
Skimming of Electronic Health Records Highlighted by an Interface
Terminology Curated with Machine Learning Mining
Mahshad Koohi H. Dehkordi
1a
, Navya Martin Kollapally
1b
, Yehoshua Perl
1c
, James Geller
1d
,
Fadi P. Deek
1e
, Hao Liu
2f
, Vipina K. Keloth
3g
, Gai Elhanan
4h
and Andrew J. Einstein
5i
I
Ying Wu College of Computing, NJIT, Dr. Martin Luther King Jr. Boulevard, Newark, NJ, U.S.A.
2
Department of Computer Science, Montclair State University, 1 Normal Avenue, Montclair, NJ, U.S.A.
3
Department of Medical Informatics, Yale University, 51 Prospect Street, New Haven, CT, U.S.A.
4
Center for Genomic Medicine, School of Medicine, University of Nevada, 1664 N. Virginia Street, Reno, NV, U.S.A.
5
Dept. of Medicine, Cardiology Division, Columbia University Irving Medical Center New York, 168th Street, U.S.A.
gelhanan@gmail.com, ae2214@cumc.columbia.edu
Keywords: Highlighting, Electronic Health Record (EHR), Interface Terminology, SNOMED, Cardiology, Machine
Learning.
Abstract: Clinical notes in Electronic Health Records (EHRs) contain large amounts of nuanced information. Healthcare
professionals, e.g., clinicians, routinely review numerous EHR notes, further burdening their busy schedules.
To capture the essential content of a note, they often quickly review its content, which can contribute to
missing critical clinical information. Highlighting important content of EHRs enable clinicians to fast skim
by reading only the highlighted words. Furthermore, effective highlighting of EHRs will support new research
and interoperability. In this paper, we design a Cardiology Interface Terminology (CIT) dedicated for the
application of highlighting cardiology EHRs to support their fast skimming. Once successful, Transfer
Learning can be used to design an interface terminology for other specialties. In EHRs, we observe phrases
of fine granularity containing SNOMED CT concepts. In our previous work, we extract such phrases from
EHR notes to be considered as CIT concepts. This early CIT serves as training data for Machine Learning
(ML) techniques, further enriching CIT and improving EHR highlighting. We describe the methodology and
results of curating CIT with ML techniques. Furthermore, we introduce the coverage and breadth metrics for
measuring the efficacy of highlighting EHRs, and discuss future improvements, enhancing the coverage of
highlighted important content.
1 INTRODUCTION
Clinical notes in Electronic Health Records (EHRs)
contain large amounts of nuanced information that is
not captured by problem lists (Agrawal et al., 2013;
Elkin et al., 2006) . Healthcare professionals,
particularly physicians and nurses, are routinely
engaged in reviewing numerous EHR notes, further
burdening their busy schedules (Apathy et al., 2023;
Dymek et al., 2021). For brevity, we will refer only to
clinicians instead of Healthcare professionals. In their
a
https://orcid.org/0000-0003-3489-0892
b
https://orcid.org/0000-0003-4004-6508
c
https://orcid.org/0000-0003-1940-9386
d
https://orcid.org/0000-0002-9120-525X
e
https://orcid.org/0009-0001-1022-4118
quest for capturing most essential content, they do not
read a whole note, but often quickly review its content
(Cui et al., 2022; Yada & Aramaki, 2023). Such
cursory review, without studying it in detail, can
contribute to missing critical clinical information,
leading to improper or risky treatment. The
prevalence of quick reviewing is higher for
overworked healthcare workers such as interns and
nurses.
Highlighting facts and important content of an
EHR note enables clinicians to fast skim EHRs, by
f
https://orcid.org/0000-0002-1975-1272
g
https://orcid.org/0000-0001-6919-1122
h
https://orcid.org/0000-0003-1518-5522
i
https://orcid.org/0000-0003-2583-9278
498
Koohi H. Dehkordi, M., Kollapally, N., Perl, Y., Geller, J., Deek, F., Liu, H., Keloth, V., Elhanan, G. and Einstein, A.
Skimming of Electronic Health Records Highlighted by an Interface Terminology Curated with Machine Learning Mining.
DOI: 10.5220/0012391600003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 498-505
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

reading only the highlighted words, since it draws
attention to the essential information of the note.
When a clinician skims only the highlighted text, time
is saved by concentrating attention on the important
content. Reading only a highlighted portion of a
larger text also saves the implicit mental effort for
selecting the essential parts to read. Currently,
unstructured EHRs are not highlighted. Clinical notes
in the broad medical domain and its specialties, such
as cardiology, are generally entered as free text using
a complex and nuanced medical dialect, including
localized and even personalized syntax, expressions,
and acronyms. This complexity is rarely captured by
terminologies such as the popular SNOMED CT
(SNOMED-CT, 2021) (SCT) clinical terminology.
Figure 1 shows an example of highlighting with SCT
concepts, which captures only part of the important
content.
In addition to fast skimming, effective
highlighting of cardiology EHR notes will enable new
research and interoperability. For example, “severe
fluid retention (edema)” is not listed as adverse effect
of Tacrolimus, an anti-rejection drug. Out of more
than 72,000 adverse events reported in the FDA’s
FAERS (Dashboard.) repository about Tacrolimus,
only 135 are relate to edema. Hypothesizing that this
is an under-reported adverse event, this association
may be missed, since edema is not likely to be
recorded in the structured EHR, especially because it
is not relevant to billing. However, the highlighting
of clinical notes of post-transplant patients who were
prescribed Tacrolimus, can enable research into
identifying the terms that might be used to describe
Figure 1: An excerpt of a MIMIC III EHR note highlighted
by SNOMED.
edema in various forms, and also establish the
association with the offending medication(s).
Since highlighting with the best reference clinical
terminology, SCT, does not provide sufficient
highlighting of EHRs, as demonstrated in Figure 1,
we propose to design an interface terminology
dedicated for highlighting. An interface terminology
is, by definition, designed to maximize utilization of
a specific application by end users, including
software applications(Jonquet et al., 2009). We select
the domain of Cardiology to illustrate our technique,
by curating the Cardiology Interface Terminology
(CIT) for highlighting of EHRs of cardiology patients
(cardiology EHRs). CIT will initially contain the
Cardiology Component of SNOMED (CCS). We
observe that EHRs contain phrases of finer
granularity than SCT concepts, and such phrases
could provide insights if highlighted in fast
skimming. Therefore, we include such phrases as
concepts in CIT by introducing a Machine Learning
(ML) technique to automatically mine EHRs for fine
granularity phrases not-currently-recognized as
concepts to enrich CIT, since the best source for
concepts to highlight EHRs are the notes themselves.
Training ML models requires high quality
training data. Manual mining of EHRs for additional
CIT concepts, which will serve for preparing training
data, is expensive and time consuming. We observe
that high granularity phrases found in EHRs typically
contain shorter SCT concepts. In (Koohi H. Dehkordi
et al., 2023), we leveraged this observation to develop
an initial version of CIT utilizing a semi-automatic
mining method, which serves as the training data for
this study. Earlier versions of such research about
EHRs of Covid patients were described in (Keloth et
al., 2021; Keloth et al., 2020).
A concept classification ML method was trained
with phrases accepted (concepts) in Phase 1 as
positive samples, and phrases rejected (non-concepts)
as negative samples. The CIT yielding from this
process presumably captures some of the important
aspects of an EHR note needed for fast skimming and
research of EHRs. After validating with cardiology
domain, Transfer Learning (Francis et al., 2019;
Giorgi & Bader, 2018; Sun & Yang, 2019; Weiss et
al., 2016) can be used for curating interface
terminologies for highlighting EHRs in other
specialties, e.g., pulmonology.
Our research hypothesis, based on a review of
many highlighted EHRs, is that an average
highlighting coverage of about 75% captures almost
all important information of EHR notes. Such
coverage could be obtained after the ML-based Phase
2 adds to CIT the concepts that were missed in Phase
Skimming of Electronic Health Records Highlighted by an Interface Terminology Curated with Machine Learning Mining
499

1. A trained ML classifier is used to identify extracted
concepts from build dataset B, which will be added to
CIT to obtain CIT
ML
. A ML classifier method was
trained with CIT concepts of Phase 1 as positive
samples, and phrases rejected as negative samples.
Phrases classified as CIT concepts by the ML
classifier presumably will capture most of the
important aspects of an EHR note needed for fast
skimming and research of EHRs.
2 METHODS
In this section, we describe Phase 2 of the curation of
the Cardiology Interface Terminology (CIT) for the
highlighting of Cardiology EHRs. Phase 2
concentrates on enriching CIT using ML techniques.
Phase 1, described in Background Section, resulted in
a training dataset for Phase 2, encompassing a) The
Cardiology Interface Terminology (CIT) as positive
samples, b) The reject list R which includes rejected
phrases that either violated our predefined rules or
failed in manual review, as negative samples. The
phrases of the training dataset were embedded
(Kusner et al., 2015) using Clinical BioBERT.
We trained a Neural Network (NN) model to
classify phrases based on positive and negative
samples. For a newly extracted phrase from EHRs,
we generated word embeddings (Kusner et al., 2015)
using Clinical BioBERT, and then employed the
trained NN model to classify whether this phrase fits
as a concept of CIT or not. The phrases that were
assigned a label “1” (positive instances) were added
to CIT, forming CIT
ML
. This new terminology was
then used to highlight the dataset B. We further
enriched CIT
ML
with concepts of SNOMED that did
not appear in CIT
ML
, using a DIFF operation, yielding
CIT
ML+
. CIT
ML+
is then used to highlight the test
dataset T. We evaluated the efficacy of the proposed
process by calculating coverage and breadth metrics
for both highlighted datasets B and T. A flowchart
describing the components of Phase 2 is displayed in
Figure 2. Section headers, in red, are added manually
to improve readability of an EHR note. This is a
general description for orientation of the flowchart.
The internal steps within Phase 2 are described as
follows.
We define two performance metrics: Coverage,
as the percentage of words being highlighted (using
(1)). Assuming that the CIT concepts reflect
meaningful phrases in EHRs, the coverage is
typically correlated with the extent the highlighting
captures the important content of the note. Breadth,
is the average number of words per highlighted
concept (using (2)). For example, for the highlighted
note shown in Figure 1, the coverage is 35% and the
breadth is 1.21. Note that using high granularity
concepts, rather than SCT concepts, increases
breadth.
𝐶𝑜𝑣𝑒𝑟𝑎𝑔𝑒 =
# 𝑎𝑛𝑛𝑜𝑡𝑎𝑡𝑒𝑑 𝑤𝑜𝑟𝑑𝑠
#𝑎𝑙𝑙𝑤𝑜𝑟𝑑𝑠
× 100
(1)
𝐵𝑟𝑒𝑎𝑑𝑡ℎ =
# 𝑎𝑛𝑛𝑜𝑡𝑎𝑡𝑒𝑑 𝑤𝑜𝑟𝑑𝑠
# 𝑎𝑛𝑛𝑜𝑡𝑎𝑡𝑒𝑑 𝑐𝑜𝑛𝑐𝑒𝑝𝑡𝑠
(2)
2.1 Training Ml Model
Embedding the training dataset using Clinical
BioBERT: We employed word tokenization (Huang
et al., 2007) on the phrases of the training dataset to
generate input sequences with a maximum length of
16 tokens.
There are several pretrained language models such as
BERT(Devlin et al., 2018) , BioBERT(Lee et al.,
2020), RoBERTa(Liu et al., 2019), ALBERT(Lan et
al., 2019), and Clinical BioBERT(Alsentzer et al.,
2019). Given our target dataset consists of EHR
notes, we chose the Clinical BioBERT model to
generate word embeddings for the tokenized phrases
of training dataset. The reason is that Clinical
BioBERT has been pretrained on a large corpus of
clinical text, including approximately two million
notes of the MIMIC-III v1.4 database (Alimova &
Tutubalina, 2020). The list of the embedded phrases,
generated using Clinical BioBERT, was converted to
PyTorch tensors as the input for the ML model.
Training the ML model: The training dataset is
randomly split into 80% for training and validation,
and 20% for testing. We opted for a Neural Network
to be trained. Following a grid search (Liashchynskyi
& Liashchynskyi, 2019) to fine-tune the
hyperparameters, we ended up having an NN model
with one hidden layer, 100 neurons, Relu (Agarap,
2018) as the activation function, and Adam (Jais et
al., 2019) as the optimization algorithm. To optimize
our model’s performance, 5-fold cross-validation
(Fushiki, 2011) process is employed while training
the model. Also, to address overfitting, a dropout rate
of 0.2 has been implemented. The NN model
achieved an accuracy of 86%, precision of 88%,
recall of 88%, and F1 of 88% on the test set.
2.2 Enriching CIT and CIT
ML
The average length of the concepts added to CIT in
Phase 1 is 3.58 words. Out of 18,749 CIT concepts
extracted form EHRs, only 1081 concepts (5.7%)
have 7-9 words, and only 191 (1%) have more than
HEALTHINF 2024 - 17th International Conference on Health Informatics
500

Figure 2: The flowchart of Phase 2. It consists of three parts: (1) Training the ML model. (2) Enriching the terminology using
the trained ML model to obtain CIT
ML
and CIT
ML+
, respectively, where CIT
ML+
is obtained by enriching CIT
ML
with SCT
concepts of T which are missing in B. (3) Evaluating the highlighting with terminologies CIT
ML
and CIT
ML+
of datasets B
and T, respectively.
nine words. We chose a parameter L for the
maximum number of words in a phrase extracted
from dataset B for assessing their suitability for CIT.
In this study, we experimented with the values 6 and
9 for L (see Discussion Section for the trade-off in
results). Hence, to enrich CIT, all subsequences of
one to L words within each sentence of the 500 EHRs
in dataset B are extracted. If a sentence has X words,
where X<L, then the subsequences will have only up
to X words, denoted by “n” number of words in the
dataset B. The total number of phrases generated by
all subsequences is less than L*n. Hence, the
complexity of the number of generated phrases is
linear with the number of words in B.
These newly generated phrases were compared
with our existing training dataset. Many of these
phrases were eliminated since they appeared in the
training dataset as positive (CIT) and negative (R)
samples. Additional rules, such as removing phrases
ending with adjectives or stop words, were applied to
remove further phrases. The remaining phrases were
embedded using Clinical BioBERT. The previously
trained NN model was applied to classify these
embedded phrases. Phrases predicted to be labelled
“1” (valid concepts) were added into CIT, resulting in
a new interface terminology, CIT
ML
.
To test the effectiveness of the highlighting of a
dataset by CIT, we evaluate its coverage and breadth
metrics, which were introduced in the Background
section. The dataset B is highlighted by CIT and
CIT
ML
for Phase 1 and Phase 2, respectively.
However, for the evaluation of the dataset T, we need
to enrich CIT
ML
with concepts of SCT which appear
in T, but not in B. The reason is that such concepts are
not included in CIT in Phase 1 unless they were in
CCS.
Skimming of Electronic Health Records Highlighted by an Interface Terminology Curated with Machine Learning Mining
501

For adding such concepts, we first define a DIFF
operation as a set difference. The DIFF operation is
calculated using (3).
𝐷𝐼𝐹𝐹
𝑇 ,𝑆𝐶𝑇, 𝐶𝐼𝑇
=
{
𝑇 𝑎𝑛𝑛𝑜𝑡𝑎𝑡𝑒𝑑 𝑤𝑖𝑡ℎ 𝑆𝐶𝑇
}
− {𝑇 𝑎𝑛𝑛𝑜𝑡𝑎𝑡𝑒𝑑 𝑤𝑖𝑡ℎ 𝐶𝐼𝑇
}
(3)
Let DIFF’ (DIFF prime) be the set resulting from
the DIFF operation. We obtained a new interface
terminology CIT
ML+
for highlighting of the dataset T.
CIT
ML+
is calculated using the union operation (4).
𝐶𝐼𝑇
=𝐶𝐼𝑇
∪ 𝐷𝐼𝐹𝐹′
(4)
3 RESULTS
Table 1 displays the results of coverage and breadth
metrics for highlighting the build dataset B and test
dataset T for the various terminologies: SCT, CIT,
CIT
ML
and CIT
ML+
. The results for SCT and for CIT
relate to Phase 1 in the Background section, described
here to enable comparison with current results. We
have two versions for CIT
ML
and CIT
ML+
denoted by
(6) and (9), according to the maximum number of
words per concept allowed to be added in Phase 2.
For build dataset B, the average coverage
increased by 16% (relatively 47%) for CIT vs SCT,
and further, increased by 16% (relatively 31%) with
CIT
ML+
(6) or 18% (relatively 35%) with CIT
ML
(9).
All in all, the average coverage with CIT
ML
(9) is
about double of the one with SCT. The average
breadth increased by 1.14 for CIT vs SCT, but then
decreased by 0.29 for CIT
ML+
(6) or by 0.22 for
CIT
ML
(9).
Similarly, for test dataset T, the average coverage
increased by 9% (relatively 26%) for CIT vs SCT,
and further increased by 15% (relatively 37%) with
CIT
ML+
(6) or 16% (relatively 41%) with CIT
ML+
(9).
All in all, the average coverage with CIT
ML+
(9) is
78% higher than with SCT. The average breadth
increased by 0.55 for CIT vs SCT, but then decreased
by 0.18 for CIT
ML+
( 6) or by 0.16 for CIT
ML+
(9).
Nevertheless, we have chosen to display in Figure
3 the highlighting of a note from T because the
purpose of our work is not only to highlight the build
dataset, but any other unseen datasets. T is a sample
of such unseen datasets. Figure 3(a) displays the
highlighting of a note in testdata highlighted with CIT
following the process of Phase 1 which is described
in (Koohi H. Dehkordi et al., 2023). Figure 3(b)
displays the highlighting of the same note using
CIT
ML+
which is obtained in Phase 2. Comparing
Figure 3(a) and Figure 3(b) demonstrates the progress
obtained by the ML technique. The Discussion
Section contains more detailed analysis.
4 DISCUSSION
In this paper, we have reported on a project that uses
Machine Learning techniques to curate an interface
terminology dedicated to highlighting EHRs of
patients of a specific medical specialty. The reason
for this is that using a single interface terminology to
highlight general EHRs, similar to SCT, would be too
unwieldy to manage. Although our project
concentrates on cardiology, we plan on using
Transfer Learning in order to curate interface
terminologies for other disciplines e.g.,
Pulmonology.
Limitations: On one side, Figure 3 demonstrates
the progress we made in achieving the ultimate target
of highlighting all and only the important content of
a clinical note required for a clinician in order to be
informed of a patient’s condition and the course of
his/her disease. On the other side, Figure 3(b) exposes
a shortcoming of the current process. Elements which
are not highlighted include numbers (e.g., 49),
abbreviations (e.g., MPGN, PO, PTSD, ROS), and
different verb tenses (e.g., discharged, feels, reports,
etc). Highlighting them is required for understanding
the important content. For example, the difference
between continued and discontinued, or between
reports and denies, is critical. Another important issue
is identifying all negation expressions. For example,
in Figure 3(b), we highlighted “No”, while we missed
“neg”. Regarding medications, they are highlighted in
Figure 3(b), but not in Figure 3(a). We will address
those issues in our Future Work Section.
Table 1: Average metrics of highlighting with various terminologies for datasets B and T of EHR notes.
Build Dataset B Test Dataset T
SCT CIT CIT
ML
(6) CIT
ML
(9) SCT CIT CIT
ML+
(6)
CIT
ML+
(9)
Coverage 34.5% 50.7% 66.4% 68.3% 35.3% 44.7% 61.4% 62.9%
Breadth 1.24 2.38 2.09 2.16 1.25 1.8 1.62 1.64
HEALTHINF 2024 - 17th International Conference on Health Informatics
502
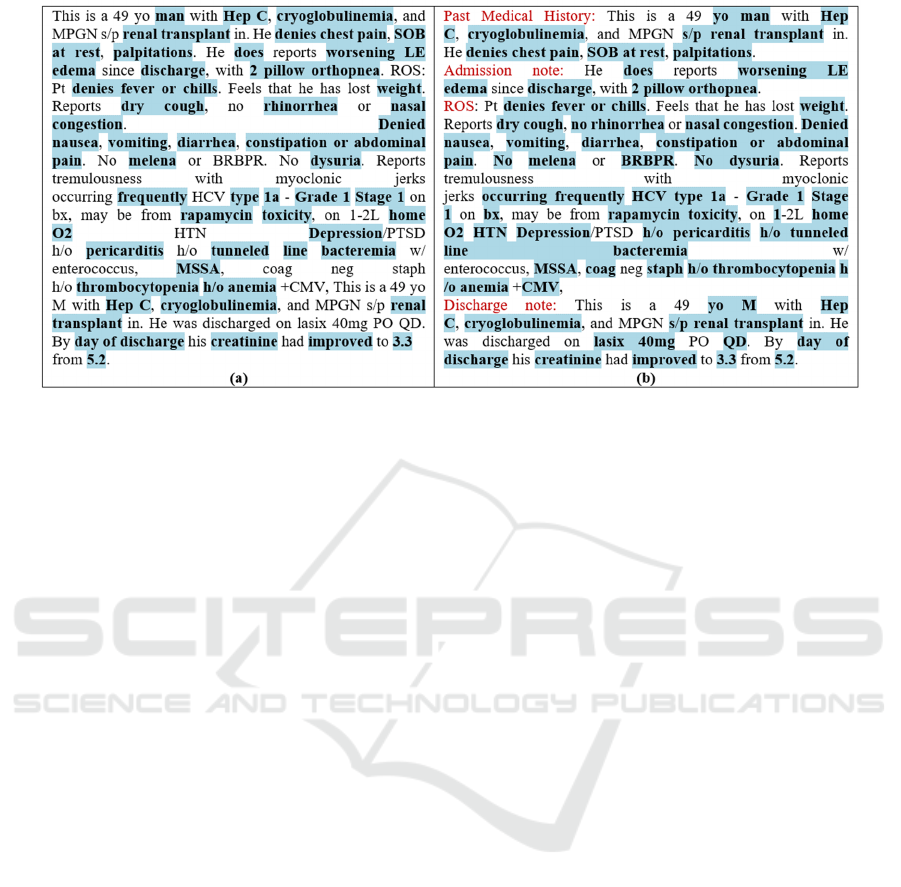
Figure 3: An excerpt of the MIMIC III EHR note highlighted by (a) CIT with 43.7% coverage and 1.59 breadth. (b) CIT
ML+
with 63.3% coverage and 1.39 breadth. Figure 1, the same note highlighted by SCT, is with 35% coverage and 1.2 breadth.
The efficiency of the highlighting depends on the
size of CIT, which in turn depends on the size of the
dataset from which the additional CIT concepts are
extracted. The proposed method was trained and
validated using 500 notes while more human
highlighted notes are warranted to improve the
performance of our ML models.
Data Analysis: Let us first demonstrate the
performance of ML techniques in identifying
concepts which were totally unhighlighted by CIT,
whereas highlighted by CIT
ML+
. For example "lasix
40mg PO QD". In other examples a phrase was
partially highlighted by CIT but fully highlighted by
CIT
ML+
. For example, “No melena or BRBPR”, “s/p
renal transplant”, and “h/o pericarditis”. Note that the
additional highlighting with CIT
ML+
is clarifying the
context of the partial highlighting with CIT.
In contrast, we have examples where CIT
ML+
missed certain highlighting important content which
the reader of a note should see. For example, in
“Reports tremulousness with myoclonic jerks
occurring frequently”. In “Feels that he has lost
weight”, the word “lost” is important to understand
the message conveyed by the sentence. Note that by
adding lists of verbs and medications to CIT, as
described in Future Work Section, the last two cases
will be properly highlighted.
Performance: We experimented with two values (6
and 9), for the maximum length of a newly extracted
phrase from the dataset B. The resulting coverage
differs slightly for those two values. The number for
CIT concepts with length of 7-9 is small (15%). We
expect a similar percentage for the number of
concepts of length of 7-9 in CIT
ML
. Indeed, in CIT
ML
the number of concepts of length 7-9 is 17,046 (15%).
Furthermore, the contribution of an L>9 is expected
to be small due to the small number of concepts of
such length in CIT (5%). Therefore, we set the length
of phrases fed into the NN model to 9, such that a
concept longer than 9 words is not obtained.
Comparing the coverage for B and T, we observe
that the values for T are smaller than for B, for CIT as
well as for CIT
ML+
vs CIT
ML
. The reason is that the
concepts of the interface terminologies CIT and
CIT
ML
are solely extracted from the build dataset B.
Hence, the likelihood of those concepts to appear in
T is lower than in B.
The breadth for SCT is low since most of the
concepts in ICIT are of only one word. For B, the
breadth for CIT is almost double than for SCT. The
reason is that the additional concepts (extracted in
Phase 1) are longer since they are obtained by the
concatenation and anchoring operations. However,
the breadth for CIT
ML
is smaller because most of the
additional concepts extracted in the ML process are
typically short.
For T, the breadth for CIT is lower than for B
because the likelihood of a concept extracted from B
to appear in T decreases as the number of words for
the concepts increases. The reason is the longer the
phrase, the more granular it is, and subsequently, its
frequency in a different dataset is lower. The reason
for the decrease for CIT
ML+
vs CIT
ML
in B is that SCT
concepts from T which did not appear in B are added
by DIFF operation to CIT
ML+
, and they lower the
Skimming of Electronic Health Records Highlighted by an Interface Terminology Curated with Machine Learning Mining
503

average since none of them contain another concept
(similar to the argument above).
We experimented with the NN model with one
and multiple hidden layers. We found that deepening
NNs did not further improve the performance
compared with simple structure NNs. A possible
explanation for this is that for deeper NNs to yield
better performance, more training data from larger
datasets is required. We will revisit those experiments
using larger datasets in the future.
Future Work: In the next stage of our project, as a
remedy to the shortcomings described in Limitations
Section, we plan to take the following actions. In
Phase 1, we will insert into the Initial CIT (ICIT)
these components:
1. Existing abbreviations in Cardiology
(Heart.org, u.d; Utah, u.d) and in Medicine in
general (Wikipedia, 2015).
2. Numbers from the range expected in
Cardiology EHRs.
3. Verbs with different tenses
(worldclasslearning, u.d).
4. Medications used in Cardiology (Heart.org,
u.d).
5. Common forms of negation (learngrammar,
u.d).
6. We will enrich the low synonym coverage of
CCS concepts (0.606) by migrating synonyms
from UMLS(Bodenreider, 2004).
As a result, we expect a more accurate
highlighting of EHRs.
5 CONCLUSIONS
We describe a research project to curate a Cardiology
Interface Terminology (CIT) dedicated for
highlighting EHRs of patients. The purpose is to
highlight all and only the important content of an
EHR note which a clinician need to review.
Highlighted EHRs will enable healthcare
professionals to read only the highlighted important
information of an EHR note rather than cursorily
review it, risking missing critical medical
information. Machine Learning techniques are
utilized for the design of CIT for the Cardiology
specialty. Transfer Learning will be used to design
interface terminologies for other specialties. As the
training data required for machine learning, an early
version of CIT (Koohi H. Dehkordi et al., 2023)
designed with a semi-automatic mining method rather
than slow manual mining is used. The results
demonstrate significant progress over highlighting
with SNOMED CT and with the early version of CIT.
We discussed ideas to further improve the coverage
of highlighting the important content of EHR to
achieve a satisfactory highlighting.
REFERENCES
Agarap, A. F. (2018). Deep learning using rectified linear
units (relu). arXiv preprint arXiv:1803.08375.
Agrawal, A., He, Z., Perl, Y., Wei, D., Halper, M., Elhanan,
G., & Chen, Y. (2013). The readiness of SNOMED
problem list concepts for meaningful use of electronic
health records. Artif Intell Med, 58(2), 73-80.
https://doi.org/10.1016/j.artmed.2013.03.008
Alimova, I., & Tutubalina, E. (2020). Multiple features for
clinical relation extraction: A machine learning
approach. Journal of biomedical informatics, 103,
103382.
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H., Jin,
D., Naumann, T., & McDermott, M. (2019). Publicly
available clinical BERT embeddings. arXiv preprint
arXiv:1904.03323.
Apathy, N. C., Rotenstein, L., Bates, D. W., & Holmgren,
A. J. (2023). Documentation dynamics: note
composition, burden, and physician efficiency. Health
Services Research, 58(3), 674-685.
Bodenreider, O. (2004). The unified medical language
system (UMLS): integrating biomedical terminology.
Nucleic acids research, 32(suppl_1), D267-D270.
Cui, S., Luo, J., Ye, M., Wang, J., Wang, T., & Ma, F.
(2022). MedSkim: Denoised Health Risk Prediction via
Skimming Medical Claims Data. 2022 IEEE
International Conference on Data Mining (ICDM),
Dashboard., F. A. E. R. S. F. P. https://fis.fda.gov/sense/
app/95239e26-e0be-42d9-a960-9a5f7f1c25ee/sheet/45
beeb74-30ab-46be-8267-5756582633b4/state/analysis
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2018).
Bert: Pre-training of deep bidirectional transformers
for language understanding. arXiv preprint
arXiv:1810.04805.
Dymek, C., Kim, B., Melton, G. B., Payne, T. H., Singh,
H., & Hsiao, C.-J. (2021). Building the evidence-base
to reduce electronic health record–related clinician
burden. Journal of the American Medical Informatics
Association, 28(5), 1057-1061.
Elkin, P. L., Brown, S. H., Husser, C. S., Bauer, B. A.,
Wahner-Roedler, D., Rosenbloom, S. T., & Speroff, T.
(2006). Evaluation of the content coverage of
SNOMED CT: ability of SNOMED clinical terms to
represent clinical problem lists. Mayo Clinic
Proceedings,
Francis, S., Van Landeghem, J., & Moens, M.-F. (2019).
Transfer learning for named entity recognition in
financial and biomedical documents. Information,
10(8), 248.
HEALTHINF 2024 - 17th International Conference on Health Informatics
504

Fushiki, T. (2011). Estimation of prediction error by using
K-fold cross-validation. Statistics and Computing, 21,
137-146.
Giorgi, J., & Bader, G. D. (2018). Transfer learning for
biomedical named entity recognition with neural
networks. Bioinformatics, 34, 4087 - 4094.
Heart.org. (u.d). Heart Medications. https://www.heart.org/
en/health-topics/heart-attack/treatment-of-a-heart-attac
k/cardiac-medications
Huang, C.-R., Šimon, P., Hsieh, S.-K., & Prévot, L. (2007).
Rethinking chinese word segmentation: tokenization,
character classification, or wordbreak identification.
Proceedings of the 45th Annual Meeting of the
Association for Computational Linguistics Companion
Volume Proceedings of the Demo and Poster Sessions,
Jais, I. K. M., Ismail, A. R., & Nisa, S. Q. (2019). Adam
optimization algorithm for wide and deep neural
network. Knowledge Engineering and Data Science,
2(1), 41-46.
Jonquet, C., Shah, N., & Musen, M. (2009). The open
biomedical annotator. Summit Transl Bioinform. 2009
Mar 01; 2009: 56–60. In.
Keloth, V. K., Zhou, S., Lindemann, L., Elhanan, G.,
Einstein, A., Geller, J., & Perl, Y. (2021). EHR Mining
for Interface Terminology Concepts for Annotating
EHRs of COVID patients. BMC Medical Informatics
and Decision Making.
Keloth, V. K., Zhou, S., Lindemann, L., Elhanan, G.,
Einstein, A. J., Geller, J., & Perl, Y. (2020). Mining
Concepts for a COVID Interface Terminology for
Annotation of EHRs. 2020 IEEE International
Conference on Big Data.
Koohi H. Dehkordi, M., Zhou, S., Perl, Y., Geller, J.,
Einstein, A., Elhanan, G., Keloth, V., & Liu, H. (2023).
Using annotation for computerized support for fast
skimming of cardiology electronic health record notes.
In proceedings of the IEEE International Conference on
Bioinformatics and Biomedicine (BIBM) 2023
Workshop: Artificial Intelligence Techniques for
BioMedicine and HealthCare, Istanbul, Turkey.
Kusner, M., Sun, Y., Kolkin, N., & Weinberger, K. (2015).
From word embeddings to document distances.
International conference on machine learning,
Lan, Z., Chen, M., Goodman, S., Gimpel, K., Sharma, P.,
& Soricut, R. (2019). Albert: A lite bert for self-
supervised learning of language representations. arXiv
preprint arXiv:1909.11942.
learngrammar. (u.d). https://www.learngrammar.net/
english-grammar/negation
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., &
Kang, J. (2020). BioBERT: a pre-trained biomedical
language representation model for biomedical text
mining. Bioinformatics, 36(4), 1234-1240.
Liashchynskyi, P., & Liashchynskyi, P. (2019). Grid
search, random search, genetic algorithm: a big
comparison for NAS. arXiv preprint
arXiv:1912.06059.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen, D.,
Levy, O., Lewis, M., Zettlemoyer, L., & Stoyanov, V.
(2019). Roberta: A robustly optimized bert pretraining
approach. arXiv preprint arXiv:1907.11692.
SNOMED-CT. (2021).
SNOMED CT https://www.sno
med.org/
Sun, C., & Yang, Z. (2019). Transfer learning in biomedical
named entity recognition: an evaluation of BERT in the
PharmaCoNER task. Proceedings of The 5th Workshop
on BioNLP Open Shared Tasks,
Utah, U. o. (u.d). Cardiology Abbreviations and Diagnosis.
http://www.ped.med.utah.edu/pedsintranet/outpatient/t
riage/team_red/cardio_abbreviations_diagnosis.pdf
Weiss, K., Khoshgoftaar, T. M., & Wang, D. (2016). A
survey of transfer learning. Journal of Big data, 3(1), 1-
40.
Wikipedia. (2015). List of medical abbreviations.
https://en.wikipedia.org/wiki/List_of_medical_abbrevi
ations
worldclasslearning. (u.d). https://www.worldclasslearning.
com/english/five-verb-forms.html
Yada, S., & Aramaki, E. (2023). HeaRT: Health Record
Timeliner to visualise patients' medical history from
health record text. arXiv preprint arXiv:2306.14379.
Skimming of Electronic Health Records Highlighted by an Interface Terminology Curated with Machine Learning Mining
505
