
GREED: Graph Learning Based Relation Extraction with Entity and
Dependency Relations
Mohamed Yassine Landolsi
a
, Lobna Hlaoua and Lotfi Ben Romdhane
MARS Research Lab LR17ES05, SDM Research Group, ISITCom, University of Sousse, Hammam Sousse, Tunisia
fi
Keywords:
Natural Language Processing, Medical Information Extraction, Relation Extraction, Clinical Named Entities,
Graph Convolutional Network.
Abstract:
A large number of electronic medical documents are generated by specialists, containing valuable information
for various medical tasks such as medical prescriptions. Extracting this information from extensive natural lan-
guage text can be challenging. Named Entity Recognition (NER) and Relation Extraction (RE) are key tasks
in clinical information extraction. Systems often rely on machine learning and rule-based techniques. Modern
methods involve dependency parsing and graph-based deep learning algorithms. However, the effectiveness
of these techniques and certain features is not thoroughly studied. Additionally, it would be advantageous to
properly integrate rules with deep learning models. In this paper, we introduce GREED (Graph learning based
Relation Extraction with Entity and Dependency relations). GREED is based on graph classification using
Graph Convolutional Network (GCN). We transform each sentence into a weighted graph via dependency
parsing. Words are represented with features that capture co-occurrence, dependency type, entities, and rela-
tion verbs, with focus on the entity pair. Experiments on clinical records (i2b2/VA 2010) show that relevant
features efficiently integrated with GCN achieve higher performance.
1 INTRODUCTION
There is a large amount of unstructured text in Elec-
tronic Health Records (EHR) which contain rich in-
formation about patients and clinical events. In
healthcare, there is more than 80% of the total data is
unstructured and it is created by hospitals, healthcare
clinics, or biomedical labs (Kong, 2019). The text in
this data is written in natural language and must be
read carefully by a medical expert to extract the de-
sired information. However, this operation is costly
and time-consuming given the huge amount of avail-
able documents.
In fact, clinical records, such as discharge sum-
maries and progress reports, are useful for many com-
puterized clinical applications such as decision sup-
port systems. Accordingly, the application of natural
language processing (NLP) (Quevedo and Chicaiza,
2023) technologies in the automatic extraction of in-
formation from narratives is becoming increasingly
interesting. Named entity recognition (Landolsi et al.,
2022b) and Relation Extraction (RE) are key compo-
nents of information extraction tasks in the clinical
a
https://orcid.org/0000-0001-8323-8943
domain (Landolsi et al., 2022a). Detecting and clas-
sifying the annotated semantic relationships between
medical named entities mentioned in biomedical texts
has numerous applications that range from advanc-
ing basic science to improving clinical practice (Luo
et al., 2017).
Significant research on RE has been carried out
on unstructured text. Most of the systems are relied
on rule-based and machine learning-based approaches
(Yang et al., 2021). Machine learning-based meth-
ods (Yang et al., 2021; Mahendran et al., 2022; Ed-
drissiya El-allaly et al., 2022) usually generate high-
quality features for sentences or words based on NLP
to train a deep learning classifier. The rule-based
methods (Chikka and Karlapalem, 2018; Ben Ab-
dessalem Karaa et al., 2021; Kim et al., 2021) consist
in constructing and applying rules which often rely
on dependency parsing, entity co-occurrence detec-
tion, and pattern matching. Some recent methods use
graph-based deep learning algorithms to incorporate
dependency parsing and other features (Ed-drissiya
El-allaly et al., 2022). However, this technique and
the use of some types of features are not well ex-
plored although they can be of greater benefit for this
task. The preparation of rules often requires a man-
360
Landolsi, M., Hlaoua, L. and Ben Romdhane, L.
GREED: Graph Learning Based Relation Extraction with Entity and Dependency Relations.
DOI: 10.5220/0012349000003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 3, pages 360-367
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

ual effort by medical experts (Ben Abdessalem Karaa
et al., 2021). Also, better integration of the rules with
the deep learning models would be of better benefit
(Chikka and Karlapalem, 2018).
In our work, we have trained a Graph Convolu-
tional Network (GCN) model (Yuan et al., 2022) to
perform the RE based on graph classification. For
that, we used dependency parsing to transform each
sentence into a weighted graph.
The remainder of this paper is organized as fol-
lows. Section 2 discusses state-of-the-art methods
for relation extraction. Section 3 presents our model
named GREED; whereas section 4 conducts an exten-
sive experimentation of GREED using standard crite-
ria. The final section concludes this paper and dis-
cusses future research directions.
2 RELATED WORK
We can categorize the relation extraction methods of
the literature into two main categories according to
the used techniques: rule-based methods and machine
learning-based methods (Yang et al., 2021).
The rule-based approach consists of preparing
rules to be inferred from the text to precisely extract
information about relations. Usually, supplemental
knowledge data can be used with these rules. As an
advantage, useful patterns for precise extraction can
be explored by the syntactical analysis of the sen-
tence. In addition, pertinent knowledge data can be
efficiently exploited by rules. However, a huge ef-
fort from medical experts is usually required to define
the rules and the knowledge data to cover most of the
possible cases. Also, these rules are usually domain-
specific and not adaptable to other types of data. For
example, Chikka and Karlapalem (2018) use some
post-processing rules based on dependency parsing to
improve the results of a BiLSTM model trained on
word and sentence features. Ben Abdessalem Karaa
et al. (2021) relied on the matching tool MetaMap
(Aronson and Lang, 2010) to search semantic types
and concepts from the UMLS meta-thesaurus (Bo-
denreider, 2004). Thus, they have applied some NLP
techniques to generate sentence features and train an
SVM (Hearst et al., 1998) model. The method of Kim
et al. (2021) applies some syntactical rules on the enti-
ties to generate lexical features to train an online gra-
dient descent model. Then, this method uses some
post-processing rules based on some trigger phrases
and terms prepared manually.
The principle of machine learning-based methods
is to learn to classify each sentence with an entity pair
into the corresponding relation class. As an advan-
tage, this approach can efficiently incorporate several
types of features, especially by using deep learning
models such as GCN. In addition, it can capture the
contextual dependency between words during classi-
fication. However, it requires a big number of training
samples which are manually annotated by medical ex-
perts. Also, relevant features and suitable techniques
must be chosen carefully for high-quality learning.
For example, the method of Li et al. (2019) is based
on a BiLSTM model with an embedding of the short-
est dependency path between the entities generated by
CNN in addition to dependency type features. Zhou
et al. (2020) have generated weighted dependency
graphs based on shortest path edges to extract generic
relations by using a GCN model. Patel et al. (2021)
use a CNN model to generate lexical and sentence
features. Ed-drissiya El-allaly et al. (2022) have com-
bined BERT with GCN in a joint model to perform an
N-level sequence labeling task.
In this paper, we propose a hybrid method named
GREED (Graph learning based Relation Extraction
with Entity and Dependency relations) which is
based on GCN to classify each sentence into rela-
tion classes. For that, the sentence is transformed
into a weighted dependency graph which is well pre-
processed to be suitable for GCN. Also, we have ex-
tracted several crucial features for each word which
take into account: word co-occurrence, dependency
types, entities, and relation verbs. The relation verbs
feature represents relevant side information. For that,
we have used a simple rule to match the verbs after
collecting them automatically from the training set.
The features, edge weights, and the GCN model pay
attention to the selected entity pair to focus on the
most important information. Our model is detailed
in the next section.
3 OUR PROPOSAL
3.1 Motivation
In our work, we have chosen the GCN model to pro-
cess a sentence weighted dependency graph and in-
corporate several word features. This model was used
by some recent methods (Mahendran et al., 2022; Li
et al., 2022; Yuan et al., 2022) and is known to be
suitable for this task. This approach is better than
analyzing the words sequentially since predicates are
significant for our task (Zhou et al., 2020). Usu-
ally, most methods give an uniform weight for the de-
pendency edges. However, we have added weighted
edges based on the shortest paths to be more adequate
for the GCN layer (Zhou et al., 2020). In addition,
GREED: Graph Learning Based Relation Extraction with Entity and Dependency Relations
361

we were inspired by the work of Yuan et al. (2022)
by paying more attention to the edges that are closer
to the entity pair. Therefore, we have assigned higher
weights to these edges. To focus on the entity pair-
related information, we have added an entity-attention
mechanism after the GCN layers based on the mask
pooling (Zhou et al., 2020). Moreover, we have used
the relative positions of the entities as word features
(Li et al., 2022). While the word co-occurrence is im-
portant information for our task (Shahab, 2017; Per-
era et al., 2020; Mahendran et al., 2022), we have
used a clinical GloVe model (Flamholz et al., 2022)
to generate word embedding. Usually, the difference
in the dependency relation types is usually ignored.
However, we have added the dependency relation type
to the word features. Relevant side information is
usually ignored while it add knowledge beyond the
sentence (Yuan et al., 2022; Mahendran et al., 2022).
Thus, we have automatically constructed a dictionary
of relation verbs. These verbs are matched based on
a syntactic dependency parsing to generate features
(Chikka and Karlapalem, 2018). In addition, we have
used other essential features such as the part of speech
and the entity types. To make a contextual word rep-
resentation and take into account the sequential de-
pendency, we have added a BiLSTM layer after the
input features layer (Yuan et al., 2022).
3.2 Architecture
The principle of our method GREED is to classify the
weighted dependency graph of the sentence by us-
ing a GCN model with the incorporation of several
word features. Thus, GREED has to detect which re-
lation can exist between a given entity pair. Firstly,
the sentence is transformed into a dependency graph
where the edges between words are weighted accord-
ing to the shortest paths and the proximity to the en-
tity pair. Furthermore, each word is represented by a
features vector which includes: clinical GloVe, PoS,
entity type, dependency type, relation verb matching,
and distance to the entity pair. To make the features
capture the contextual and sequential information, a
BiLSTM layer is added after the input features. Thus,
the graph and the features are fed into the GCN model
with an entity-attention mechanism based on mask
pooling. Thus, the output is the corresponding rela-
tion class of the input sentence and entity pair. The
main steps of our proposal are depicted in Figure 1.
3.3 Graph Construction
The sentence is transformed into a graph G = (V, E)
where V is the number of nodes (words) and E is the
Figure 1: The general architecture of our proposal.
number of edges that represent the dependency rela-
tions. Each edge has a weight and each word has a
feature vector. The weights are based on the distance
between words with the distance to the entity pair.
The features are based on sentence-level and global
information and take into account the entity pair.
3.4 Edge Weighting
We have syntactically parsed the sentence to get its
dependency tree and define the edges between words.
This parsing is very important for the relation extrac-
tion that is essentially based on the predicates (Ma-
hendran et al., 2022; Li et al., 2022; Yuan et al.,
2022). Most of the methods based on this idea use
unweighted edges. Indeed, we have followed the
method of Zhou et al. (2020) by adding weighted
edges based on the shortest paths to enhance the de-
pendency propagation made by the GCN layers. The
method of Yuan et al. (2022) focuses on some parts of
edges that are close to the entity pair to get the most
relevant information. Based on this idea, we have
modified the weights to take into account the prox-
imity to the entity pair. In addition, we have taken
into account the sequential distance between words
by exploiting a standard metric (Liu, 2008). In order
to compute the edge weights between nodes, we pro-
ceed as follows. First, we use the phrase to compute
the distance between two words using their order of
appearance in the same phrase (say i and j) using the
Manhatten distance:
dSeq
i j
= |i − j| (1)
Thereafter, we use the dependency graph to compute
another form of distance between nodes i and j by
summing the distances dSeq of the edges along the
shortest path between i and j as follows:
d
i j
=
∑
k∈Π
i j
dSeq(src
k
, dst
k
) (2)
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
362

where Π
i j
is the shortest between nodes i and j;
src
k
, dst
k
are the source and destination for each edge
k ∈ Π
i j
; and dSeq(.) is the distance computed by
equation 1. Doing this, we fuse between two impor-
tant informations: sequence distance and dependency
distance. Hence, given a node i, we compute the dis-
tance between the corresponding word and the exist-
ing entities in the relation as:
dEnt
i
= min
e
d
ie
(3)
where e is the set of entity words. Now, we compute
the weight between nodes i and j by the following
formula:
w
i j
=
1
e
d
i j
+min(dEnt
i
,dEnt
j
)−2
(4)
Note that we use the exponential to amplify the dis-
tance between nodes. We will describe in the next
section, the features used for graph nodes.
3.5 Node Features
In order to describe the nodes in the dependency
graph, we have adopted two categories of features
judged to be of great importance for relation ex-
traction: entity features and dependency features de-
scribed by:
3.5.1 Word Embedding:
We have used a GloVe model pre-trained on clinical
text to generate a static embedding of size 100 for
each word (Flamholz et al., 2022). This embedding
takes into account the global co-occurrence between
words which is a significant factor in determining the
relations (Shahab, 2017; Perera et al., 2020; Mahen-
dran et al., 2022). Since we have fed all the input fea-
tures into a BiLSTM layer, we can capture contextual
information rather than using a contextual embedding
like BERT (Mahendran et al., 2022; Ed-drissiya El-
allaly et al., 2022).
3.5.2 Part of Speech:
The PoS is essential information to the relation extrac-
tion since it is highly dependent on grammatical prop-
erties (Xu et al., 2018; Li et al., 2022). Thus, we have
used a trainable embedding layer of size N
pos
= 10
to represent 45 fine-grained PoS tags such as singular
noun, past participle verb, and determiner.
3.5.3 Entity Type:
Since we have to detect the relations between entities,
the entity types are essential information for this task
(Patel et al., 2021; Yuan et al., 2022). Thus, we have
used the entity types as lexical word features repre-
sented by a trainable embedding layer of size 10.
3.5.4 Dependency Type:
Most methods ignore the dependency types and con-
sider that the different dependency relations are iden-
tical (Zhou et al., 2020; Li et al., 2022; Yuan et al.,
2022). However, we have used a trainable embedding
layer of size 10 to represent the type of the incoming
dependency relation for each word. Thus, we are able
to distinguish 40 types such as nominal subject, object
predicate, particle, etc.
3.5.5 Matching a Relation Verb:
Relevant side information is usually ignored while it
enables to benefit from knowledge about the relations
(Yuan et al., 2022). To get information beyond the
sentence, we have automatically collected verbs for
each relation type from the training set. Thus, the
feature vector indicates the types which have a verb
matching the word. We were inspired by the post-
processing rules of Chikka and Karlapalem (2018) to
select a verb that connects a pair of entities. For that,
the verb should appear in the shortest dependency
path that relies these entities, otherwise it should be
in sequence between them. Each verb is normalized
by a stemming process.
3.5.6 Distance to the Entities:
Many methods have included information about the
entity pair position in their features (Chikka and Kar-
lapalem, 2018; Patel et al., 2021; Yuan et al., 2022).
Since it is essential to know which words are sup-
posed to belong to the relation entities. To represent
the relative position of the subject and the object, we
have used a trainable embedding layer of size 10 for
each one. We have used 21 classes to represent each
position where the direction and belonging to an en-
tity are taken into account.
3.6 Graph Convolutional Network
In our work, we have used the GCN to perform deep
learning on graph-structured data. The GCN layer
represents the first-order neighborhood dependency
by propagating the node features to the direct neigh-
bor nodes. Usually, a larger number of GCN layers
is required to represent higher order which increases
the model complexity (Mahendran et al., 2022; Yuan
et al., 2022). By adding the shortest path edges
(Zhou et al., 2020), one layer is able to perform long-
distance propagation, especially with edge weights.
GREED: Graph Learning Based Relation Extraction with Entity and Dependency Relations
363
In addition, we have used the mask pooling method
as an entity-attention mechanism to resume the GCN
output into three parts: subject entity, sentence, and
object entity (Zhou et al., 2020). By this mechanism,
the model is able to focus deeply on the relation en-
tities during classification. The next section analyzes
experimentally the performance of our proposal.
4 EXPERIMENTATION
We have chosen the benchmark dataset of the
i2b2/VA 2010 challenge (Uzuner et al., 2011). This
dataset contains patient reports which are divided into
170 for training and 256 for testing. It embeds sen-
tences annotated by 8 types of relations that hold be-
tween 3 types of entities: medical problem, treatment,
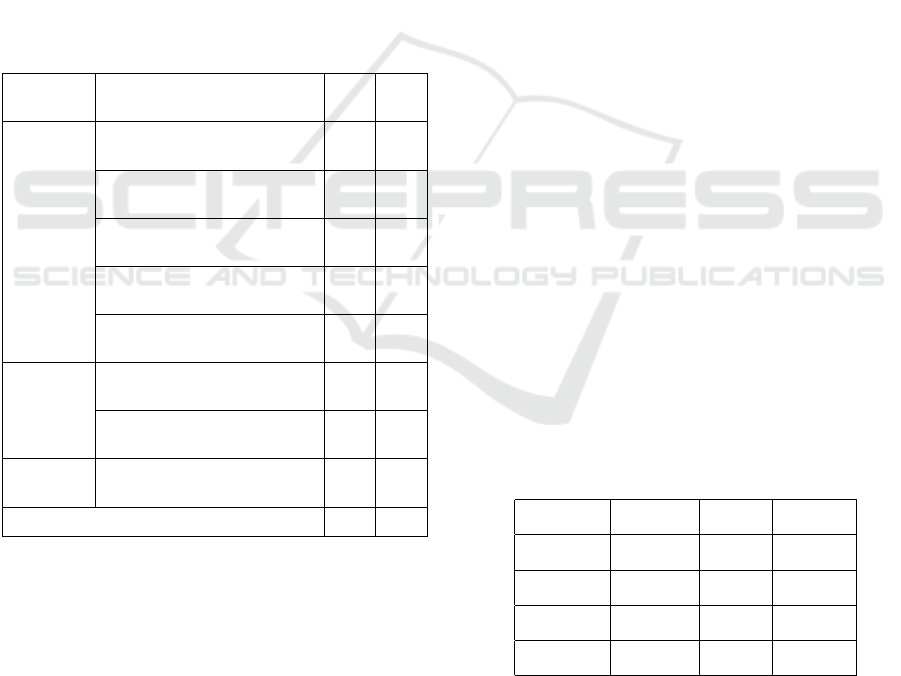
and test. More information is shown in Table 1.
Table 1: Information about relations in i2b2/VA 2010
dataset.
Entity
types
Relation type Train
set
Valid
set
Treatment-
Problem
Treatment improves problem
(TrIP)
107 198
Treatment worsens problem
(TrWP)
56 143
Treatment causes medical prob-
lem (TrCP)
296 444
Treatment is administered for
medical problem (TrAP)
1 422 2 487
Treatment is not administered
because of problem (TrNAP)
106 191
Test-
Problem
Test reveals medical problem
(TeRP)
1 733 3 033
Test conducted to investigate
medical problem (TeCP)
303 588
Problem-
Problem
Medical problem indicates
medical problem (PIP)
1 239 1 986
Total 5 262 9 070
In our research, we used a BiLSTM hidden layer
of size 64 and a WGCN layer of size 128, enabling
effective residual computation. Our model consists of
2 WGCN layers with a dropout rate of 0.5 and was
trained over 150 epochs with batch sizes of 50. The
computations were carried out on a system with 8 GB
of RAM and an Intel(R) Core(TM) i5-9300H CPU
@ 2.40GHz processor. We implemented our work us-
ing the Python programming language with the Spacy
module for natural language preprocessing and Py-
Torch for deep learning models.
For the evaluation, we have adapted the standard
metrics: precision, recall, and F1-score. For that, we
use 9 classes: a ”Non-relation” class and a class for
each relation type. Each sample represents a sentence
with a selected entity pair. In order to analyze the im-
portance of each category of features, we have imple-
mented different versions of our proposal as follows:
• GCN+WE: The GCN model with our edge
weighting method and essential features.
• +DEP: Adding the dependency type features.
• +TYRE: Adding the relation verb features.
• +DIST: Adding the entity pair position features
(full set of features).
The overall and the relation type-level results are
shown in the tables 2 and 3, respectively. In the ta-
ble 2, all components contribute to reaching high re-
sults since the minimal result is ≈ 81%. The added
features contribute to enhancing the F1-score results
by +0.71%. The best recall improvement is achieved
by adding the dependency type features by +0.67%.
Hence, the model can cover well the relations by dis-
tinguishing the different dependency types. The best
precision improvement (+0.30%) was made by the
entity distance features. Thus, recognizing the words
of the relation entities is useful to precisely classify
the relation.
In table 2, all the features contribute to increas-
ing the results for almost all the relation types with an
average improvement of +4% for 7 different classes.
However, the TrWP relations have the lowest result
(6.30%) due to the lack of data since there are only 56
training samples for this type. After adding our fea-
tures, the result for this type is improved by +4, 48%
and especially after adding the relation verbs features.
Thus, our model benefits from the automatically con-
structed dictionary to cover well the different relation
types and to deal with the lack of data.
Table 2: Overall evaluation of the GREED components.
Model Precision Recall F1-score
GCN+WE 81.87% 81.83% 81.38%
+DEP 81.77% 82.50% 81.97%
+TYRE 81.99% 82.67% 81.97%
+DIST 82.29% 82.40% 82.09%
In order to assess the effectiveness of GREED, we
have compared it with four other recent models: BiL-
STM (Li et al., 2019), WGCN (Zhou et al., 2020),
LSTM+R (Chikka and Karlapalem, 2018), and CNN
(Patel et al., 2021). The overall and the type-level
results of all models are shown in tables 4 and 5,
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
364
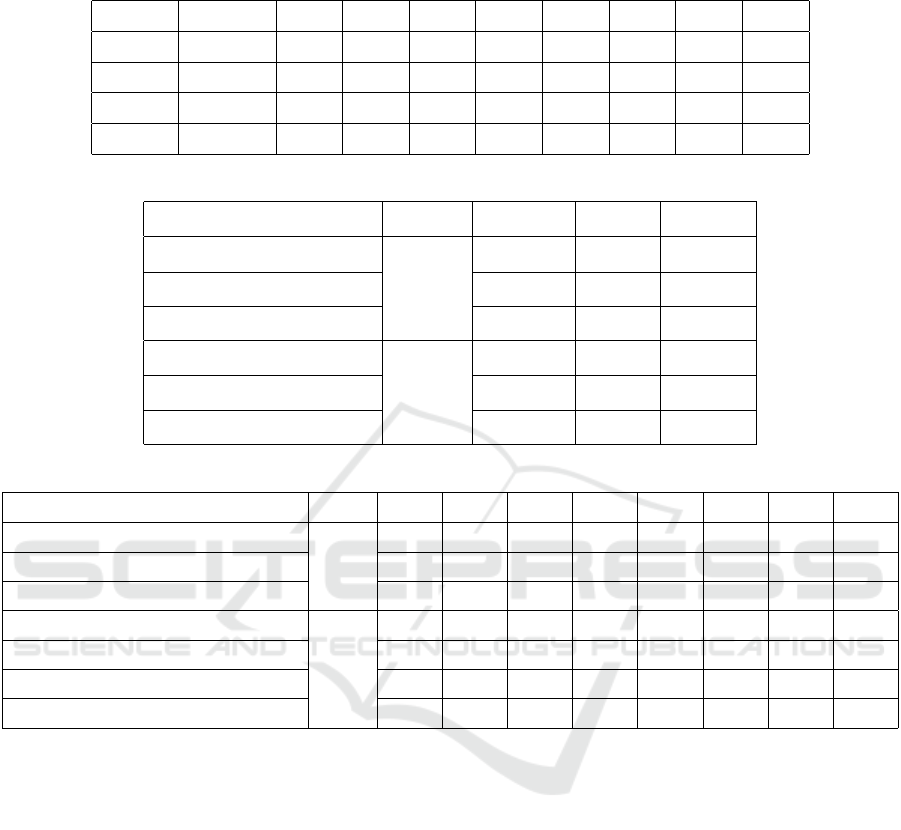
Table 3: Evaluation of the GREED components in type-level according to F1-score. The ”No relation” class means that there
is no defined relation between the entity pair.
Model No relation TrWP TrAP TrNAP TrCP TrIP TeCP TeRP PIP
GCN+WE 88.90% 01.82% 73.63% 26.09% 53.37% 26.40% 52.48% 84.14% 43.26%
+DEP 89.27% 00.00% 74.53% 16.90% 57.31% 27.72% 55.87% 85.39% 44.19%
+TYRE 89.32% 06.61% 74.87% 32.94% 57.09% 29.17% 51.34% 85.19% 42.77%
+DIST 89.21% 06.30% 75.26% 36.00% 58.10% 29.17% 49.73% 83.43% 47.58%
Table 4: Overall comparison of GREED with state-of-the-art models.
Model Method Precision Recall F1-score
BiLSTM (Li et al., 2019)
ET
75.69% 73.03% 74.34%
WGCN (Zhou et al., 2020) 80.94% 82.44% 81.14%
GREED 82.29% 82.40% 82.09%
WGCN (Zhou et al., 2020)
T
86.20% 87.91% 86.60%
CNN (Patel et al., 2021) 75.00% 72.00% 74.00%
GREED 87.88% 89.08% 87.72%
Table 5: Comparison of GREED with state-of-the-art models according to type-level F1-score.
Model Method TrWP TrAP TrNAP TrCP TrIP TeCP TeRP PIP
BiLSTM (Li et al., 2019)
ET
44.57% 79.74% 42.27% 62.13% 61.59% 61.17% 84.44% 63.33%
WGCN (Zhou et al., 2020) 00.00% 72.72% 30.28% 49.02% 34.07% 42.13% 81.73% 40.85%
GREED 06.30% 75.26% 36.00% 58.10% 29.17% 49.73% 83.43% 47.58%
LSTM+R (Chikka and Karlapalem, 2018)
T
58.00% 47.00% 42.00% 30.00% 17.00% NA NA NA
WGCN (Zhou et al., 2020) 07.35% 88.55% 33.20% 65.73% 43.39% 55.81% 93.63% 100.0%
CNN (Patel et al., 2021) 16.66% 78.54% 17.10% 04.00% 28.57% 46.66% 83.95% 92.43%
GREED 11.11% 89.02% 38.16% 65.64% 37.40% 68.20% 95.36% 99.97%
respectively. The abbreviation ”ET” in these tables
means that the model should extract the relation be-
fore identifying its type; the abbreviation ”T” means
that we have only to identify the type of relation be-
tween a pair of related entities; while the abbreviation
”NA” means Not Available. In table 4, our GREED
model outperforms all the state-of-the-art models ac-
cording to all metrics. WGCN (Zhou et al., 2020)
provides the closest results to ours with F1-score dif-
ference of −0.95% and −1.12% according to ”ET”
and ”T” methods, respectively. Note that this is the
only model based on GCN to process weighted de-
pendency graphs and it has not been applied in the
medical field before (Zhou et al., 2020). However, by
our features and edge weighting method, GREED is
able to achieve higher results. These two GCN-based
models have significantly outperformed the others
with a difference of more than +6.8 according to the
overall F1-score.
We can see in Table 5 that our proposal GREED is
more stable than the rest of the models and provides
the best results in most cases for all relation types.
However, GREED is outperformed by the LSTM+R
(Chikka and Karlapalem, 2018) model in the TrWP
relation type with a difference of −46.89%, while
we significantly outperform this model by an average
of +32.68% on three other types: TrAP, TrCP, and
TrIP. Note that LSTM-R used only 5 classes to clas-
sify ”treatment-problem” relations. This reduces the
confusion to focus more on the TrWP class, but the re-
sults are significantly decreased for other classes. We
have outperformed the CNN (Patel et al., 2021) model
with an average of +20.35% on 7 relation classes.
Indeed, CNN provides higher results than GREED
for the TrWP relations, with a difference of +5.55%.
In fact, this class has the lowest number of training
samples (56), but CNN benefited from a big man-
ually constructed meta-thesaurus called UMLS (Bo-
GREED: Graph Learning Based Relation Extraction with Entity and Dependency Relations
365

denreider, 2004) to make lexical features. Although
we don’t make any manual effort to construct our dic-
tionary, we significantly outperform LSTM+R in the
other classes. We have outperformed the two versions
of WGCN on all the classes, especially with the ”ET”
method, with an average increase of +5.66%. WGCN
provides a result of 0% for the TrWP class while we
obtain a result of 6.30%. Thus, our added features
and edge weighting, and pre-processing technique al-
low GREED to cover more relations. Although we
outperformed BiLSTM (Li et al., 2019) by +7.75%
in terms of overall F1-score, this model achieved the
highest results on all relation type classes. Note that
BiLSTM uses the ”non-relation” class but its result is
not available. Thus, this class must have a very low re-
sult and this means that many entity pairs without re-
lations are misclassified. This model uses dependency
relations and their types with CNN and BiLSTM lay-
ers. Thus, we can conclude that the use of GREED
of this information with GCN leads to higher overall
results without affecting the ”non-relation” class.
5 CONCLUSION
In this paper, we have proposed a hybrid method
named GREED (Graph learning based Relation Ex-
traction with Entity and Dependency relations) which
is able to extract clinical relations between entities in
sentences by using GCN on dependency graphs. Our
model processes the dependency relations efficiently
by appropriately weighting and filtering the edges,
taking into account the entity pair. GREED outper-
forms four state-of-the-art models especially which
are not based on graphs, without the need for a big
manual effort. Moreover, it can deal with the lack
of data. However, the time complexity of extracting
relations in one sentence may be unreasonable since
every possible entity pair needs to be processed. For
that, we need to find a way to extract all relations in a
sentence with only one process.
REFERENCES
Aronson, A. R. and Lang, F.-M. (2010). An overview
of metamap: historical perspective and recent advances.
Journal of the American Medical Informatics Associa-
tion, 17(3):229–236.
Ben Abdessalem Karaa, W., Alkhammash, E. H., and Bchir,
A. (2021). Drug disease relation extraction from biomed-
ical literature using nlp and machine learning. Mobile
Information Systems, 2021.
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nucleic
acids research, 32(suppl 1):D267–D270.
Chikka, V. R. and Karlapalem, K. (2018). A hybrid deep
learning approach for medical relation extraction. arXiv
preprint arXiv:1806.11189.
Ed-drissiya El-allaly, Sarrouti, M., En-Nahnahi, N., and
Ouatik El Alaoui, S. (2022). An attentive joint model
with transformer-based weighted graph convolutional
network for extracting adverse drug event relation. Jour-
nal of Biomedical Informatics, 125:103968.
Flamholz, Z. N., Crane-Droesch, A., Ungar, L. H., and
Weissman, G. E. (2022). Word embeddings trained on
published case reports are lightweight, effective for clin-
ical tasks, and free of protected health information. Jour-
nal of Biomedical Informatics, 125:103971.
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J., and
Scholkopf, B. (1998). Support vector machines. IEEE
Intelligent Systems and their applications, 13(4):18–28.
Kim, Y., Heider, P. M., Lally, I. R., and Meystre, S. M.
(2021). A hybrid model for family history information
identification and relation extraction: Development and
evaluation of an end-to-end information extraction sys-
tem. JMIR Medical Informatics, 9(4):e22797.
Kong, H.-J. (2019). Managing unstructured big data in
healthcare system. Healthcare informatics research,
25(1):1–2.
Landolsi, M. Y., Hlaoua, L., and Ben Romdhane, L.
(2022a). Information extraction from electronic medical
documents: state of the art and future research directions.
Knowledge and Information Systems, pages 1–54.
Landolsi, M. Y., Romdhane, L. B., and Hlaoua, L.
(2022b). Medical named entity recognition using sur-
rounding sequences matching. Procedia Computer Sci-
ence, 207:674–683.
Li, T., Ma, L., Qin, J., and Ren, W. (2022). Dtgcn: a method
combining dependency tree and graph convolutional net-
works for chinese long-interval named entity relationship
extraction. Journal of Ambient Intelligence and Human-
ized Computing, pages 1–13.
Li, Z., Yang, Z., Shen, C., Xu, J., Zhang, Y., and Xu, H.
(2019). Integrating shortest dependency path and sen-
tence sequence into a deep learning framework for rela-
tion extraction in clinical text. BMC medical informatics
and decision making, 19(1):1–8.
Liu, H. (2008). Dependency distance as a metric of lan-
guage comprehension difficulty. Journal of Cognitive
Science, 9(2):159–191.
Luo, Y., Uzuner,
¨
O., and Szolovits, P. (2017). Bridging
semantics and syntax with graph algorithms—state-of-
the-art of extracting biomedical relations. Briefings in
bioinformatics, 18(1):160–178.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
366

Mahendran, D., Tang, C., and McInnes, B. (2022). Graph
convolutional networks for chemical relation extraction.
Proceedings of the Semantics-enabled Biomedical Liter-
ature Analytics (SeBiLAn).
Patel, R., Tanwani, S., and Patidar, C. (2021). Relation ex-
traction between medical entities using deep learning ap-
proach. Informatica, 45(3).
Perera, N., Dehmer, M., and Emmert-Streib, F. (2020).
Named entity recognition and relation detection for
biomedical information extraction. Frontiers in cell and
developmental biology, page 673.
Quevedo, X. and Chicaiza, J. (2023). Using nlp to enrich
scientific knowledge graphs: A case study to find similar
papers. In ICAART (2), pages 190–198.
Shahab, E. (2017). A short survey of biomedical relation
extraction techniques. arXiv preprint arXiv:1707.05850.
Uzuner,
¨
O., South, B. R., Shen, S., and DuVall, S. L. (2011).
2010 i2b2/va challenge on concepts, assertions, and re-
lations in clinical text. Journal of the American Medical
Informatics Association, 18(5):552–556.
Xu, J., Gan, L., Cheng, M., and Wu, Q. (2018). Unsuper-
vised medical entity recognition and linking in chinese
online medical text. Journal of healthcare engineering,
2018.
Yang, X., Yu, Z., Guo, Y., Bian, J., and Wu, Y. (2021). Clin-
ical relation extraction using transformer-based models.
CoRR, abs/2107.08957.
Yuan, C., Huang, H., Feng, C., and Cao, Q. (2022). Piece-
wise graph convolutional network with edge-level atten-
tion for relation extraction. Neural Computing and Ap-
plications, pages 1–13.
Zhou, L., Wang, T., Qu, H., Huang, L., and Liu, Y. (2020).
A weighted gcn with logical adjacency matrix for rela-
tion extraction. In ECAI 2020, pages 2314–2321. IOS
Press.
GREED: Graph Learning Based Relation Extraction with Entity and Dependency Relations
367
