
Evaluating Data Augmentation Techniques for Coffee Leaf Disease
Classification
Adrian Gheorghiu
1,∗
, Iulian-Marius T
˘
aiatu
1,∗
, Dumitru-Clementin Cercel
1, †
, Iuliana Marin
2
and Florin Pop
1,3,4
1
Computer Science and Engineering Department, National University of Science and Technology Politehnica Bucharest,
Romania
2
Faculty of Engineering in Foreign Languages, National University of Science and Technology Politehnica Bucharest,
Romania
3
National Institute for Research and Development in Informatics - ICI Bucharest, Romania
4
Academy of Romanian Scientists, Bucharest, Romania
fl
Keywords:
pix2pix, CycleGAN, Augmentations, Image Classification, Vision Transformers, Leaf Diseases.
Abstract:
The detection and classification of diseases in Robusta coffee leaves are essential to ensure that plants are healthy
and the crop yield is kept high. However, this job requires extensive botanical knowledge and much wasted time.
Therefore, this task and others similar to it have been extensively researched subjects in image classification.
Regarding leaf disease classification, most approaches have used the more popular PlantVillage dataset while
completely disregarding other datasets, like the Robusta Coffee Leaf (RoCoLe) dataset. As the RoCoLe dataset
is imbalanced and does not have many samples, fine-tuning of pre-trained models and multiple augmentation
techniques need to be used. The current paper uses the RoCoLe dataset and approaches based on deep learning
for classifying coffee leaf diseases from images, incorporating the pix2pix model for segmentation and cycle-
generative adversarial network (CycleGAN) for augmentation. Our study demonstrates the effectiveness of
Transformer-based models, online augmentations, and CycleGAN augmentation in improving leaf disease
classification. While synthetic data has limitations, it complements real data, enhancing model performance.
These findings contribute to developing robust techniques for plant disease detection and classification.
1 INTRODUCTION
The Robusta coffee plant (also called Coffea
Canephora) is a species of coffee that is susceptible
to many diseases. Whether those diseases are caused
by insects or fungi, they can have a major impact on
crop yields and even cause complete crop destruction
if left untreated. The detection of diseases from im-
ages has been a significant focus in the research of
classification tasks for many years, starting with dis-
eases in humans (Ding et al., 2023; Sun et al., 2023;
Lungu-Stan et al., 2023) and then moving to animals
(Stauber et al., 2008; Nääs et al., 2020; Nam and Dong,
2023) and plants (Dawod and Dobre, 2022a; Dawod
and Dobre, 2022b; Dawod and Dobre, 2022c; Echim
et al., 2023).
Generally, detecting diseases requires expert
knowledge and a lot of time spent analyzing images to
*
Equal contributions.
†
Corresponding author.
determine the severity of the disease. For this reason,
there have been many research interests in developing
machine learning tools that anyone can use to detect
and classify diseases (Kamal et al., 2019; Dawod and
Dobre, 2021). These tools must be easy to use, ac-
curate, and not overly confident when making wrong
predictions.
In this paper, we use the Robusta Coffee Leaf
(RoCoLe) dataset (Parraga-Alava et al., 2019) for
which the main issues are the small number of avail-
able images and the class imbalance. These are both
widespread problems that arise in the field of ma-
chine learning and for which multiple approaches exist.
Therefore, several methods were tested and compared
to determine which solution fits the RoCoLe dataset
the best, with the main contributions of our work being
as follows:
•
We test offline and online augmentations of the
dataset conjointly with different combinations of
models and hyperparameters. The main focuses of
Gheorghiu, A., T
ˇ
aiatu, I., Cercel, D., Marin, I. and Pop, F.
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification.
DOI: 10.5220/0012466300003636
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Conference on Agents and Artificial Intelligence (ICAART 2024) - Volume 2, pages 549-560
ISBN: 978-989-758-680-4; ISSN: 2184-433X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
549

these comparisons are the performance evaluations
of the augmentations, followed by the assessment
of Transformer (Vaswani et al., 2017)-based mod-
els compared to a state-of-the-art convolutional
model.
•
We employ different visualization and explain-
ability techniques (Zhou et al., 2016; Hinton and
Roweis, 2002) to better understand why the models
perform in certain ways.
•
To the best of our knowledge, we are the first to
augment the RoCoLe dataset and use it for training
and testing Transformer-based models.
2 RELATED WORK
Most leaf disease classification approaches used large
datasets comprising tens of thousands of images (Mo-
hameth et al., 2020; Thakur et al., 2023). Only some
works used the RoCoLe dataset; if they do, it is not
the primary training dataset and is mainly used for
evaluation (Tassis et al., 2021; Rodriguez-Gallo et al.,
2023; Faisal et al., 2023). The approaches used vary,
with both deep learning and classical machine learning
models being employed (Brahimi et al., 2017; Tassis
et al., 2021).
Therefore, Brahimi et al. (Brahimi et al., 2017)
used a deep learning approach by testing two tra-
ditional convolutional neural network (CNN) (Kim,
2014) architectures: AlexNet (Krizhevsky et al., 2017)
and GoogLeNet (Szegedy et al., 2015). Traditional
machine learning models, specifically support vector
machines (SVMs) and Random Forest, are also tested.
The work compares pre-trained models to models with-
out pre-training, as well as deep models trained on raw
data to shallow models trained on manually extracted
features. Feature activation visualization is also used
as a more rudimentary technique. For this, regions of
the image are sequentially occluded, after which the
classification model is used to get the prediction. The
negative log-likelihood is then used to estimate the
importance of the occluded region. This method has
the disadvantage of inefficiency and is only feasible
when generating extremely low dimensional activation
maps comprised of only a few pixels. For the SVM
and Random Forest shallow models, the images are
first transformed from the RGB color space to another
color space, after which manual features such as the
color moment, wavelet transform, and gray level co-
occurrence matrix are extracted and used in training
the classifiers.
Tassis et al.(Tassis et al., 2021) used a multi-stage
pipeline of three different models to segment and clas-
sify the leaf diseases. Thus, a Mask R-CNN model
(He et al., 2017) is first used for instance segmenta-
tion. This model is trained to mask the background
and only highlight the leaves in images. After that,
either a U-Net (Ronneberger et al., 2015) or a PSPNet
(Zhao et al., 2017) is used for semantic segmentation.
This model highlights the relevant areas of the studied
image, specifically diseased regions. Those regions
are then cropped and fed into a ResNet classifier (He
et al., 2016), and the predictions are used to estimate
the severity of the disease. The models were trained
using random rotations and color variations as aug-
mentations. As a training dataset, the authors used
images scraped from the web. The RoCoLe dataset
was also used to evaluate the instance segmentation
model.
Mohameth et al. (Mohameth et al., 2020) em-
ployed the popular PlantVillage dataset (Mohanty
et al., 2016) to train their proposed models. For train-
ing, both transfer learning and deep feature extraction
methods are used. For transfer learning, only the clas-
sification head of a pre-trained model is trained from
scratch, with the rest of the layers having their weights
frozen. Using this method, the authors test multiple
CNN-based models: VGG16 (Simonyan and Zisser-
man, 2015), GoogLeNet, and ResNet. When deep
feature extraction is utilized, these models only ex-
tract the features before the classification head. Those
features are then used to train an SVM or a k-nearest
neighbor classifier.
3 DATASET
3.1 Relabeling
By performing exploratory data analysis on the Ro-
CoLe dataset, it becomes evident that the classes are
imbalanced. This is a significant problem for any
classifier architecture, as the model will overfit the
most frequent class and rarely predict the less frequent
classes.
It is easy to notice that the labels corresponding
to the two most severe cases of rust are also the least
frequent. The rust_level_3 and rust_level_4 labels
both represent high levels of rust, and even by com-
bining them, they still make up fewer samples than
any other label. Therefore, the first step towards al-
leviating class imbalance is to relabel the samples as
follows: healthy and red_spider_mite stay the same,
rust_level_1 becomes rust_level_low, rust_level_2 be-
comes rust_level_medium, and both rust_level_3 and
rust_level_4 become rust_level_high.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
550
3.2 Preprocessing
Before further addressing the class imbalance, the next
step was to analyze the images from the RoCoLe
dataset. We notice that the photos were taken with
a smartphone camera and, therefore, have a high res-
olution, 1152x2048, to be more precise. Using im-
ages at this resolution for deep learning models would
consume many computational resources without the
models benefiting much from the increased resolution.
Therefore, the images were rescaled to the less de-
manding and more common 256x256 resolution. The
images in the dataset also come with a mask, repre-
sented by the points that comprise the mask polygon.
This mask was plotted, rescaled, and saved with its
related image.
3.3 Split
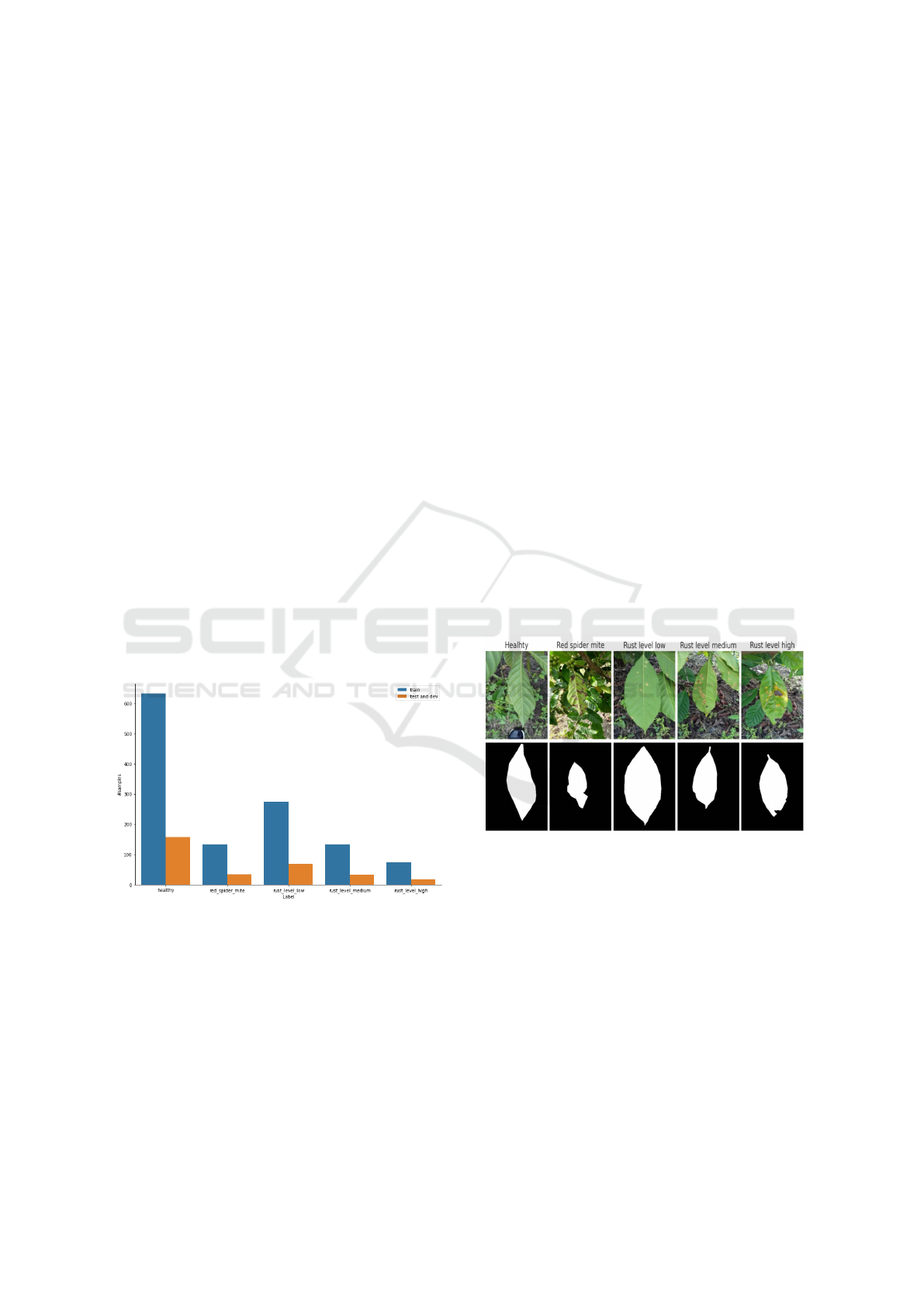
The initial dataset is randomly split into train, devel-
opment (dev), and test sets as shown in Figure 1. The
split is done as follows: 80% goes to the train set, with
the rest of 20% being split equally between the dev
and test sets. After the split, the train and dev sets
are augmented together and then split back with an
80%-10% ratio. We do not augment the test set since
the model metrics computed on the test set must only
reflect the performance on data from the real input
distribution.
Figure 1: Statistics of the number of samples in the relabeled
dataset.
4 METHOD
4.1 Segmentation
As can be seen from the examples in Figure 2, the
backgrounds of the images are very random and do
not provide any additional information. Therefore, it
only makes sense to use masked images to train classi-
fiers. For training a classifier, it is trivial to apply the
mask that comes with the image. Unfortunately, the
issue is for the examples that do not already come seg-
mented. The segmentation problem involves learning
a mapping from an input distribution to the distribu-
tion that represents the mask of the inputs. Because of
the pairwise nature of the segmentation problem, the
pix2pix model (Isola et al., 2017) is the perfect fit for
it. Furthermore, pix2pix is based on the U-Net archi-
tecture, which is state of the art in image segmentation
(Siddique et al., 2021). The pix2pix model was thus
trained on image-mask pairs from the RoCoLe dataset.
To improve the quality of the predicted segmenta-
tion masks, online augmentation was also used to crop
and flip the image randomly. After training, the mask
was inferred for each image in the dataset and then ap-
plied to the image to only contain the area of interest,
namely the leaf in the foreground. The reason segmen-
tation of the dataset is done using the trained pix2pix
model and not using the already provided masks is
because the inferred masks are slightly different from
the provided ones. Training a classifier on images seg-
mented with the provided masks but segmenting the
test images with pix2pix will affect the quality of the
predictions as the input distribution for the classifier
will be slightly different.
Figure 2: Examples of rescaled images from each class and
the associated masks.
4.2 Offline Augmentations
In order to alleviate the class imbalance, image gener-
ation was used to supplement the less frequent classes,
thus making the dataset perfectly balanced. The class
frequencies show that the healthy class is the most fre-
quent. This means that it can augment the other classes
by transferring the style of the images with diseased
leaves onto the images with healthy leaves. The cycle-
generative adversarial network (CycleGAN) model
(Zhu et al., 2017) has been proven to be effective for
style transfer tasks (Liu et al., 2020). Also, CycleGAN
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification
551

is a good fit for the augmentation task since it does
not require paired inputs and works just as well with
unpaired inputs, which is the case for the augmentation
task.
In addition to learning the mapping from the source
domain
X
to the target domain
Y
, CycleGAN also
learns the inverse mapping from the target domain to
the source domain (Zhu et al., 2017). In this sense,
two generators and two discriminators are used, one
of each type for each mapping:
G
and
D
Y
for the di-
rect mapping
G : X → Y
, as well as
F
and
D
X
for the
inverse mapping
F : Y → X
. Furthermore, CycleGAN
introduces another objective, called cycle consistency
loss, in addition to the classic GAN objectives (Good-
fellow et al., 2014). Intuitively, this novel objective
uses the L1 norm to enforce the consistency of the in-
verse mapping, canceling the direct mapping, as shown
in Eq. (1).
L
cyc
(G, F) = E
x∼p
data
(x)
[||F(G(x)) − x||
1
]+
+ E
y∼p
data
(y)
[||G(F(y)) − y||
1
] (1)
When passing an image from
X
through generator
F
or an image from
Y
through generator
G
, the out-
put is expected to be the same provided image since
the input already comes from the output distribution.
Therefore, a second regularization loss called the iden-
tity loss is used to constrain the model. This loss uses
the same L1 norm as the cycle consistency loss and
can be expressed through Eq. (2).
L
identity
(G, F) = E
x∼p
data
(x)
[||F(x) − x||
1
]+
+ E
y∼p
data
(y)
[||G(y) − y||
1
] (2)
By combining the identity and cycle consistency
losses with the standard GAN objectives, the final loss
becomes as described by Eq. (3).
L (G, F, D
X
, D
Y
) = L
GAN
(G, D
Y
, X, Y )+
+ L
GAN
(F, D
X
, Y, X ) + λ
1
L
cyc
(G, F)+
+ λ
2
L
identity
(G, F) (3)
Therefore, we train the CycleGAN model on our
dataset of segmented images for each combination
of healthy and diseased classes. This means that four
CycleGAN models were trained in total. After training,
the models were used on every segmented image of the
healthy class to generate its diseased counterpart. The
dataset was augmented using those generated diseased
images by supplementing every diseased class such
that the number of samples in each class is equal. For
this, the required number of images was randomly
picked from the generated images.
4.3 Online Augmentations
During training, online augmentation of the dataset
was also tested. The most basic augmentations that
have been tested are horizontal and vertical flips, as
well as random rotations. Therefore, an image is ro-
tated with a random angle between 0 and 180 degrees,
after which random horizontal and vertical flips are
applied with a probability of 25% each.
More advanced techniques include MixUp (Zhang
et al., 2018), CutMix (Yun et al., 2019), Cutout (De-
Vries and Taylor, 2017), and FMix (Harris et al., 2020).
These augmentations are applied during the batching
process so that the training dataset will differ with
each epoch. For the batched augmentations, there is a
50% probability that the augmentation will be applied;
otherwise, the batch is left unmodified. Furthermore,
all batched augmentations use a random parameter
λ
,
sampled from the beta distribution each time the aug-
mentation is applied. For FMix, the beta distribution’s
α
and
β
parameters are set to 1, while for the other
batched augmentations, both parameters are set to 0.8.
With CutMix and Cutout, the square that is cut out
is chosen to have the center at least one-quarter away
from the edge of the image, as most images have the
leaf centered in the middle of the image.
4.4 Classification Models
After segmentation and augmentation, the dataset
is ready for training classification models. Thus,
Transformer-based architectures (i.e., ViT (Dosovit-
skiy et al., 2020) and CvT (Wu et al., 2021)) were
tested using different hyperparameters, sizes, and aug-
mentation techniques. In order to compare these mod-
els to convolutional state-of-the-art models, ResNet
was tested in some scenarios.
5 EXPERIMENTAL SETUP
5.1 Performance Metrics
For model evaluation, macro-averaging was used to
combine accuracy, precision, recall, and F1-score bi-
nary classification metrics into multiclass metrics.
Apart from the initial ViT and ResNet tests, all
tests also feature the top-k accuracy metric. Top-k
accuracy means that when calculating the accuracy
score, a prediction is considered accurate if any of the
top-k outputs with the highest confidence is correct.
For testing, the top-2 accuracy was used in addition to
the classic accuracy metric.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
552

5.2 Hyperparameters
5.2.1 Pix2pix Setup
The pix2pix model was trained using the Adam opti-
mizer (Kingma and Ba, 2014) with a learning rate of
2 ∗ 10
−4
and the momentum parameters
β
1
and
β
2
set
to 0.5 and 0.999, respectively. These parameters are
the same as in the pix2pix paper (Isola et al., 2017).
Other parameters taken from the paper are the batch
size of 1, a value shown to work best for the pix2pix
model, and the L1 loss weight,
λ
, set to 100. For the
discriminator, we used the PatchGAN (Li and Wand,
2016; Isola et al., 2017) of output size 30x30. The
model was trained on the whole dataset for 25 epochs.
5.2.2 CycleGAN Setup
For training the CycleGAN models, the Adam opti-
mizer was used along with the same learning rate, mo-
mentum parameters, and batch size as for the pix2pix
model. The weights for the cycle consistency loss and
identity loss were also taken from the CycleGAN paper
(Zhu et al., 2017), specifically from the “Monet paint-
ings to photos” experiment, and were set to 10 and 5,
respectively. From an architectural point of view, the
generator with nine residual blocks from the paper was
used, along with the 70x70 PatchGAN discriminator,
also from the paper (Zhu et al., 2017). Each model
was trained on the whole segmented dataset for 100
epochs.
5.2.3 Classification Model Setup
All models were trained using a batch size of 32, with a
few exceptions where batch sizes of 16 were used due
to limited resources. The Adam optimizer was used,
along with a learning rate scheduler that multiplies the
learning rate by 0.25 every 15 epochs.
6 QUANTITATIVE RESULTS
6.1 Results for Offline Augmentations
Initially, we tested a ViT-small model with a patch size
of 16 and an input size of 224. The tests aim to com-
pare different models and augmentation techniques, so
the first tests involve finding the optimum hyperparam-
eters at which to train the following models. Therefore,
the model is first tested with and without dropout and
learning rates of 0.001 and 0.0002 for 50 epochs to
observe how hyperparameters affect the scores and the
evolution of train and dev losses.
The Augmentations Improve Performance in
Almost All Cases. As can be observed from Ta-
ble 1, the scores in most cases are higher when the
model is trained on the augmented dataset than the
non-augmented dataset. In the case of the fifth and
sixth rows, the model has higher accuracy when trained
on the non-augmented dataset, but all the other scores
are low. The explanation is that the model overfits
the most frequent class because of the imbalanced na-
ture of the non-augmented dataset. However, for the
third and fourth rows, all the scores are higher for the
non-augmented model than all the others. As the differ-
ences are not that big, this anomaly can be explained
by the small dimension of the test set.
The ViT-Small Model Performs Better When
Trained Using a Lower Learning Rate. In Ta-
ble 1, in the case of no dropout and a learning rate of
0.001, the model presents worse performance on both
augmented and non-augmented datasets compared to
the cases when a learning rate of 0.0002 is used. This
means the lower learning rate is the correct choice,
as the model presents better learning abilities regard-
less of the dataset. Another observation in our tests
is that introducing dropout reduces overfitting mostly
on the augmented dataset, with the losses on the non-
augmented dataset remaining almost unchanged. Us-
ing a learning rate of 0.0002 and enabling dropout, the
overfitting on the augmented dataset almost disappears,
while it is greatly improved on the non-augmented
dataset.
The ResNet-50V2 and ResNet-101V2 models were
tested to compare how augmentation affects traditional
convolutional neural networks. Like the ViT, these
models are pre-trained on the ImageNet datasets (Deng
et al., 2009). A learning rate of 0.001 was used to test
both augmented and non-augmented datasets, while
all other settings were kept as before.
Bigger Models Encourage Overfitting. In Table
2, it can be noticed how ResNet performs worse in
the case of the bigger model, as the higher number
of parameters worsens overfitting. Furthermore, per-
formance is better on the augmented dataset for both
versions of ResNet, as the larger dataset discourages
overfitting.
ViT Has Better Performance Than ResNet.
When comparing the scores of the ViT-small with en-
abled dropout, augmentation, and a learning rate of
0.0002 to the ResNet-50V2 model with augmentation,
it can be observed that the Transformer-based model
performs better compared to the convolutional-based
model.
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification
553
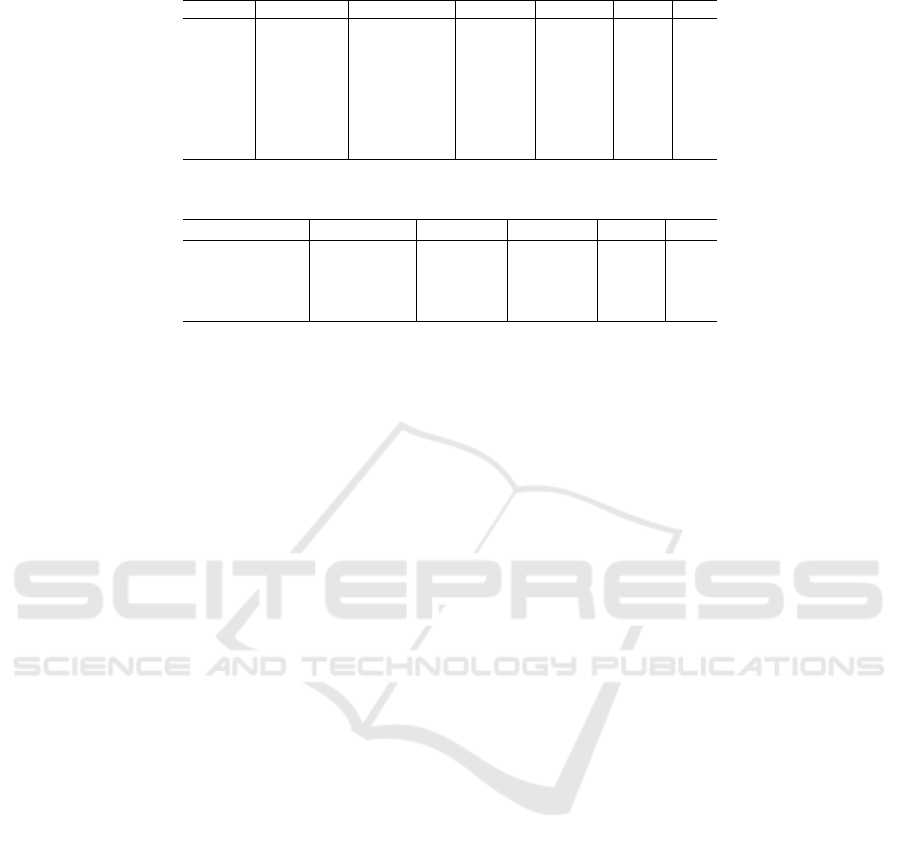
Table 1: ViT-small scores for different combinations of hyperparameters.
Dropout Augmented Learning rate Accuracy Precision Recall F1
No Yes 0.001 64.1 54.4 56.9 55.0
No No 0.001 51.9 32.3 32 32.1
No Yes 0.0002 73.1 58.5 54.9 55.7
No No 0.0002 75.0 61.6 58.3 59.4
Yes Yes 0.001 57.1 42.3 43.5 42.0
Yes No 0.001 59.6 36.2 36.6 36.2
Yes Yes 0.0002 73.7 59.1 56.5 57.2
Yes No 0.0002 64.7 47.3 47.1 46.3
Table 2: ResNet scores for different variants and augmentations.
Model Augmented Accuracy Precision Recall F1
ResNet-50V2 Yes 69.2 48.7 47.9 47.7
ResNet-50V2 No 62.8 43.8 44.5 43.9
ResNet-101V2 Yes 61.5 43.6 42.3 42.6
ResNet-101V2 No 48.7 31.9 31.3 31.5
By analyzing the loss evolution of the models, it
could be noticed how the models started overfitting
mostly around 20 to 30 epochs. For this reason, all
tests described from now on will be done using 25
epochs. Regarding the other hyperparameters, enabled
dropout, a learning rate of 0.0002, and the augmented
dataset will be used. Until another model is tested, the
ViT-small model will be used.
In order to further test how well the synthetically
generated data captures the distribution of the real data,
we compared the Train on Real - Test on Real (TRTR),
Train on Real - Test on Synthetic (TRTS), Train on
Synthetic - Test on Real (TSTR), and Train on Syn-
thetic - Test on Synthetic (TSTS) scenarios (Fekri et al.,
2019). As the names suggest, these methods train and
test the model on combinations of either the original
dataset with real images or a dataset consisting of only
synthetic images with no real images in the augmented
classes. The synthetic data was thus extracted and split
into train, dev, and test using the same ratios as before.
Synthetic Data Poorly Captures the Distribution of
the Real Data. Table 3 shows how, to a certain
extent, the synthetic data captures the distribution of
the real data rather poorly. The discrepancy between
TRTR and TSTS especially shows this. The model
performs much better on synthetic data and learns by
overfitting the specific properties of each generated
class. As there is still one real class among the syn-
thetic images, the healthy class, as well as all other
synthetic classes being generated with different trained
instances of CycleGAN, the model learns to distin-
guish between the classes by learning the different
footprints left in the generated images.
Models Trained Only on Synthetic Data General-
ize Poorly to Real Data. The slight difference
between TRTR and TRTS shows how a model trained
on real data generalizes just as well to synthetic data,
as the model learns the actual distribution without
overfitting. The poor performance of TSTR compared
to TSTS further emphasizes how the model overfits
specific properties in the synthetic data and does not
generalize well to real data.
6.2 Results for Online Augmentations
Online Augmentations Improve Performance in Al-
most All Cases. It can be observed in Table 4 how,
with a few exceptions, online augmentations further
improve the performance when compared to the equiv-
alent configuration in Table 1, specifically the second
to last row. One of the exceptions is Cutout, which has
no other augmentations.
Cutout with No Augmentations Offers the Worst
Performance. Because the segmented images
have most pixels set to 0, cutting out additional parts of
the image erases essential information. Adding other
augmentations seems to counteract this, as the Cutout
model with random rotations and flips is among the
better-performing models.
FMix Features the Best All-Around Performance.
CutMix and FMix have similar performances, with
FMix taking the lead, especially when adding rotations
and flips. Rotations and flips seem to improve all
metrics except accuracy, while batched augmentations
have more of an effect on the standard accuracy metric.
Finally, we tested the CvT model (Wu et al., 2021)
on the augmented dataset, non-augmented dataset, and
the various combinations of online augmentations also
tested up until now. By analyzing the loss evolution,
we observed that CvT is more prone to overfitting,
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
554

Table 3: ViT-small scores for TRTR, TRTS, TSTR, and TSTS.
Method Accuracy Top-2 Accuracy Precision Recall F1
TRTR 62.8 86.5 45.9 45.7 44.1
TRTS 62.3 84.0 60.1 55.7 55.4
TSTR 55.1 76.3 34.2 30.6 29.0
TSTS 96.9 99.6 97.1 96.2 96.6
Table 4: ViT-small scores for different online augmentations.
Augmentation Accuracy Top-2 Accuracy Precision Recall F1
Rotation+Flips 73.7 91.7 68.3 65.1 62.7
MixUp 73.7 88.5 60.1 57.4 57.0
CutMix 76.3 88.5 62.9 59.9 59.7
Cutout 69.2 89.1 53.7 51.6 51.3
FMix 74.4 89.7 64.2 57.8 59.1
Rotation+Flips+MixUp 69.9 93.6 52.2 50.1 49.4
Rotation+Flips+CutMix 73.7 88.5 66.4 57.5 59.3
Rotation+Flips+Cutout 76.3 92.3 68.1 65.5 63.3
Rotation+Flips+FMix 77.6 91.0 71.6 67.1 67.7
being a bigger model than ViT. The augmented dataset
helped with overfitting, as did the augmentations with
random rotations and flips. The choice of batched
augmentation did not impact overfitting much, but
CutMix and FMix had slightly better performance in
that aspect.
CvT Has Similar Performance to ViT. As seen
in Table 5, the performances of CvT are similar to ViT,
with rotation and flip augmentations making the model
carry out better. In the case of CvT, the versions where
Cutout is used have similar performance and perform
better than the other batched augmentations. While
ViT favored CutMix over MixUp, the performances
are flipped with CvT, such that CutMix outperforms
MixUp. FMix seems to be the most robust, with its
performance having smaller fluctuations than the rest
of the batched augmentations, regardless of the model
or the other online augmentations used. Using the non-
augmented dataset has the same performance impact
on CvT as it had on ViT.
7 QUALITATIVE RESULTS
7.1 Pix2pix Examples
Predicted Masks Are Smoother Than Hand-Drawn
Masks. As can be seen in the first and second ex-
amples of Figure 3, the predicted mask is smoother
around the edges compared to the ground truth mask,
albeit less close to the outline of the leaf. The fourth
example shows that the model could generate a mask
that surpasses the ground truth in the correct condi-
tions.
Predicted Masks Sometimes Include Background
Leaves. The third image shows how certain back-
ground leaves hinder the model’s ability to find an
outline that follows the exact shape of the leaf, out-
putting a smoother segmentation instead. Therefore,
in areas where the outline is less visible, the model
also includes the background leaves when performing
segmentation.
The Model Has Trouble Segmenting Leaves Against
Specific Backgrounds. Because the pix2pix
model was trained on images that mostly contain back-
grounds of other leaves combined with grass and dirt, it
has trouble segmenting images of leaves taken against
backgrounds such as asphalt and concrete. Surpris-
ingly, the model is good at segmenting images of
leaves taken against plain or solid color backgrounds.
Figure 3: Examples of pix2pix predicted masks compared to
the ground truth masks.
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification
555
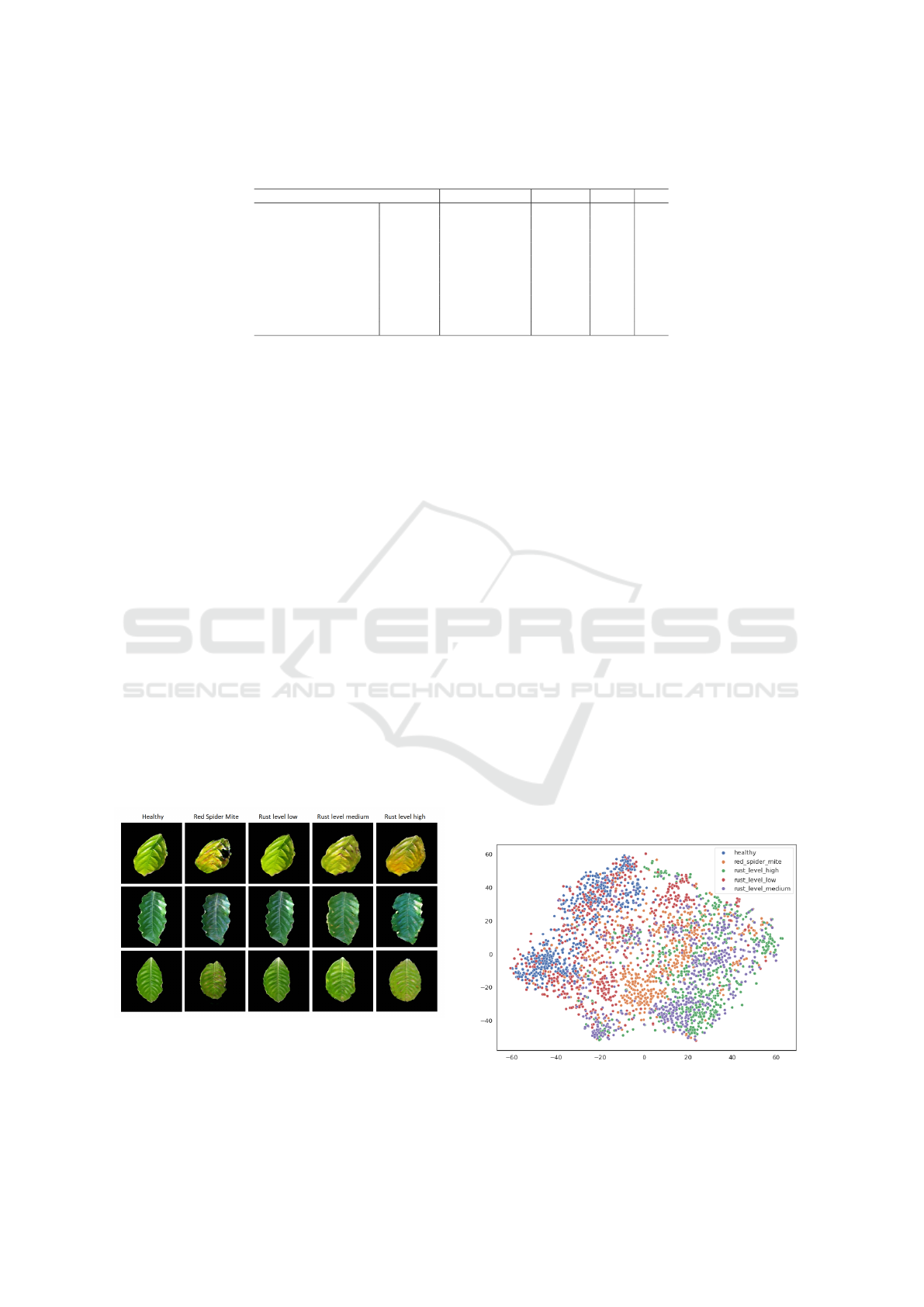
Table 5: CvT scores for the offline augmented dataset (first row), the non-augmented dataset (second row), and the augmented
dataset with combinations of online augmentations (from third to last row).
Augmentation Accuracy Top-2 Accuracy Precision Recall F1
Augmented 76.3 91.7 62.1 58.9 60.2
Non-augmented 59.0 80.8 43.4 44.2 43.4
MixUp 75.0 89.7 55.6 53.6 54.1
CutMix 71.2 84.6 55.9 54.3 54.8
Cutout 75.0 92.9 61.1 60.6 60.6
FMix 75.0 85.9 60.5 60.8 59.8
Rotation+Flips+MixUp 76.3 89.7 63.2 62.6 62.8
Rotation+Flips+CutMix 75.0 91.0 58.9 60.4 58.9
Rotation+Flips+Cutout 78.2 92.9 66.1 64.3 63.4
Rotation+Flips+FMix 76.9 91.0 61.5 63.0 61.0
7.2 CycleGAN Examples
Some Synthetic Images Contain Noisy Holes. As
shown in the first example of Figure 4, the generated
red_spider_mite image features a big hole in the leaf.
While the hole is an excellent example of leaf dam-
age, there is also a lot of noise surrounding it, which
decays the quality of the sample. Better-looking hole
examples are found in the high rust-level image gener-
ated from the second healthy leaf, the synthetic image
featuring small holes around the edges of the leaf.
The Low Rust-Level Synthetic Images Feature Al-
most No Discernible Modifications. For the low
rust level, there is little discernible difference between
the healthy and diseased images. This might be be-
cause this level of rust has a minimal impact on the
appearance of the leaf, making it look almost identical
to a healthy leaf in most cases. In comparison, the
medium and high levels of rust have the most visible
effects. As expected, the first and third examples show
more severe rust stains for the high level than for the
medium level, while the second example shows more
holes.
Figure 4: Examples of diseased leaf images generated from
healthy leaf images.
7.3 T-SNE Visualizations
In order to show how the generated diseased leaf im-
ages are close to the actual images in the input distri-
bution space, latent features are first extracted from
the images by passing them through a ViT. This vision
Transformer is pre-trained on the ImageNet-21k and
ImageNet-1k datasets (Deng et al., 2009), so it should
extract distinctive features out of the leaf images with-
out prior knowledge about the input distribution. In
order to visualize those latent features, the t-distributed
stochastic neighbor embedding (t-SNE) dimensional-
ity reduction technique (Hinton and Roweis, 2002) is
used.
In Figure 5, it is easy to notice how the different
classes are each spread into their clusters, with the
clusters also overlapping each other a bit. This is ex-
pected from a model pre-trained on a generic image
dataset without prior knowledge of the plant leaf clas-
sification task. It can still be observed how there are
no evident outliers for any class, with the synthetic
images seemingly blending into the real images.
In Figure 6, for the most part, the synthetic image
representations surround the real images representa-
tions, except for the red_spider_mite class, where the
real and synthetic images overlap almost perfectly.
Figure 5: 2D t-SNE representations of each class’s images,
both synthetic and real.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
556
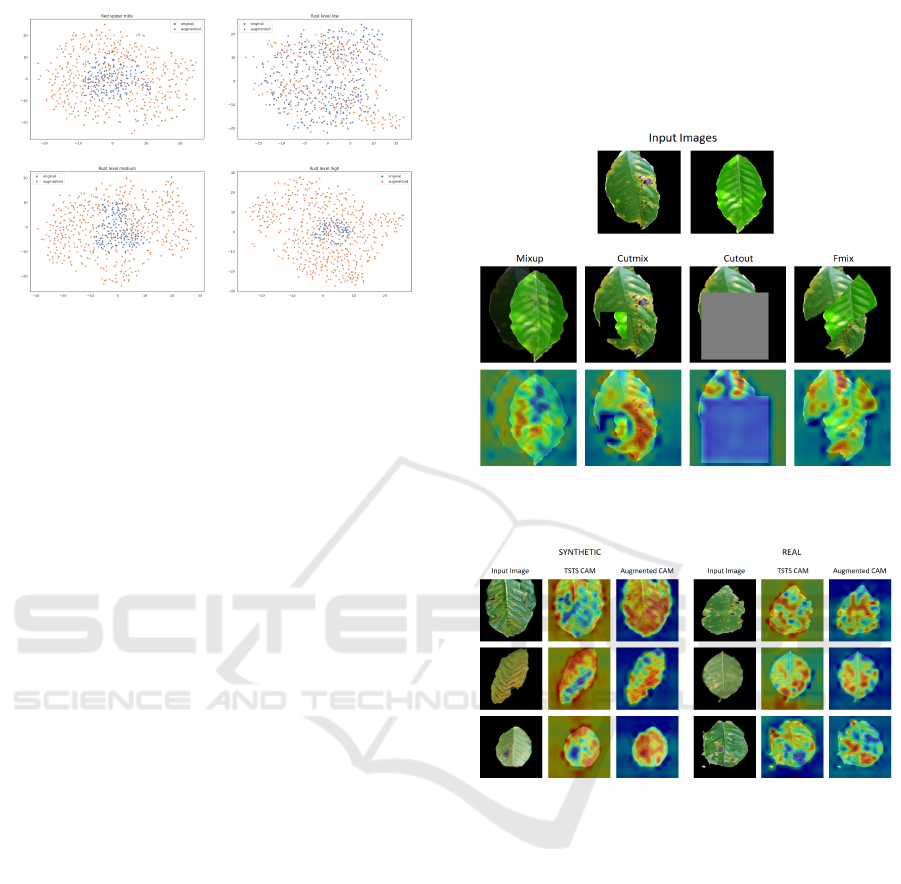
Figure 6: 2D t-SNE visualizations of each class, comparing
real and synthetic images.
7.4 CAM Visualizations
Models Trained with Certain Online Augmenta-
tions Focus Better on the Critical Parts of Leaves.
For the MixUp strategy, the model focuses more on the
area where the two images overlap, and there are no
rust spots (see Figure 7). Interestingly, the big rust re-
gion is mainly ignored, with the model focusing more
on the small rust spots on the left side of the rusty
leaf. For CutMix, the model learned to ignore the ar-
eas that encompass the transitions from one image to
another while also focusing on the essential elements
in both images. With Cutout, even when a big area
is cut out in the image, the model learns to ignore it
and focuses instead on the unobstructed regions. FMix
CAM is similar to CutMix CAM, the model ignoring
the transitions, especially in regions where one image
transitions to the background of the other image.
Models Trained On Synthetic Data Focus More
on the Background and Edges of Leaves. The
CAMs in Figure 8 show how the model trained on
the synthetic dataset focuses the most on the edges
of the leaves, where more artifacts were left by the
CycleGAN model when generating the images. Also,
the TSTS model seems to focus surprisingly much
on the background, which can be explained because
the CycleGAN model leaves noise in the background
of the generated image. The model trained on the
augmented dataset focuses the most on areas where
the leaf is affected by rust.
Models Trained Using Online Augmentations Focus
on the Image as a Whole. By comparing the vi-
sualizations in Figure 7 to the visualizations in Figure
8, it can be observed that the models trained using on-
line augmentations direct their attention more toward
the background of the image compared to the model
trained without online augmentations. This is because
the models trained using online augmentations learn to
focus on the image as a whole, not only on the center
part.
Figure 7: CAM visualizations for batched augmentation
techniques.
Figure 8: CAM visualizations for synthetic (left) and real
(right) for the TSTS model (TSTS CAM) and a model trained
normally on the augmented dataset (Augmented CAM). All
images are part of the rust_level_high class.
8 CONCLUSIONS AND FUTURE
WORK
This paper presented a deep learning pipeline for
the task of leaf disease classification on the RoCoLe
dataset. Consequently, image leaf segmentation was
performed using a pix2pix model, and CycleGAN was
used to augment the diseased classes in the dataset.
Also, Transformer-based image classification models
were trained on the augmented dataset using various
online augmentations.
This approach of using augmentations combined
with Transformer models increased the classification
performance compared to approaches using convolu-
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification
557

tional models or without augmentations. Even though
the results of the TRTR, TRTS, TSTR, and TSTS sce-
narios showed that the synthetic data only vaguely
captures the distribution of the real data, the increased
performance of the models trained on the augmented
dataset proved that the CycleGAN augmentations were
helpful.
The presented approach can still be improved, with
better alternatives being viable in most parts of the
architecture. Therefore, the main improvement that
can be implemented is the use of the StarGAN (Choi
et al., 2018) multi-domain image translation model for
augmenting the RoCoLe dataset. This approach would
benefit from needing only one model to be trained for
augmenting all diseased classes. Other GAN varia-
tions can also be tested, such as the Wasserstein GAN
(Arjovsky et al., 2017), which has proven more stable
than other GANs.
The leaf segmentation could also be improved
by using semantic segmentation (Tassis et al., 2021).
However, this would increase computational costs. A
cheaper alternative would be to implement semantic
segmentation using either the previously presented
CAM method or the better GradCAM variant (Sel-
varaju et al., 2017).
Finally, a Swin Transformer (Liu et al., 2021) can
be used as a classifier instead of the ViT or CvT. An-
other alternative would be to use vision-language mod-
els (Yu et al., 2022), which are the current state of
the art in image classification. Models like the con-
trastive captioner (Yu et al., 2022) use encoder-decoder
architectures to learn captions for images, so the en-
coder could be fine-tuned to classify images from the
RoCoLe dataset.
ACKNOWLEDGEMENTS
This work was supported by GNAC ARUT 2023.
REFERENCES
Arjovsky, M., Chintala, S., and Bottou, L. (2017). Wasser-
stein generative adversarial networks. In Interna-
tional conference on machine learning, pages 214–223.
PMLR.
Brahimi, M., Boukhalfa, K., and Moussaoui, A. (2017).
Deep learning for tomato diseases: classification and
symptoms visualization. Applied Artificial Intelligence,
31(4):299–315.
Choi, Y., Choi, M., Kim, M., Ha, J.-W., Kim, S., and Choo,
J. (2018). Stargan: Unified generative adversarial net-
works for multi-domain image-to-image translation.
In Proceedings of the IEEE conference on computer
vision and pattern recognition, pages 8789–8797.
Dawod, R. G. and Dobre, C. (2021). Classification of
sunflower foliar diseases using convolutional neural
network. In 2021 23rd International Conference on
Control Systems and Computer Science (CSCS), pages
476–481. IEEE.
Dawod, R. G. and Dobre, C. (2022a). Automatic segmen-
tation and classification system for foliar diseases in
sunflower. Sustainability, 14(18):11312.
Dawod, R. G. and Dobre, C. (2022b). Resnet interpretation
methods applied to the classification of foliar diseases
in sunflower. Journal of Agriculture and Food Re-
search, 9:100323.
Dawod, R. G. and Dobre, C. (2022c). Upper and lower leaf
side detection with machine learning methods. Sensors,
22(7):2696.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei,
L. (2009). Imagenet: A large-scale hierarchical image
database. In 2009 IEEE conference on computer vision
and pattern recognition, pages 248–255. Ieee.
DeVries, T. and Taylor, G. W. (2017). Improved regular-
ization of convolutional neural networks with cutout.
arXiv preprint arXiv:1708.04552.
Ding, S., Wang, J., Li, J., and Shi, J. (2023). Multi-scale
prototypical transformer for whole slide image classifi-
cation. In International conference on medical image
computing and computer-assisted intervention, pages
602–611. Springer.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. In International Conference on Learning
Representations.
Echim, S.-V., T
˘
aiatu, I.-M., Cercel, D.-C., and Pop, F. (2023).
Explainability-driven leaf disease classification using
adversarial training and knowledge distillation.
Faisal, M., Leu, J.-S., and Darmawan, J. T. (2023). Model
selection of hybrid feature fusion for coffee leaf disease
classification. IEEE Access.
Fekri, M. N., Ghosh, A. M., and Grolinger, K. (2019). Gen-
erating energy data for machine learning with recurrent
generative adversarial networks. Energies, 13(1):130.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-
Farley, D., Ozair, S., Courville, A., and Bengio, Y.
(2014). Generative adversarial nets. Advances in neural
information processing systems, 27.
Harris, E., Marcu, A., Painter, M., Niranjan, M., Prügel-
Bennett, A., and Hare, J. (2020). Fmix: Enhanc-
ing mixed sample data augmentation. arXiv preprint
arXiv:2002.12047.
He, K., Gkioxari, G., Dollár, P., and Girshick, R. (2017).
Mask r-cnn. arxiv e-prints, page. arXiv preprint
arXiv:1703.06870.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
558

Hinton, G. E. and Roweis, S. (2002). Stochastic neighbor
embedding. In Becker, S., Thrun, S., and Obermayer,
K., editors, Advances in Neural Information Process-
ing Systems, volume 15. MIT Press.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017). Image-
to-image translation with conditional adversarial net-
works. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 1125–
1134.
Kamal, K., Yin, Z., Wu, M., and Wu, Z. (2019). Depthwise
separable convolution architectures for plant disease
classification. Computers and electronics in agricul-
ture, 165:104948.
Kim, Y. (2014). Convolutional neural networks for sentence
classification. In Proceedings of the 2014 Conference
on Empirical Methods in Natural Language Processing
(EMNLP). Association for Computational Linguistics.
Kingma, D. P. and Ba, J. (2014). Adam: A
method for stochastic optimization. arXiv preprint
arXiv:1412.6980.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2017).
Imagenet classification with deep convolutional neural
networks. Communications of the ACM, 60(6):84–90.
Li, C. and Wand, M. (2016). Combining markov random
fields and convolutional neural networks for image
synthesis. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2479–
2486.
Liu, C., Chang, X., and Shen, Y.-D. (2020). Unity style
transfer for person re-identification. In Proceedings
of the IEEE/CVF conference on computer vision and
pattern recognition, pages 6887–6896.
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin,
S., and Guo, B. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF international conference
on computer vision, pages 10012–10022.
Lungu-Stan, V.-C., Cercel, D.-C., and Pop, F. (2023).
Skindistilvit: Lightweight vision transformer for skin
lesion classification. In International Conference on
Artificial Neural Networks, pages 268–280. Springer.
Mohameth, F., Bingcai, C., and Sada, K. A. (2020). Plant
disease detection with deep learning and feature ex-
traction using plant village. Journal of Computer and
Communications, 8(6):10–22.
Mohanty, S. P., Hughes, D. P., and Salathé, M. (2016). Using
deep learning for image-based plant disease detection.
Frontiers in Plant Science, 7.
Nääs, I. A., Garcia, R. G., and Caldara, F. R. (2020). In-
frared thermal image for assessing animal health and
welfare. Journal of Animal Behaviour and Biometeo-
rology, 2(3):66–72.
Nam, M. G. and Dong, S.-Y. (2023). Classification of com-
panion animals’ ocular diseases: Domain adversarial
learning for imbalanced data. IEEE Access.
Parraga-Alava, J., Cusme, K., Loor, A., and Santander, E.
(2019). Rocole: A robusta coffee leaf images dataset
for evaluation of machine learning based methods in
plant diseases recognition. Data in brief, 25:104414.
Rodriguez-Gallo, Y., Escobar-Benitez, B., and Rodriguez-
Lainez, J. (2023). Robust coffee rust detection us-
ing uav-based aerial rgb imagery. AgriEngineering,
5(3):1415–1431.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18, pages 234–241.
Springer.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2017). Grad-cam: Visual
explanations from deep networks via gradient-based
localization. In 2017 IEEE International Conference
on Computer Vision (ICCV), pages 618–626.
Siddique, N., Paheding, S., Elkin, C. P., and Devabhaktuni,
V. (2021). U-net and its variants for medical image
segmentation: A review of theory and applications.
Ieee Access, 9:82031–82057.
Simonyan, K. and Zisserman, A. (2015). Very deep con-
volutional networks for large-scale image recognition.
In 3rd International Conference on Learning Repre-
sentations (ICLR 2015). Computational and Biological
Learning Society.
Stauber, J., Lemaire, R., Franck, J., Bonnel, D., Croix,
D., Day, R., Wisztorski, M., Fournier, I., and Salzet,
M. (2008). Maldi imaging of formalin-fixed paraffin-
embedded tissues: application to model animals of
parkinson disease for biomarker hunting. Journal of
Proteome Research, 7(3):969–978.
Sun, M., Huang, W., and Zheng, Y. (2023). Instance-aware
diffusion model for gland segmentation in colon histol-
ogy images. In International Conference on Medical
Image Computing and Computer-Assisted Intervention,
pages 662–672. Springer.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions.
In Proceedings of the IEEE conference on computer
vision and pattern recognition, pages 1–9.
Tassis, L. M., de Souza, J. E. T., and Krohling, R. A. (2021).
A deep learning approach combining instance and se-
mantic segmentation to identify diseases and pests of
coffee leaves from in-field images. Computers and
Electronics in Agriculture, 186:106191.
Thakur, P. S., Sheorey, T., and Ojha, A. (2023). Vgg-icnn: A
lightweight cnn model for crop disease identification.
Multimedia Tools and Applications, 82(1):497–520.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I. (2017).
Attention is all you need. Advances in neural informa-
tion processing systems, 30.
Wu, H., Xiao, B., Codella, N., Liu, M., Dai, X., Yuan, L.,
and Zhang, L. (2021). Cvt: Introducing convolutions to
vision transformers. In Proceedings of the IEEE/CVF
International Conference on Computer Vision, pages
22–31.
Yu, J., Wang, Z., Vasudevan, V., Yeung, L., Seyedhosseini,
M., and Wu, Y. (2022). Coca: Contrastive caption-
Evaluating Data Augmentation Techniques for Coffee Leaf Disease Classification
559

ers are image-text foundation models. arXiv preprint
arXiv:2205.01917.
Yun, S., Han, D., Oh, S. J., Chun, S., Choe, J., and Yoo, Y.
(2019). Cutmix: Regularization strategy to train strong
classifiers with localizable features. In Proceedings of
the IEEE/CVF international conference on computer
vision, pages 6023–6032.
Zhang, H., Cisse, M., Dauphin, Y. N., and Lopez-Paz, D.
(2018). mixup: Beyond empirical risk minimization.
In International Conference on Learning Representa-
tions.
Zhao, H., Shi, J., Qi, X., Wang, X., and Jia, J. (2017). Pyra-
mid scene parsing network. In Proceedings of the IEEE
conference on computer vision and pattern recognition,
pages 2881–2890.
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., and Torralba,
A. (2016). Learning deep features for discriminative
localization. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2921–
2929.
Zhu, J.-Y., Park, T., Isola, P., and Efros, A. A. (2017).
Unpaired image-to-image translation using cycle-
consistent adversarial networks. In Proceedings of
the IEEE international conference on computer vision,
pages 2223–2232.
ICAART 2024 - 16th International Conference on Agents and Artificial Intelligence
560
