
Compositional Techniques for Asynchronous Boolean Networks
Maram Alshahrani
1,2 a
and Jason Steggles
1 b
1
School of Computing, Newcastle University, Newcastle upon Tyne, U.K.
2
Faculty of Computer Science, King Khalid University, Abha, Saudi Arabia
Keywords:
Asynchronous Boolean Networks, Model Composition, Behaviour Preservation, Attractor Identification.
Abstract:
Asynchronous Boolean networks are an important qualitative modelling approach for analysing and engineer-
ing biological systems. However, their practical application is limited by the state space explosion problem
and lack of engineering tools. To help address these limitations we develop new compositional techniques
for constructing and analysing asynchronous Boolean networks based on the idea of merging entities using
Boolean operators. We propose a novel asynchronous interference state graph to model the interference that
occurs in a composition and develop a range of important new asynchronous compositional techniques for
analysing behavioural preservation and identifying point attractors.
1 INTRODUCTION
Boolean networks (Kauffman, 1969) are a widely
studied qualitative modelling approach (Barbuti et al.,
2020) that has been successfully applied to a range of
biological systems (for example, see (Pandey et al.,
2010; Dahlhaus et al., 2016)). A Boolean network
abstractly represents the state of regulatory entities as
Boolean values and provides update rules to capture
how these entities interact. They can provide key in-
sights into the behaviour of a regulatory system by
considering the attractors (state cycles) they exhibit
(for example, see (Huang and Ingber, 2000; Saadat-
pour et al., 2011)). The global state of a Boolean net-
work can be updated either synchronously (all entities
update their state simultaneously) or asynchronously
(entities update their state independently and non-
deterministically) (Schwab et al., 2020). While the
synchronous update scheme provides more tractable
analysis, the asynchronous update scheme is gener-
ally seen to allow more realistic dynamic behaviour
(Barbuti et al., 2020).
The practical application of Boolean networks is
limited by the state space explosion problem (Groote
et al., 2015) (exponential growth of state space) and
by the lack of engineering techniques. In this paper
we make an important contribution to supporting the
practical application of asynchronous Boolean net-
a
https://orcid.org/0009-0005-0793-0504
b
https://orcid.org/0000-0001-9174-5531
works by developing formal techniques for their com-
positional construction and analysis. We take as our
starting point an approach for composing syn-
chronous Boolean networks based on merging en-
tities between submodels using Boolean operators
(Alkhudhayr and Steggles, 2019; Abdulrahman and
Steggles, 2023). Adapting this approach to the asyn-
chronous setting involves dealing with the fundamen-
tal semantic differences between synchronous and
asynchronous updating schemes and has led to a
range of important new insights and results.
We begin by developing a new form of interfer-
ence state graph (Alkhudhayr and Steggles, 2019)
for the asynchronous setting which captures the inter-
ference that can occur to a Boolean network’s asyn-
chronous behaviour in a composition. We prove this
interference state graph bounds the asynchronous be-
haviour possible for a submodel in a composition and
then develop key results about the preservation of a
submodel’s behaviour in a composition.
We further strengthen our asynchronous compo-
sitional framework by developing new compositional
techniques for identifying point attractors (Hopfen-
sitz et al., 2013) (global states with no outgoing transi-
tions) in an asynchronous composition. We formulate
an approach based on identifying and combining po-
tential self-loops in the submodels, taking account of
the impact that interference can have. The results here
are promising and importantly provide a foundation
for developing general techniques and tools for com-
positionally identifying cyclic and complex attractors.
Alshahrani, M. and Steggles, J.
Compositional Techniques for Asynchronous Boolean Networks.
DOI: 10.5220/0012324300003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 429-437
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
429

2 BACKGROUND
2.1 Boolean Networks
A Boolean network (Kauffman, 1969) is a qualitative
modelling approach which consists of a set of regula-
tory entities that have a Boolean state. Each entity has
a next-state update function that defines its behaviour
based on the state of its related entities.
Definition 1. A Boolean network BN is a tuple
BN = (G,N,F) where:
i) G = {g
1
,g
2
,...,g
n
} is a non-empty, finite set of en-
tities;
ii) N = (N(g
1
),N(g
2
),...,N(g
n
)) is a tuple of neigh-
bourhoods, such that N(g
i
) ⊆ G is the neighbourhood
of g
i
;
iii) F = (F(g
1
),F(g
2
),..., F(g
n
)) is a tuple of next-
state functions, where F(g
i
) : B
|N(g
i
)|
→ B defines the
next state of g
i
.
We define a global state of a Boolean network BN
to be a function S : G → B, where S(g) represents the
state of entity g ∈ G, and we let S
BN
= [G → B] repre-
sent the set of all global states. If a Boolean network
has n ∈ N entities then S
BN
will contain 2
n
states
and this indicates that Boolean networks are impacted
by the state space explosion problem (Groote et al.,
2015). Given a global state S ∈ S
BN
and a subset of
entities X ⊆ G we define the projection of S over X to
be the function S[X] : X → B, where S[X](g) = S[g],
for any g ∈ X. For any S ∈ S
BN
, g ∈ G and b ∈ B we
let S[g → b] denote the state update which results in
a new global state where S[g → b](h) = S(h), for all
h ∈ G, h ̸= g, and S[g → b](g) = b.
A range of update semantics exist for Boolean
networks and two key approaches are: synchronous,
where all entities update their state simultaneously;
and asynchronous, where each entity updates its state
independently and non-deterministically (Schwab
et al., 2020). The asynchronous semantics can be seen
to capture more realistic dynamic behaviour (Barbuti
et al., 2020) and we focus on this.
Definition 2. Given global states S,S
′
∈ S
BN
, we
say S
BN
−−→ S
′
is an asynchronous update step iff ex-
ists g ∈ G such that S
′
(g) = ¬S(g) = F(g)(S[N(g)])
and S(h) = S
′
(h), for all h ∈ G, h ̸= g. We let
U
BN
(S) = {S
′
| S
′
∈ S
BN
, and S
BN
−−→ S
′
}. The com-
plete asynchronous behaviour of a Boolean network is
concisely captured by the (asynchronous) state graph
SG(BN ) = (S
BN
,
BN
−−→).
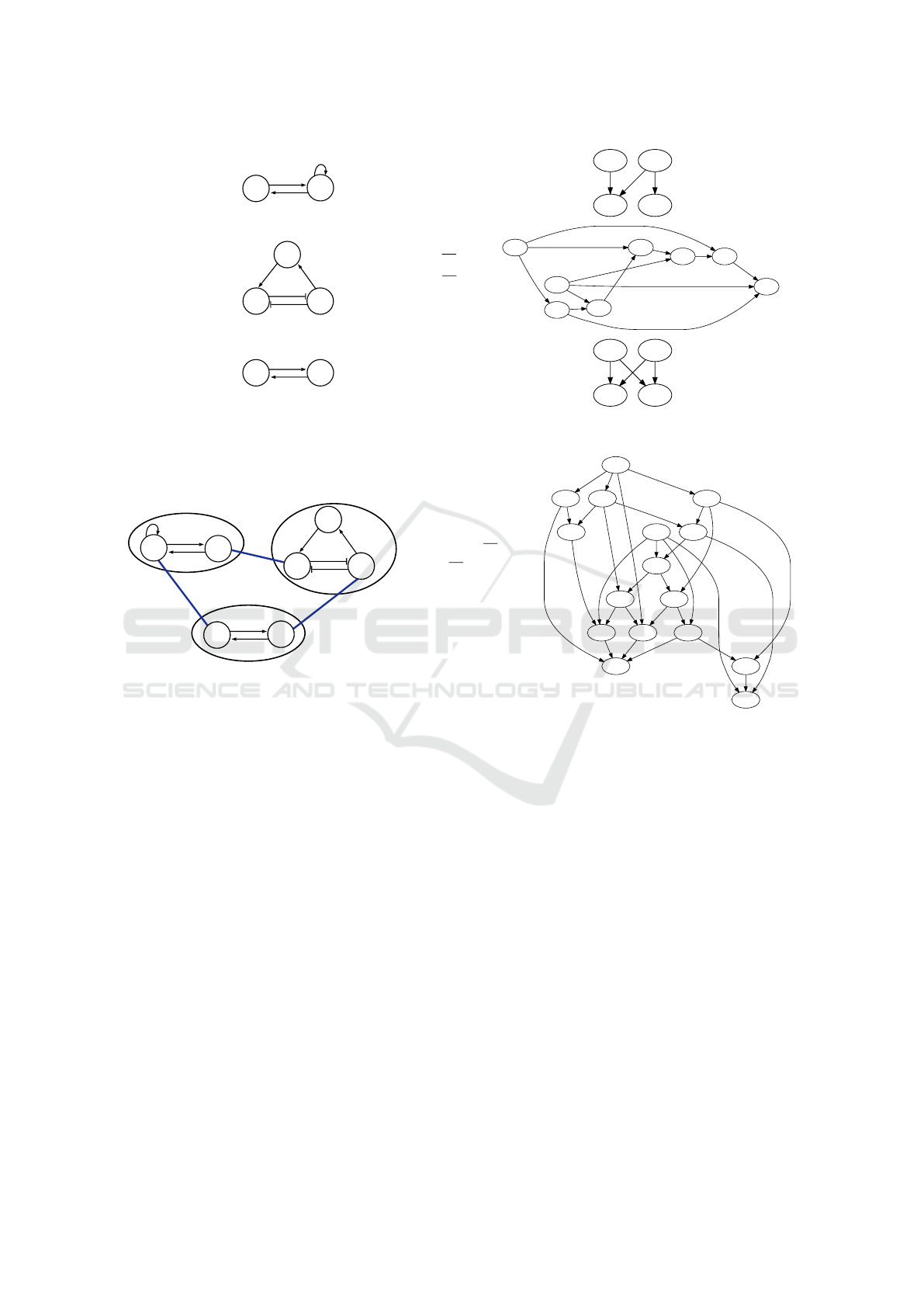
For illustrative examples, consider the Boolean
networks presented in Figure 1.
A path in the asynchronous state graph SG(BN )
represents one possible sequence of behaviour and
can be finite or infinite (this contrasts with the syn-
chronous setting where all paths are infinite (Kauff-
man et al., 1993)). We let Path(SG(BN )) denote
the set of all paths over SG(BN ). Given α ∈
Path(SG(BN )) we let SD(α) represent the step do-
main of α, where SD(α) = {0, ..., k − 1} if α :
{0,.. .,k} → S
BN
is a finite path and SD(α) = N if
α : N → S
BN
is an infinite path.
A global state with no update steps is known as
a point attractor and other complex, cyclic attractors
can be considered (Schwab et al., 2020; Hopfensitz
et al., 2013). Attractors provide important insights
into a model’s behaviour and can be associated with
biological phenomena (Huang and Ingber, 2000).
2.2 A Compositional Framework
A range of approaches for composing and decom-
posing Boolean networks for analysis have been pro-
posed (for example, see (Alkhudhayr and Steggles,
2019; Zhao et al., 2013; Mizera et al., 2017)). We fo-
cus on recent work which presents a novel approach
to composing synchronous Boolean networks based
on merging entities between Boolean networks using
Boolean operators (Alkhudhayr and Steggles, 2019;
Abdulrahman and Steggles, 2023). They focused
solely on the synchronous update semantics and de-
veloped a range of interesting techniques and tools for
analysing composed synchronous Boolean networks.
We briefly introduce key concepts from this com-
positional framework needed in the sequel. We only
consider using conjunction for merging entities in the
sequel but note that all the results presented also hold
for disjunction. We begin by recalling the definition
of a composition (Abdulrahman and Steggles, 2023).
Definition 3. A composition Σ = (M,E), where M =
{BN
1
,BN
2
,...,BN
n
} is the set of Boolean networks
(submodels) that are used in the composition Σ, for
some n ∈ N, n > 1, and E defines merged entities
E ⊆ {{g
1
,g
2
} | BN
i
,BN
j
∈ M, BN
i
̸= BN
j
and
g
1
∈ G
i
, g
2
∈ G
j
}
As an illustrative example, consider the com-
position Σ
Ex
= (M
Ex
,E
Ex
) presented in Figure 2,
where M
Ex
= {BN
Ex1
,BN
Ex2
,BN
Ex3
}, and E
Ex
=
{{g
1
1
,g
2
1
},{g
1
2
,g
3
2
},{g
2
2
,g
3
1
}}.
In order to reason about a composition we intro-
duce the following important notations and defini-
tions. We let gc(Σ,BN
i
) represent the set of enti-
ties from BN
i
that are composed in Σ and define it
as gc(Σ,BN
i
) = {g | g ∈ G
i
and {g,g
′
} ∈ E}. We let
gc(Σ) = gc(Σ,BN
1
) ∪ ... ∪ gc(Σ, BN
n
) represent the
set of all entities that are composed in Σ.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
430
BN
Ex1
g
1
1
g
1
2
[g
1
1
] = g
1
2
[g
1
2
] = g
1
1
∨ g
1
2
01
11
10
00
BN
Ex2
g
2
1
g
2
2
g
2
3
[g
2
1
] = g
2
2
∧ g
2
3
[g
2
2
] = g
2
1
[g
2
3
] = g
2
2
000 010
011
001
101
100110
111
BN
Ex3
g
3
1
g
3
2
[g
3
1
] = g
3
2
[g
3
2
] = g
3
1
01
11 00
10
Figure 1: The interaction diagram, next-state functions and the state graph for three example Boolean networks BN
Ex1
,
BN
Ex2
and BN
Ex3
.
g
2
1
g
2
2
g
2
3
g
3
2
g
3
1
BN
Ex1
BN
Ex2
BN
Ex3
g
2
3
g
2
2
g
1
2
g
2
1
g
3
1
g
3
2
g
1
1
[g
c
1
] = g
c
3
∧ (g
c
2
∧ g
2
3
)
[g
c
2
] = g
c
1
∧ g
c
3
[g
c
3
] = (g
c
1
∨ g
c
3
) ∧ g
c
2
[g
2
3
] = g
c
2
0001
0000
0010
0110
0111
0011
1011
1001 1010
0100
0101
1000
1100
11101101
1111
Figure 2: An example composition Σ
Ex
in which three Boolean networks BN
Ex1
, BN
Ex2
and BN
Ex3
are composed, where
the resulting compositional entities (represented by thick blue edges) are g
c
1
= {g
1
1
,g
2
1
}, g
c
2
= {g
2
2
,g
3
1
} and g
c
3
= {g
1
2
,g
3
2
}. The
asynchronous state graph is also depicted for the resulting Boolean network BN (Σ
Ex
).
For any g ∈ gc(Σ) we let ∆(g) denote the set of all
entities that are composed with g (including g itself).
We let λ(g) denote the index of the Boolean net-
work that g belongs to. For any i ∈ {1, ... , n} and any
g ∈ G
i
we define λ(g) = i.
We let Σ(g) be the renaming used to move
between a submodel and the composition defined
for any entity g ∈ (G
1
∪ ... ∪ G
n
) by Σ(g) = g, if
g /∈ gc(Σ); or Σ(g) = ∆(g), otherwise.
A composition Σ results in a Boolean network
BN (Σ) (Abdulrahman and Steggles, 2023).
Definition 4. Let Σ = (M,E) be a composition.
Then we define the Boolean network BN (Σ) =
(G(Σ),N(Σ),F(Σ)) that results from Σ as follows.
1. Entities: G(Σ) = Σ(G
1
∪ ... ∪ G
n
).
2. Neighbourhood: for any entity h ∈ G(Σ), we define
the neighbourhood N(Σ)(h) by
N(Σ)(h) =
S
g∈h
Σ(N
λ(g)
(g)),
if h = ∆(g
′
), for some g
′
∈ gc(Σ); or N(Σ)(h) =
Σ(N
λ(h)
(h)), otherwise.
3. Functions: For any entity g ∈ G(Σ), we define the
next–state function F(Σ)(g) on any S ∈ S
BN (Σ)
by
F(Σ)(g)(S[N(Σ)(g)]) =
V
h∈∆(g
′
)
F
λ(h)
(h)(S[Σ(N
λ(h)
(h))]),
if g = ∆(g
′
), for some g
′
∈ gc(Σ); or
F(Σ)(g)(S[N(Σ)(g)]) = F
λ(g)
(g)(S[Σ(N
λ(g)
(g))]),
otherwise.
For an example, see Figure 2 where the composed
model BN (Σ
Ex
) resulting from Σ
Ex
is shown. In a
slight abuse of notation we use the set of entities com-
posed as the name of the resulting composed entity.
In the sequel we use the above definitions for our
asynchronous compositional theory but place an im-
portant restriction on a composition’s graph structure
Compositional Techniques for Asynchronous Boolean Networks
431

to ensure that no two entities in an asynchronous sub-
model are composed together (since this would result
in synchronous behaviour).
2.3 Related Work
The compositional framework for asynchronous
Boolean networks we present is based on the novel
idea of composing synchronous Boolean networks by
merging entities using a Boolean operator (Alkhud-
hayr and Steggles, 2019; Abdulrahman and Steggles,
2023). However, the work presented here takes a sig-
nificantly different approach as required by our focus
on asynchronous models.
There are some interesting studies on behaviour
preservation and subnetwork embeddings in the liter-
ature (for example, see (Thomas and d’Ari, 1990)) but
the approach we take is new.
A range of research into the composition and de-
composition of Boolean networks can be found in the
literature, e.g. (Tournier and Chaves, 2013; Dubrova
and Teslenko, 2005; Cheng et al., 2012; Zhao et al.,
2013). The approaches taken by these papers appear
to be significantly different to our work and focus on
applying decomposition/composition to aid identify-
ing the attractors of a Boolean network. They ap-
pear not to consider developing a general composi-
tional framework for constructing and analysing asyn-
chronous Boolean networks as we do here.
3 BEHAVIOUR PRESERVATION
3.1 Asynchronous Interference
A key concept in the synchronous compositional
framework (Alkhudhayr and Steggles, 2019) is the
behavioural interference that can occur when entities
are merged in a composition. To illustrate this idea,
consider the situation where two entities g
1
and g
2
are
merged using conjunction. Then interference would
occur if g
1
wanted to transition to 1 but g
2
to 0 since
the composed entity would transition to 1∧ 0 = 0 (i.e.
the behaviour of entity g
1
has been interfered with to
become 0). To capture the impact interference can
have on a submodel’s behaviour the interference state
graph was developed in (Alkhudhayr and Steggles,
2019) by extending the state graph for a Boolean net-
work with edges that could result due to interference.
We formulate a new definition for an interference
state graph for asynchronous compositions by extend-
ing the asynchronous state graph SG(BN
i
) with two
new types of edges to reflect the new asynchronous
behaviour possible in a composition. Firstly, we add
self-loops to every global state in the state graph
SG(BN
i
) to capture BN
i
needing to remain at a state
while an entity in another submodel of the composi-
tion updates. Secondly, we add interference edges to
capture when the state of an entity in BN
i
involved in
a composition is 1 and so can be forced to update to 0
by interference from another submodel.
Definition 5. Let BN
i
= {G
i
,N
i
,F
i
} and let X ⊆
G
i
. Then an asynchronous interference state graph
AISG
X
(BN
i
) = (S
BN
i
,
BN
i
−−→
X
), where
BN
i
−−→
X
=
BN
i
−−→
∪ ε ∪ κ, and ε = {(S,S) | S ∈ S
BN
i
} and κ =
{(S,S[h → 0]) | h ∈ X, S ∈ S
BN
i
, S(h) = 1}.
For any i ∈ {1,..., n}, we let I
i
denote
the asynchronous interference state graph
AISG
gc(Σ,BN
i
)
(BN
i
) for BN
i
over Σ. As an example,
consider Figure 3 which presents the asynchronous
interference state graph I
Ex1
= AISG
{g
1
1
,g
1
2
}
(BN
Ex1
)
for BN
Ex1
over Σ
Ex
.
01 11
00
10
Figure 3: The asynchronous interference state graph of
BN
Ex1
in composition Σ
Ex
, where dashed edges represent
new transitions arising from interference.
The following theorem shows the asynchronous
interference state graph bounds the behaviour a
Boolean network can exhibit in a composition.
Theorem 1. Let Σ = (M,E), where M =
{BN
1
,...,BN
n
}. Then for any i = {1,..., n},
we have Path(SG(BN (Σ)))[Σ(G
i
)] ⊆ Path(I
i
).
Proof. Let i ∈ {1,...,n} and let path β ∈
Path(SG(BN (Σ))). Then we have to show that
there is a path α ∈ Path(I
i
) such that β[Σ(G
i
)] = α. It
suffices to show that for any k ∈ SD(β) there exists
an edge
β(k)[Σ(G
i
)]
I
i
−−→ β(k + 1)[Σ(G
i
)] (1)
in the interference state graph I
i
. The transition (1)
must have resulted from updating an entity g ∈ G(Σ)
and we therefor have three possible cases to consider
based on entity g.
Case 1: Suppose g /∈ Σ(G
i
) (i.e. the state update step
involved an entity unassociated with BN
i
). It follows
that β(k)[Σ(G
i
)] = β(k + 1)[Σ(G
i
)]. By the definition
of the asynchronous interference state graph (Defi-
nition 5), we know that β(k)[Σ(G
i
)]
I
i
−−→ β(k)[Σ(G
i
)]
holds (self-loop) and so we must have (1) holds as
required.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
432

Case 2: Suppose g ∈ (G
i
\gc(Σ,BN
i
)) (i.e. the state
update step involved an entity in G
i
that is not used
in the composition). It must follow by the definition
of BN (Σ) that β(k)[Σ(G
i
)]
BN
i
−−→ β(k +1)[Σ(G
i
)] and
so (1) must hold by the definition of asynchronous
interference state graph.
Case 3: Suppose g = ∆(h), for some h ∈ gc(Σ,BN
i
)
(i.e. the state update step involved a composed entity
associated with BN
i
). Then, there are two sub cases
to consider based on if entity h updates or not in BN
i
.
The proof of these cases are based on the definition
of the asynchronous interference state graph and are
omitted here for brevity (see (Alshahrani, 2024) for
details).
3.2 Behaviour Preservation
Often when analysing a compositional model we want
to check whether the behaviour of a submodel has
been preserved. In this section we reformulate and ex-
tend important behavioural preservation results from
(Alkhudhayr and Steggles, 2019) to asynchronous
compositions. We begin by recalling the definition of
compatibility (Alkhudhayr and Steggles, 2019) which
formalises the idea of preserving a submodel’s be-
haviour in a composition.
Definition 6. Let Σ = (M,E) be a composition
with M = {BN
1
,...,BN
n
}, and let i ∈ {1,...,n}.
Then, BN
i
is said to be compatible with Σ iff
Path(SG(BN
i
)) ⊆ Path(SG(BN (Σ)))[Σ(G
i
)].
As an example, consider the composition Σ
Ex
(Figure 2); it can be shown that BN
Ex2
is compati-
ble with Σ
Ex
but that BN
Ex1
and BN
Ex3
are not.
In order to check compatibility compositionally
a property called weak alignment was proposed in
(Alkhudhayr and Steggles, 2019) which composi-
tionally characterises compatibility using interference
state graphs. We update and extend this approach to
the asynchronous case. This provides important in-
sights into the semantic differences that exist between
the synchronous and asynchronous update semantics.
We begin by recalling how submodel states and paths
can be merged (Abdulrahman and Steggles, 2023).
Definition 7. Let S
i
∈ S
BN
i
, for i ∈ {1,...,n}. We
let ∧
Σ
(S
1
,...,S
n
) ∈ S
BN (Σ)
be the global state that re-
sults from merging S
1
,.. .,S
n
defined for any g ∈ G(Σ)
by ∧
Σ
(S
1
,...,S
n
)(g) =
V
h∈g
S
λ(h)
(h), if g = ∆(g
′
),
for some g
′
∈ gc(Σ); or ∧
Σ
(S
1
,...,S
n
)(g) = S
λ(g)
(g),
otherwise. Let α
i
∈ Path(I
i
), for i ∈ {1, ...,n},
such that SD(α
1
) = ··· = SD(α
n
). Then we let
∧
Σ
(α
1
,...,α
n
) represent the path resulting from merg-
ing paths α
1
,.. .,α
n
defined for any m ∈ SD(α
1
) by
∧
Σ
(α
1
,...,α
n
)(m) = ∧
Σ
(α
1
(m),. . .,α
n
(m)).
Next we introduce what it means for submodel
paths to align such that they can be merged to form
behaviour in the composed model.
Definition 8. Let α
i
∈ Path(I
i
), for i ∈ {1, ...,n}. We
say α
1
,.. .,α
n
align (on Σ) iff we have
1) SD(α
1
) = SD(α
2
) = ··· = SD(α
n
); and
2) for every g ∈ gc(Σ), every h ∈ ∆(g) and every m ∈
SD(α
1
), we have α
λ(g)
(m)(g) = α
λ(h)
(m)(h).
We say α
1
,.. .,α
n
update align (on Σ) iff they align
and for each m ∈ SD(α
1
) there exists i ∈ {1,...,n}
and g ∈ G
i
such that α
i
(m + 1)(g) = ¬α
i
(m)(g) and
α
i
(m)
BN
i
−−→ α
i
(m+ 1), and for all k ∈ {1, ...,n}, k ̸= i,
and all h ∈ G
k
we have α
k
(m)(h) = α
k
(m + 1)(h), if
g ̸∈ gc(Σ,BN
i
) or h ̸∈ ∆(g).
Update alignment is an important new property
which captures when a collection of paths can be
merged asynchronously to produce a well-defined
composed path. The following are important results
about merging aligned and update aligned paths.
Lemma 1. Let α
1
∈ Path(I
1
),.. .,α
n
∈ Path(I
n
), such
that α
1
,.. .,α
n
align. Then, for every k ∈ {1, ...,n},
we have ∧
Σ
(α
1
,...,α
n
)[Σ(G
k
)] = α
k
.
Proof. This is straightforward to prove (see (Al-
shahrani, 2024) for details).
Lemma 2. Let α
1
∈ Path(I
1
),. . .,α
n
∈ Path(I
n
) be
paths such that α
1
,.. .,α
n
update align. Then we have
∧
Σ
(α
1
,...,α
n
) ∈ Path(SG(BN (Σ))).
Proof. Let α
1
∈ Path(I
1
),. . .,α
n
∈ Path(I
n
) such that
α
1
,.. .,α
n
update align. To show that ∧
Σ
(α
1
,...,α
n
) ∈
Path(SG(BN (Σ))) it suffices to show that for each
m ∈ SD(α
1
), there exists g ∈ G(Σ) such that
∧
Σ
(α
1
(m + 1),... ,α
n
(m + 1))(g) =
F(Σ)(g)(∧
Σ
(α
1
(m),. . .,α
n
(m))[N(Σ)(g)]) =
¬ ∧
Σ
(α
1
(m),. . .,α
n
(m))(g)
(1)
and for all other g
′
∈ G(Σ), g ̸= g
′
, we have
∧
Σ
(α
1
(m + 1),... ,α
n
(m + 1))(g
′
) =
∧
Σ
(α
1
(m),. . .,α
n
(m))(g
′
)
(2)
By the assumption of update alignment we know
there exists i ∈ {1,...,n} and h ∈ G
i
such that α
i
(m +
1)(h) = ¬α
i
(m)(h). Then to show (1) and (2), there
are two cases to consider based on whether entity
h is used in the composition or not. The proof of
these cases is based on using the definition of BN (Σ)
and Lemma 1, and are omitted for brevity (see (Al-
shahrani, 2024) for details).
Compositional Techniques for Asynchronous Boolean Networks
433

We can now formulate the asynchronous version
of the weak alignment property.
Definition 9. For any i ∈ {1,.. .,n}, we say BN
i
is update weakly aligned (on Σ) iff for every α
i
∈
Path(SG(BN
i
)), there exists α
k
∈ Path(I
k
), for each
k ∈ {1,...,n}, k ̸= i, such that α
1
,.. .,α
n
update align.
We now prove that update weak alignment com-
positionally characterises compatibility.
Theorem 2. For i ∈ {1,...,n}, we have BN
i
is com-
patible on Σ iff BN
i
is update weakly aligned on Σ.
Proof. Let i ∈ {1,...,n}.
1) Suppose BN
i
is compatible on Σ. We need to show
that BN
i
is update weakly aligned on Σ. Let α
i
∈
Path(SG(BN
i
)). Then by assumption of compati-
bility there must exist a path β ∈ Path(SG(BN (Σ)))
such that α
i
= β[Σ(G
i
)]. For each k ∈ {1,...,n}, k ̸= i,
let α
k
= β[Σ(G
k
)]. Then, by Theorem 1 we know
that α
k
∈ Path(I
k
), for each k ∈ {1, ...,n},k ̸= i. It
remains to show that α
1
,.. .,α
n
update align. Since
α
1
,.. .,α
n
are all projected from the same path β it
follows that α
1
,.. .,α
n
must align. Furthermore, as
α
i
∈ Path(SG(BN
i
)) we know that for every m ∈
SD(α
1
) there must exist an entity h ∈ G
i
which up-
dates such that α
i
(m + 1)(h) = ¬α
i
(m)(h). There are
now two cases to consider based on if entity h is used
in the composition or not. These cases are proved us-
ing the definition of BN (Σ) and the definition of an
asynchronous update (Definition 2). We omit them
for brevity (see (Alshahrani, 2024) for details).
2) Suppose BN
i
is weakly aligned on Σ. Then we
need to show that BN
i
is compatible on Σ, that
is Path(SG(BN
i
)) ⊆ Path(SG(BN (Σ)))[Σ(G
i
)]. To
prove this, consider any path α
i
∈ Path(SG(BN
i
)).
Then, by update weak alignment we know there must
exist paths α
k
∈ Path(I
k
), for k ∈ {1, ...,n}, k ̸= i, such
that α
1
,.. .,α
n
update align. It follows by Lemma 1
that ∧
Σ
(α
1
,...,α
n
)[Σ(G
i
)] = α
i
and by Lemma 2 that
∧
Σ
(α
1
,...,α
n
) ∈ Path(SG(BN (Σ))) as required.
3.3 Behaviour Preservation Based on
Asynchronous Sequences
The concept of compatibility introduced previously
was based on ideas formulated for synchronous mod-
els (Alkhudhayr and Steggles, 2019). In the asyn-
chronous case it is possible for a submodel to pause
it’s behaviour while another submodel updates and
this can result in a submodel’s projected path in
a composition containing repeated states. To take
account of this we consider reducing paths by re-
moving any duplicated consecutive states. We for-
malise this using red(α) which takes a path α ∈
Path(SG(BN (Σ)))[Σ(G
i
)] and removes any dupli-
cated consecutive states. These ideas lead to a new
notion of compatibility called sequence compatible.
Definition 10. Let Σ = (M,E) be a composition with
M = {BN
1
,...,BN
n
}, and let i ∈ {1, ...,n}. Then,
BN
i
is sequence compatible (on Σ) iff
Path(SG(BN
i
)) ⊆ red(Path(SG(BN (Σ)))[Σ(G
i
)])
It can be seen that BN
Ex3
is sequence compatible
on Σ
Ex
even though it was not compatible. We can
also see that BN
Ex2
is both compatible and sequence
compatible. Note it can be shown that compatibility
implies sequence compatibility.
In order to compositionally characterise sequence
compatibility we adapt the definition of update weak
alignment to take account of reduced paths. For any
α ∈ Path(SG(BN
i
)), we let Exp(α) = {α
′
| α
′
∈
Path(I
i
), red(α
′
) = α}.
Definition 11. For any i ∈ {1,...,n}, we say BN
i
is
sequence weakly aligned (on Σ) iff for every path α ∈
Path(SG(BN
i
)), there exists a path α
i
∈ Exp(α), and
paths α
k
∈ Path(I
k
), for each k ∈ {1,...,n}, k ̸= i, such
that α
1
,.. .,α
n
update align.
We now prove that sequence weak alignment com-
positionally characterises sequence compatibility.
Theorem 3. For any i ∈ {1,...,n}, we have BN
i
is
sequence compatible on Σ iff BN
i
is sequence weakly
aligned on Σ.
Proof. Let i ∈ {1,...,n}.
1) Suppose BN
i
is sequence compatible on Σ.
Then we need to show BN
i
is sequence weakly
aligned on Σ. Suppose α ∈ Path(SG(BN
i
)). By
sequence compatibility we know there exists β ∈
Path(SG(BN (Σ))) such that red(β[Σ(G
i
)]) = α. Let
α
k
= β[Σ(G
k
)], for each k ∈ {1, ...,n}. By Theorem
1 we know α
k
∈ Path(I
k
), for each k ∈ {1,...,n},
and since red(β[Σ(G
i
)]) = α we have α
i
∈ Exp(α).
It remains to show α
1
,.. .,α
n
update align. Since
α
1
,.. .,α
n
are projected from β it follows that they
align. Furthermore, since β ∈ Path(SG(BN (Σ))) we
know that for any m ∈ SD(β) there must exist g ∈
G(Σ) such that β(m + 1)(g) = ¬β(m)(g) and for all
h ∈ G(Σ), h ̸= g, we have β(m + 1)(h) = β(m)(h).
Given above it is straightforward to show that update
alignment holds using two cases based on whether or
not g is a composed entity.
2) Suppose BN
i
is sequence weakly aligned
on Σ. Then we need to show BN
i
is se-
quence compatible on Σ. Consider any path α ∈
Path(SG(BN
i
)). Then by assumption of sequence
weak alignment, there exists α
i
∈ Exp(α), and
α
k
∈ Path(I
k
), for k ∈ {1, ...,n}, k ̸= i, such that
α
1
,.. .,α
n
update align. It follows by Lemma 2 that
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
434

∧
Σ
(α
1
,...,α
n
) ∈ Path(SG(BN (Σ))). Furthermore,
by Lemma 1, we have ∧
Σ
(α
1
,...,α
n
)[Σ(G
i
)] = α
i
,
and so by assumption α
i
∈ Exp(α), we have α =
red(∧
Σ
(α
1
,...,α
n
)[Σ(G
i
)]) holds as required.
4 POINT ATTRACTORS
Boolean networks can exhibit cyclic behaviour known
as attractors (Hopfensitz et al., 2013) and identify-
ing attractors is a crucial analysis step as they provide
important practical insights into a Boolean network’s
behaviour (for example, see (Huang and Ingber, 2000;
Saadatpour et al., 2011)).
In this section we develop an important new com-
positional technique for identifying point attractors
(global states with no update steps) in an asyn-
chronous composition. It is based on identifying po-
tential self-loop states in submodels and then merg-
ing these when they align to construct point attractors.
The approach is summarised below.
1) Identify Potential Self-Loop States in Submod-
els. We identify states in each submodel which can
remain constant either because they are point attrac-
tors or due interference. We refer to these as self-loop
states and for i ∈ {1, ...,n}, define the set of self-loop
states
SL(BN
i
) = {S | S ∈ S
BN
i
, ω(S)},
where ω(S) is true iff for every S
′
∈ U
BN
i
(S), exists
h ∈ gc(Σ,BN
i
) such that S(h) = 0, and S
′
(h) = 1.
2) Identify Aligned State Tuples. Next we group
self-loop states that align to form state tuples. Let
S
i
∈ SL(BN
i
), for i ∈ {1, ...,n}. Then (S
1
,...,S
n
) is
an aligned state tuple (over Σ) iff for every g ∈ gc(Σ),
and for all h ∈ ∆(g), we have S
λ(g)
(g) = S
λ(h)
(h). We
denote the set of all aligned states tuples over a com-
position Σ as alignST (Σ).
3) Merge Valid Aligned State Tuples. We say
(S
1
,...,S
n
) ∈ alignST (Σ) is valid iff for all g ∈ gc(Σ),
if S
λ(g)
(g) = 0, then there exists h ∈ ∆(g) such that
F
λ(h)
(h)(S
λ(h)
[N
λ(h)
(h)]) = 0. Each valid aligned state
tuple (S
1
,...,S
n
) ∈ alignST (Σ) represents a point at-
tractor ∧
Σ
(S
1
,...,S
n
) in BN (Σ).
To illustrate the approach consider composition-
ally identifying the point attractors in BN (Σ
Ex
) (Fig-
ure 2). We first identify the self-loop set for each
submodel: SL(BN
Ex1
) = {11,00,01}; SL(BN
Ex2
) =
{000,011}; and SL(BN
Ex3
) = {11,00}. Next, we
generate the aligned state tuples alignST (Σ
Ex
) =
{(00,000,00), (01,011,11)}. Both of these aligned
state tuples are valid and so are merged to form
point attractors: ∧
Σ
Ex
(00,000,00) = 0000 and
∧
Σ
Ex
(01,011,11) = 0111.
It remains to show that our approach is correct by
proving it is sound and complete.
Theorem 4. (Soundness) Let (S
1
,...,S
n
) ∈
alignST (Σ) be a valid aligned state tuple. Then
∧
Σ
(S
1
,...,S
n
) is a point attractor in BN (Σ).
Proof. Suppose (S
1
,...,S
n
) ∈ alignST (Σ). Then we
need to show that ∧
Σ
(S
1
,...,S
n
) is a point attractor in
BN (Σ). By definition of merging states (Definition
7), we know ∧
Σ
(S
1
,...,S
n
) ∈ S
BN (Σ)
. To prove that
∧
Σ
(S
1
,...,S
n
) is a point attractor in BN (Σ), we need
to show U
BN (Σ)
(∧
Σ
(S
1
,...,S
n
)) = {}. To do this we
show that for every g ∈ G(Σ), we have
F(Σ)(g)(∧
Σ
(S
1
,...,S
n
)[N(Σ)(g)]) =
∧
Σ
(S
1
,...,S
n
)(g)
(1)
There are two cases to consider based on whether en-
tity g is a composed entity or not.
Case 1: Suppose g ∈ ((G
1
∪ ... ∪ G
n
)\ gc(Σ)) (i.e. g
is not a composed entity). By the assumptions, we
know S
λ(g)
∈ SL(BN
λ(g)
). Then the result follows by
the alignment assumption, Lemma 1 and by the defi-
nition of SL(BN
λ(g)
).
Case 2: Suppose g = ∆(g
′
), for some g
′
∈ gc(Σ) (i.e.
g is a composed entity). By definition of BN (Σ) and
case assumption, we have S
λ(h)
∈ SL(BN
λ(h)
), for ev-
ery h ∈ g. Then we have two subcases to consider:
Case 2.1: Suppose ∧
Σ
(S
1
,...,S
n
)(g) = 0. Then by
assumptions, Lemma 1 and conjunction we have
V
h∈g
F
λ(h)
(h)(∧
Σ
(S
1
,...,S
n
)[Σ(N
λ(h)
(h))]) = 0,
and so result follows by the definition of BN (Σ).
Case 2.2: Suppose ∧
Σ
(S
1
,...,S
n
)(g) = 1. Then by
assumptions, Lemma 1 and conjunction we have
V
h∈g
F
λ(h)
(h)(∧
Σ
(S
1
,...,S
n
)[Σ(N
λ(h)
(h))]) = 1
and so result follows by definition of BN (Σ).
Theorem 5. (Completeness) Let S ∈ S
BN (Σ)
be
a point attractor in the composed model BN (Σ).
Then, there must exist a valid aligned state tuple
(S
1
,...,S
n
) ∈ alignST (Σ) such that ∧
Σ
(S
1
,...,S
n
) = S.
Proof. Let S ∈ S
BN (Σ)
be a point attractor in BN (Σ).
Then U
BN (Σ)
(S) = {} and F(Σ)(g)(S[N(Σ)(g)]) =
S(g), for every g ∈ G(Σ). Let S
i
= S[Σ(G
i
)], for each
i ∈ {1,...,n}. We know S
1
,...,S
n
must align and so
(S
1
,...,S
n
) ∈ alignST (Σ). Furthermore, by Lemma 1
∧
Σ
(S
1
,...,S
n
) = S. We have two properties to show:
i) Let i ∈ {1,...,n}. We need to show that S
i
∈
SL(BN
i
). If U
BN
i
(S
i
) = {} then by definition S
i
∈
SL(BN
i
). Alternatively, suppose U
BN
i
(S
i
) ̸= {} and
let S
′
i
∈ U
BN
i
(S
i
). By the definition of conjunction
and BN (Σ) it can be seen that interference must oc-
cur here to produce a self-loop and so S
i
∈ SL(BN
i
).
Compositional Techniques for Asynchronous Boolean Networks
435

ii) We must show that (S
1
,...,S
n
) is valid. Suppose
g = ∆(g
′
), for some g
′
∈ gc(Σ), and S(g) = 0. Further-
more, suppose for a contradiction there is no required
interference. Then by definition of BN (Σ) and con-
junction we can contradict S being a point attractor. It
follows that (S
1
,...,S
n
) must be valid.
5 CONCLUSIONS
In this paper we developed a range of new com-
positional techniques for constructing and analysing
asynchronous Boolean networks by building on re-
cent compositional work on synchronous Boolean
networks (Alkhudhayr and Steggles, 2019; Abdulrah-
man and Steggles, 2023). The compositional frame-
work developed provides a foundation for helping
to address the current practical limitation of apply-
ing asynchronous Boolean networks and provides in-
teresting insight into the differences between syn-
chronous and asynchronous updating.
The key contributions of the paper are:
i) Formulated a new asynchronous version of the in-
terference state graph, a key concept in the compo-
sitional framework (Alkhudhayr and Steggles, 2019;
Abdulrahman and Steggles, 2023) and proved it
bounds a submodel’s compositional behaviour.
ii) Developed range of new compositional behaviour
preservation results.
iii) Developed a new compositional technique for
identifying point attractors which we formally
showed to be correct.
We are now working to develop compositional
techniques for identifying more general types of asyn-
chronous cyclic and complex attractors (Hopfensitz
et al., 2013; Schwab et al., 2020). The aim is to then
develop tool support for compositionally analysing
asynchronous Boolean networks and undertake large
realistic case studies. We also intend to consider de-
veloping decompositional techniques.
We would like to thank Hanin Abdulrahman and
the anonymous referees for their comments. We
gratefully acknowledge the support provided by Fac-
ulty of Computer Science, King Khalid University.
REFERENCES
Abdulrahman, H. and Steggles, J. (2023). Compositional
Techniques for Boolean Networks and Attractor Anal-
ysis. LNCS 14150, Springer.
Alkhudhayr, H. and Steggles, J. (2019). A composi-
tional framework for boolean networks. Biosystems,
186:103960.
Alshahrani, M. (2024). Compositional Techniques and
Tools for Asynchronous Boolean Networks. PhD the-
sis, Newcastle University (to appear).
Barbuti, R., Gori, R., Milazzo, P., and Nasti, L. (2020). A
survey of gene regulatory networks modelling meth-
ods: from differential equations, to boolean and quali-
tative bioinspired models. Journal of Membrane Com-
puting, 2:207–226.
Cheng, D., Zhao, Y., Kim, J., and Zhao, Y. (2012). Ap-
proximation of boolean networks. In Proceedings of
the 10th World Congress on Intelligent Control and
Automation, pages 2280–2285.
Dahlhaus, M., Burkovski, A., Hertwig, F., Mussel, C., Vol-
land, R., Fischer, M., Debatin, K.-M., Kestler, H. A.,
and Beltinger, C. (2016). Boolean modeling identi-
fies greatwall/mastl as an important regulator in the
aurka network of neuroblastoma. Cancer Letters,
371(1):79–89.
Dubrova, E. and Teslenko, M. (2005). Compositional prop-
erties of random boolean networks. Physical Review
E, 71(5):056116.
Groote, J. F., Kouters, T. W., and Osaiweran, A. (2015).
Specification guidelines to avoid the state space ex-
plosion problem. Software Testing, Verification and
Reliability, 25(1):4–33.
Hopfensitz, M., M
¨
ussel, C., Maucher, M., and Kestler,
H. A. (2013). Attractors in boolean networks: a tu-
torial. Computational Statistics, 28(1):19–36.
Huang, S. and Ingber, D. E. (2000). Shape-dependent
control of cell growth, differentiation, and apoptosis:
switching between attractors in cell regulatory net-
works. Experimental cell research, 261(1):91–103.
Kauffman, S. A. (1969). Metabolic stability and epigene-
sis in randomly constructed genetic nets. Journal of
theoretical biology, 22(3):437–467.
Kauffman, S. A. et al. (1993). The origins of order: Self-
organization and selection in evolution. Oxford Uni-
versity Press, USA.
Mizera, A., Pang, J., Qu, H., and Yuan, Q. (2017). A new
decomposition method for attractor detection in large
synchronous boolean networks. In Int. Symp. on De-
pendable Software Engineering: Theories, Tools, and
Applications, pages 232–249. Springer.
Pandey, S., Wang, R.-S., Wilson, L., Li, S., Zhao, Z.,
Gookin, T. E., Assmann, S. M., and Albert, R. (2010).
Boolean modeling of transcriptome data reveals novel
modes of heterotrimeric g-protein action. Molecular
systems biology, 6(1):372.
Saadatpour, A., Wang, R.-S., Liao, A., Liu, X., Loughran,
T. P., Albert, I., and Albert, R. (2011). Dynamical and
structural analysis of a t cell survival network identi-
fies novel candidate therapeutic targets for large gran-
ular lymphocyte leukemia. PLoS computational biol-
ogy, 7(11):e1002267.
Schwab, J. D., K
¨
uhlwein, S. D., Ikonomi, N., K
¨
uhl, M., and
Kestler, H. A. (2020). Concepts in boolean network
modeling: What do they all mean? Computational
and structural biotechnology journal, 18:571–582.
Thomas, R. and d’Ari, R. (1990). Biological feedback. CRC
press.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
436

Tournier, L. and Chaves, M. (2013). Interconnection of
asynchronous boolean networks, asymptotic and tran-
sient dynamics. Automatica, 49(4):884–893.
Zhao, Y., Kim, J., and Filippone, M. (2013). Aggrega-
tion algorithm towards large-scale boolean network
analysis. IEEE Transactions on Automatic Control,
58(8):1976–1985.
Compositional Techniques for Asynchronous Boolean Networks
437
