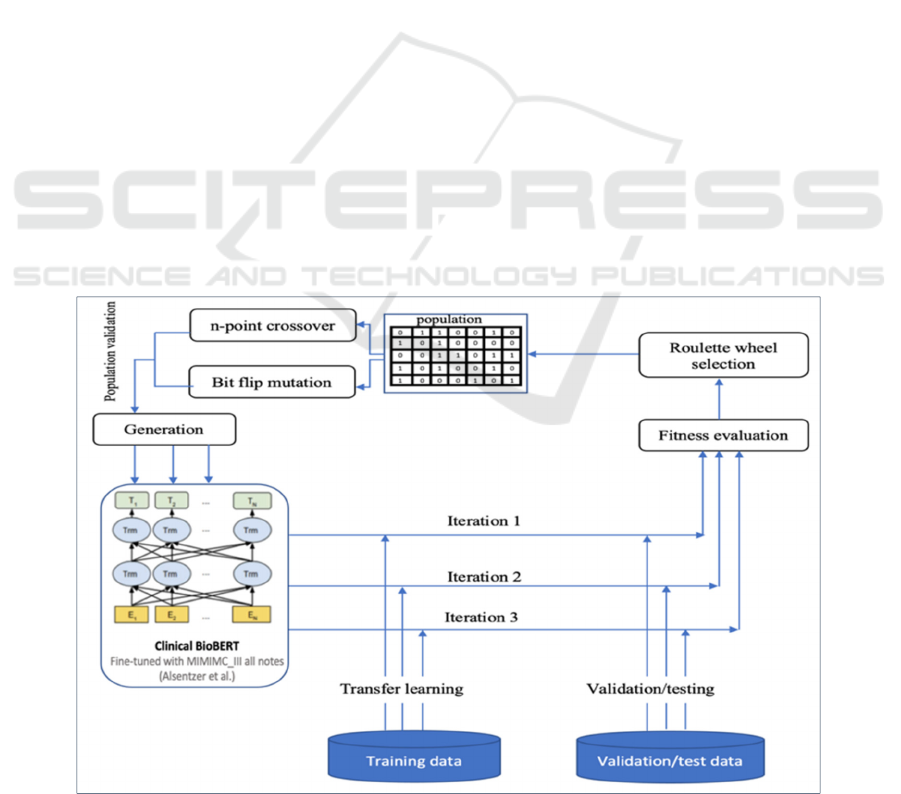
in the selection process, thus reducing the chance of
getting stuck in a local optimum in a multimodal
problem. Three iterations were performed with 25
population updates in each. The number of
generations was fixed as 25, based on the
convergence of cross entropy between consecutive
iterations. To perform recombination and mutation
operations, we used n-bit crossover and random bit
flip mutations. Figure 3 shows a 1-point crossover
operation where the crossover happens at the 7-th
locus position. At this point, the tail from parent B is
combined with the head of Parent A to generate child
1. To generate child 2, the head of parent B is
combined with the tail of parent A. We have used a
crossover probability (P
c
) of 0.75.
Recombination operations (i.e., crossover) ensure
that the best features are likely to persist into the next
generation. Mutations are a way of introducing new
features into the existing population. The mutation
probability P
m
is 0.03 in our GA. The offspring in
Figure 4 is generated by flipping the bits at loci 0, 3,
7, 18, and 19. We only choose viable offspring for the
next stage, while catastrophic offspring was
eliminated.
2.5 Fitness Evaluation
The evolutionary algorithm is guided by a fitness
evaluation representing the user’s objectives. Thus,
the formulation of an ideal fitness function is task-
specific. Accuracy is defined as the ratio of the
number of correctly classified data points to the total
number of data points.
The decoded chromosome values corresponding
to valid choices are used as hyperparameters in
training of Clinical BioBERT. The fitness of the
model is evaluated in terms of accuracy and those
hyperparameters corresponding to roulette wheel-
selected chromosomes are moved to the next
generation. Experiments were repeated three times
(denoted as three iterations) with three different
random initializations. In all three iterations, the
stopping criterion was that the accuracy did not
improve during four consecutive generations. Figure
5 represents the evolutionary approach of genetic
algorithm-based hyperparameter tuning.
3 ALGORITHM
We will now present an algorithm for optimizing the
set of hyperparameters in Clinical BioBERT, such
that the cross-entropy loss is minimal and fitness in
terms of accuracy is maximized. In Step 2 of
Algorithm 1, selected chromosome is a list of
chromosomes that have survived the selection
process. The variable counter in Step 3 is used to
escape local optima. In Step 4, elite_acc
prev
is the
accuracy of the best candidate from the previous
generation.
Figure 5: The evolutionary approach of genetic algorithm-based hyperparameter tuning.
Hyperparameter Optimization Using Genetic Algorithm for Extracting Social Determinants of Health Text
303

In Step 5, elite_error
prev
is the cross-entropy loss
of the best candidate from the previous generation. In
Step 6, elitist_acc is the accuracy of the best candidate
in the current generation. In Step 8, max_gen is the
maximum number of generational updates in an
iteration. Steps 9-20 are the core of the genetic
algorithm. It starts with choosing chromosomes with
viable combinations of traits, followed by limiting the
size of the population to 20. The first 2 bits encode
the optimizer type, as mentioned before. We use 00
to represent AdamW, 01 for Adafactor, and 10 for the
LAMB optimizer. To incorporate the fact that the best
traits from parents should persist in the offspring, we
perform n-bit crossover with probability P
c
(Step 14).
To introduce new traits, the chromosomes undergo bit
flip mutation with probability P
m
(Step 15). We
evaluate the fitness of the generation (Step 18) and
spin the roulette-wheel 20 times to choose 20
survivors to the next generation. The algorithm stops
if either 1000 evolutions have passed and the
algorithm has not converged toward an optimal
solution, or if the accuracy between successive
generations stays the same for four generations. In the
latter case, it might be stuck in a local optimum or it
already found the best global solution.
Algorithm 1: Finding optimal parameter set for Clinical BioBERT.
1 for iteration i=1 to 3: // run the experiment three times
//start with 24-bit encoded chromosomes, create a set of n random chromosomes C
1
to C
n
2 selected-chromosome= []
//list initialization to store the survivor chromosomes
3 counter=0
4 elite_acc
prev
=0 // elite_acc
prev
is the accuracy from best candidate of previous gen
5 elite_error
prev
=0// elite_error
prev
is the cross-entropy loss of best candidate of previous gen
6 elitist_acc=0
// elite_acc is the accuracy of the best candidate of current generation
7 max_gen=0
8 begin:
// start of genetic algorithm
9 max_gen +=1 // generation counter
10 for k=1 to n: // n is a random seed
11 validate viable chromosomes
12
//only valid chromosomes are captured in the list and undergo crossover and mutation
selected-chromosome. append (C
k
)
13 If len (selected-chromosome) =20:
14 break
15 apply n-bit crossover(p
c
) -> selected-chromosome
16 apply random bit flip mutation(p
m
) -> selected-chromosome
17
//P contains the viable chromosomes and their offspring
let P be the new population with parents and offspring
18 for g = 1 to len(P):
19
//decode the chromosome and run Clinical BioBERT model with hyperparameters
evaluate the fitness of chromosomes P
g
in terms of acc
g
20 apply Roulette-wheel selection and choose 20 from the new candidates
21 for g= 1 to 20:
22 if acc
g
> elitist_acc: //acc
g
is the accuracy from survivor chromosome
23 elitist_acc= acc
g
24 else if elitist_acc - elitist_acc
prev
~ 0:
25
//to make sure not stuck in local optimum we add weak chromosomes
add diverse valid weak chromosomes to selected-chromosome []
26 counter+=1
27 elitist_acc
prev
= elitist_acc
28 elitist_error
prev
= elitist_error
29 Continue to step 15 if counter < 5 or max_iter < 1000
30 end:
HEALTHINF 2024 - 17th International Conference on Health Informatics
304
4 RESULTS
For the population-based optimization, to find the
best global solution, a large size population with
diversity is a key factor. In our experiments, each
iteration performs 25 generational updates, each with
a population size of 20. Hence, in each iteration we
had a total population size of 20*25=500
chromosomes. To avoid the problem of local optima,
we considered three different initial configurations
(computed in three iterations), each with 500
chromosomes, thus totalling 500*3=1500
evaluations to derive the best hyperparameters.
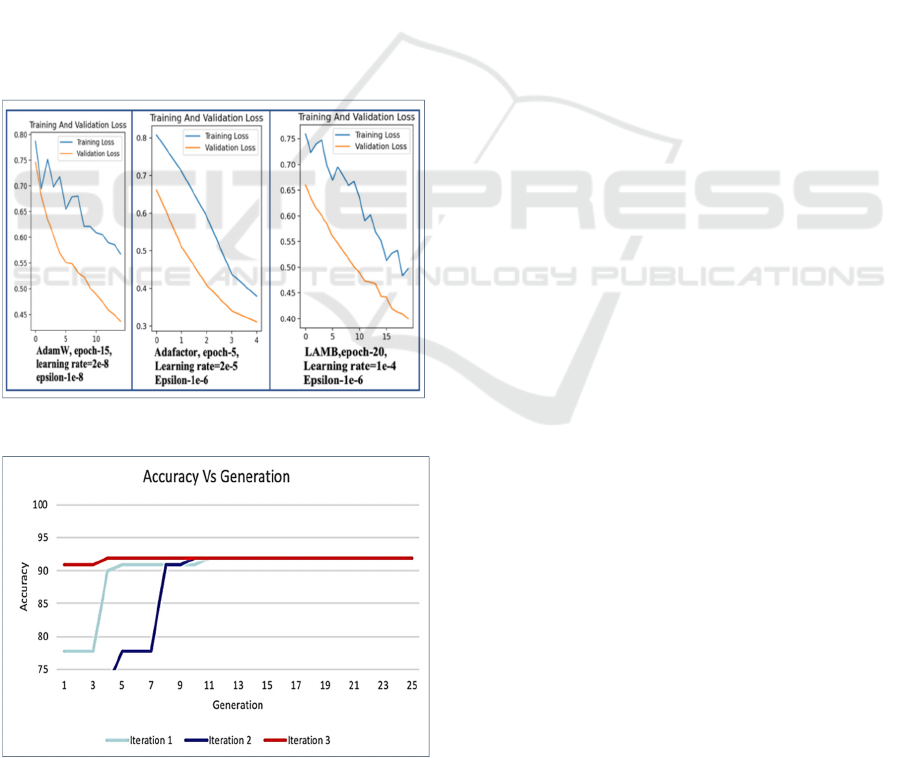
The graph in Figure 6 shows the validation vs
training loss curves for three iterations with respect
to the three optimizers. We found that the best
hyperparameter combination for Clinical BioBERT
uses the AdamW optimizer with a learning rate=2e-
8, a number of epochs=10, and epsilon=1e-08,
implemented along with a linear warmup scheduler.
This combination resulted in an accuracy of 91.91%
for the classification task.
Figure 6: Sample training vs Validation loss curve.
Figure 7: Best fitness values across all the generation.
Figure 7 above represents the fitness value of the best
candidate in each generation plotted for all three
iterations. Because the diversity of candidates was
maintained, the problem of local optima was
overcome, and our model converged to the best
global parameter set.
Table 1 shows a partial view of the decoded
chromosomes corresponding to the best candidate in
each generation. In our experiments, AdamW and
LAMB performed well, but Adafactor was never
found in any of the elite candidate solutions. The
highest accuracy with Adafactor was 63.7% for a
learning rate=1e-03, epsilon=1e-8, and epochs=25,
along with linear warmup and cosine annealing.
We observed that training with Adafactor was
also most time consuming, with a 3-fold increase in
time for Adafactor compared to AdamW. LAMB
found near optimal solutions and its time of training
was better than that of AdamW for higher epochs. For
instance, the LAMB optimizer finished the training 17
minutes faster than AdamW, when running both for
50 epochs and with equal learning rates and epsilon
values. The best accuracy was achieved by the model
with AdamW until epoch 10, at the expense of
training time, compared to the model using the
LAMB optimizer. The optimized model with the
learned parameters, i.e., weights and biases, was
stored using the Python Torch module.
5 DISCUSSION AND
LIMITATIONS
The context of SDoH text samples in clinical notes is
limited to a few sentences. In these situations, it is
important to perform an informed search for
hyperparameters. We compared the hyperparameters
obtained as part of this research with 1)
hyperparameters used in the Clinical BioBERT paper
of Alsentzer et al. (Alsentzer, Murphy, & Boag,
2019) (these are the same hyperparameters as in the
BERT paper), and 2) hyperparameters from Han’s
paper (Han, 2022) on multilabel classification of
SDoH data using BERT. To compare fairly, we
trained all three sets of hyperparameters on the BERT
model using our SDoH dataset.
In another experiment, we utilized Optuna to find
the hyperparameters for the designed model using the
SDoH dataset involving Bayesian sampling.
According to Optuna, the best hyperparameters were
AdamW, lr=2e-6, with dropout probabilities 0.1077
and 0.1763. The accuarcy and F1 scores were 0.9096
and 0.8992, respectively, for epoch value 10..
Hyperparameter Optimization Using Genetic Algorithm for Extracting Social Determinants of Health Text
305

Table 1: Decoded chromosomes with highest fitness functions across generations.
Gen Iteration 1 Iteration 2 Iteration 3
1 LAMB, 50,lr = 0.00001,eps = 1e-06 LAMB,25, lr = 0.00001,eps = 1e-05 AdamW,10, lr=2e-7, eps =1e-07
2 LAMB,50, lr = 0.00001,eps = 1e-05 LAMB ,25,lr = 0.00001,eps = 1e-05 AdamW,10, lr=2e-7, eps =1e-07
3 LAMB,50, lr = 0.00001,eps = 1e-05 LAMB,25, lr = 0.00001,eps = 1e-05 AdamW,10, lr=2e-7, eps =1e-07
4 LAMB ,25, lr = 0.001,eps = 1e-06 LAMB ,25,lr = 0.00001,eps = 1e-05 AdamW,10, lr=2e-8, eps=1e-08
………. …………. ………
23 AdamW,10, lr=2e-8, eps=1e-08 AdamW,10, lr=2e-8, eps=1e-08 AdamW,10, lr=2e-8, eps=1e-08
24 AdamW,10, lr=2e-8, eps=1e-08 AdamW,10, lr=2e-8, eps=1e-08 AdamW ,10,lr=2e-8, eps=1e-08
25 AdamW,10, lr=2e-8, eps=1e-08 AdamW,10, lr=2e-8, eps=1e-08 AdamW,10, lr=2e-8, eps=1e-08
Table 2: Hyperparameter comparison of the models.
Our results
Alsentzer et al. (Alsentzer,
Murphy, & Boag, 2019)
Han et al. (Han, 2022)
Optuna Frame work
AdamW, learning rate=2e-
08,
epochs=10, epsilon=1e-08,
batch size=16
Adam,learning rate= 5e-05,
epochs=2/3/4,
Epsilon=1e-12,
batchsize =16/32
Adam
momentum=0.9,learning
rate=1e-04, epochs=10,
batch size=32
AdamW, lr=2e-
6,D1=0.1077,D2=0.1763,e
pochs=10,batch size=16
Table 3: Performance metrics of all the considered benchmarks.
Metrics Our results
Alsentzer et al.
23
Han et al.
21
Optuna framework
Accuracy
0.91919
0.8 0.5454 0.9096
Micro F1 score 0.91919 0.8 0.5454 0.8992
Recall score 0.833333 0.666 0.8333 0.7322
Precision score 1.0 1.0 0.555 0.8372
Compared to 15 minutes to complete the entire
generation on NVIDIA GPUs with Pytorch CUDA,
it took four minutes for Optuna to find the optimal
parametes.
We used Universal NER for the phrase below.
“The patient reported an increased level of stress
the day prior to admission due to financial issues.
Also, the patient's social situation is complicated by
an impending separation and concern over the
abusive nature of her relationship with her husband.
She stated that she feels safe at home and was seen
by psychiatry and social work (please refer to OMR
notes) who believed she was safe for discharge to
home; the patient declined consultation by the
domestic violence service.”
Because Universal NER could not recognize the
entites “abusive relationship,” “Social situation,” and
“Domestic Violence Service,” it did not succed at
entity recognition for social context. Future research
will attempt to address this problem.
As noted before, not all the 72,668 rows returned
by the regular expression match expressed an SDoH
sentiment about the patient. However, it was
impossible to manually review all of them and we
limited the review to 1500 rows. Of those, only 1054
rows expressed an SDoH sentiment about the patient.
Assuming that the same ratio (about 70%) holds for
the whole dataset there would be about 51,000 rows
expressing a sentiment about the patient. Verifying
that fact was beyond the scope of this paper.
6 CONCLUSIONS AND FUTURE
WORK
We performed genetic algorithm-based hyperpara-
meter tuning of a Clinical BioBERT model trained
on SDoH data. Our analysis suggests the best
configuration for the specific problem uses an
AdamW optimizer with a learning rate=2e-8, a
number of epochs=10 and epsilon=1e-08. This
achieved an accuracy of 91.91% and minimal cross
entropy loss. We conclude that the hyperparameters
obtained by our informed search using the genetic
algorithm outperformed the other models trained on
the same dataset. The optimal hyperparameters
presented in this paper for Clinical BioBERT should
be tested with other datasets, to determine if a similar
accuracy improvement can be achieved for text
classification in other domains.
HEALTHINF 2024 - 17th International Conference on Health Informatics
306

ACKNOWLEDGMENT
Research reported in this publication was supported
by the National Center for Advancing Translational
Sciences (NCATS), a component of the National
Institute of Health (NIH) under award number
UL1TR003017. The content is solely the
responsibility of the authors and does not necessarily
represent the official views of the National Institutes
of Health.
REFERENCES
Abbas, A., Afzal, M., Hussain, J., & Ali, T. (2021). Clinical
Concept Extraction with Lexical Semantics to Support
Automatic Annotation. International Journal of
Environmental Research and Public Health, 18-20.
Akiba, T., Sano, S., & Yanase, T. (2019). Optuna: A Next-
generation Hyperparameter Optimization Framework.
ArXiv.
Alsentzer, E., Murphy, J., Boag, W., & et al. (2019).
Publicly Available Clinical Embedding. Proceedings
of the 2nd Clinical Natural Language Processing
Workshop.
Bergstra, J., & Bengio, Y. (2012). Random search for
hyper-parameter optimization. 281–305.
Bodenreider, O. (2004). The Unified Medical Language
System (UMLS): integrating biomedical terminology.
Nucleic Acids Research, 32.
Cote, R. A., & Robboy, S. (1980). Progress in medical
information management. Systematized nomenclature
of medicine (SNOMED). JAMA: The Journal of the
American Medical Association, 756–762.
Devlin, J. C. (2019). BERT: Pre-training of Deep
Bidirectional Transformers for Language
Understanding. North American Chapter of the
Association for Computational Linguistics.
E. Alsentzer, J. M.-H. (June 2019). Publicly Available
Clinical BERT Embeddings. " Proceedings of the 2nd
Clinical Natural Language Processing Workshop,, (pp.
pp. 72-78).
Han, S. Z. (2022). Classifying social determinants of health
from unstructured electronic health records using deep
learning-based natural language processing. Journal of
Biomedical Informatics, 127.
Hutter, J. N. (2018). Hyperparameter Importance Across
Datasets. Knowledge Discovery & Data Mining, 2367–
2376.
Janca, A., Bedirhan, T et al. (1993). The ICD-10 Symptom
Checklist: a companion to the ICD-10 Classification of
Mental and Behavioural Disorders. Social Psychiatry
and Psychiatric Epidemiology, 239–242.
Johnson, A. E. (2016). MIMIC-III, a freely accessible
critical care database. Scientific data.
Klein, A. F. (2017). Fast Bayesian Optimization of
Machine Learning Hyperparameters on Large
Datasets. 528–536.
Kollapally, N., Chen, Y., Xu, J., & Geller, J. (2022). An
Ontology for the Social Determinants of Health
Domain. IEEE International Conference on
Bioinformatics and Biomedicine (BIBM).
Li, L., Jamieson, K. G., DeSalvo, G., et al. (2017).
Hyperband: Bandit-Based Configuration Evaluation
for Hyperparameter Optimization. International
Conference on Learning Representations.
Loshchilov, I., & Hutter, F. (2017). Decoupled Weight
Decay Regularization. International Conference on
Learning Representations.
Noam Shazeer, &. S. (2018). Adafactor: Adaptive
Learning Rates with Sublinear Memory Cost.
International Conference on Machine Learning.
Rogers, F. (1965). Medical Subject Headings. Nature,
236–236.
US Department of Health and Human Services, U. D.
(2023). Healthy people 2023. Retrieved from
https://health.gov/healthypeople/prioriareas/socialdete
rminants-health
Vanschoren, J., Soares, C., & Brazdil, P. (2014). Meta
learning and algorithm selection. CEUR workshop
proceedings, 298–309.
You, Y. L. (2020). Large Batch Optimization for Deep
Learning: Training BERT in 76 minutes. International
Conference on Learning Representations.
Zhang, Z. (2018). Improved Adam Optimizer for Deep
Neural Networks. IEEE/ACM 26th International
Symposium on Quality of Service (IWQoS).
Zhou, W., & Zhang, S. G. (2023). UniversalNER: Targeted
Distillation from Large Language Models for Open
Named Entity Recognition. ArXiv (Cornell
University).
Hyperparameter Optimization Using Genetic Algorithm for Extracting Social Determinants of Health Text
307
