
Predicting the MGMT Promoter Methylation Status in T2-FLAIR
Magnetic Resonance Imaging Scans Using Machine Learning
Martyna Kurbiel
1
, Agata M. Wijata
2a
and Jakub Nalepa
1b
1
Department of Algorithmics and Software, Silesian University of Technology, Akademicka 16, 44-100 Gliwice, Poland
2
Faculty of Biomedical Engineering, Silesian University of Technology, Roosevelta 40, 41-800 Zabrze, Poland
Keywords: Glioblastoma, MGMT Promoter Methylation, MRI, Machine Learning, Radiomics.
Abstract: Glioblastoma is the most common form of brain cancer in adults, and is characterized by one of the worst
prognosis, with median survival being less than one year. Magnetic resonance imaging (MRI) plays a key
role in detecting and objectively tracking the disease by extracting quantifiable parameters of the tumor, such
as its volume or bidimensional measurements. However, it has been shown that the presence a specific genetic
sequence in a lesion, being the DNA repair enzyme O
6
-methylguanine-DNA methyltransferase (MGMT)
promoter methylation, may be effectively used to predict the patient’s responsiveness to chemotherapy. The
invasive process of analyzing a tissue sample to verify the MGMT promoter methylation status is time-
consuming, and may require performing multiple surgical interventions in longitudinal studies. Thus, building
non-invasive techniques of predicting the genetic subtype of glioblastoma is of utmost practical importance
to not only accelerate the overall process of determining the MGMT promoter methylation status in
glioblastoma patients, but also to minimize the number of necessary surgeries. In this paper, we tackle this
problem and propose an end-to-end machine learning classification pipeline benefitting from radiomic
features extracted from brain MRI scans, and validate it over a well-established RSNA-MICCAI Brain Tumor
Radiogenomic Classification benchmark dataset.
1 INTRODUCTION
Glioblastoma (GBM) stands out as the prevalent
malignant brain tumor among adults, and despite
extensive research spanning decades, it is still one of
the deadliest cancers, primarily attributed to its
unfavorable prognosis. Consequently, the precise
assessment of therapy response in GBM poses
significant challenges and holds immense clinical
importance (Qi et al., 2023). Although, multi-modal
magnetic resonance imaging (MRI) scans can bring
important structural information concerning such
brain lesions, their manual analysis of acquired
images is time- and cost-inefficient, it lacks
reproducibility and suffers from significant inter- and
intra-rater disagreement (Xuan et al., 2022; Hu et al.
2022). To automate the tedious process of analyzing
MRI scans, various algorithms have been emerging at
a steady pace recently. These practical challenges can
be effectively tackled by automatic brain lesions
a
https://orcid.org/0000-0001-6180-9979
b
https://orcid.org/0000-0002-4026-1569
detection and segmentation techniques. They may be
split into atlas-, image analysis-, machine learning-
based, and hybrid techniques. In the atlas-based
approaches, we exploit manually-delineated atlases to
segment unseen scans, relying on image registration
and facing challenges with diverse tumor
characteristics that are difficult to capture within an
atlas (Xing et al., 2022). Similarly, image analysis-
based algorithms, including thresholding and region-
growing techniques, are often easy to implement and
offer fast operation, but they struggle with
heterogeneous tumors and noisy images (Puttagunta
et al., 2021; Vadmal et al., 2022). Conventional
machine learning approaches offer advantages
directly related to their nature (of such methods being
data-driven), but they require heavy feature
engineering, hence elaborating manually-designed
features that would capture intrinsic brain tumor
characteristics. Finally, deep learning models
encompass a range of network architectures,
872
Kurbiel, M., Wijata, A. and Nalepa, J.
Predicting the MGMT Promoter Methylation Status in T2-FLAIR Magnetic Resonance Imaging Scans Using Machine Learning.
DOI: 10.5220/0012467400003654
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2024), pages 872-879
ISBN: 978-989-758-684-2; ISSN: 2184-4313
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

including ensembles (Shi et al., 2021), U-Net
(Bukhari et al., 2022), encoder-decoder (Yan et al.,
2022), and more (Peiris et al., 2022) that were
thoroughly validated over the Brain Tumor
Segmentation (BraTS) Challenge throughout the
recent years, and established the current state of the
art in the field (Baid et al. 2021). Although accurate
brain lesion segmentation is of paramount importance
in order to objectively assess the tumor progression
through extracting its quantifiable characteristics,
such as its volumetric or bidimensional
measurements (Hu et al. 2022), the structural
information concerning the brain may not be enough
to fully understand the patient status and benefit from
it in planning the treatment (Beyer et al., 2020).
There have been various research efforts
indicating that the identification of a particular
genetic sequence in a lesion – specifically, the DNA
repair enzyme O
6
-methylguanine-DNA
methyltransferase (MGMT) promoter methylation –
can serve as an effective predictor of a patient's
responsiveness to chemotherapy (Weller et al., 2010).
Additionally, the MGMT status has become a
stratification parameter of patients with glioblastoma
within clinical trials as well. The intrusive nature of
examining a tissue sample to confirm the MGMT
promoter methylation status is time-intensive and
may necessitate multiple surgical interventions in
longitudinal studies. Consequently, the development
of non-invasive techniques for predicting the genetic
subtype of glioblastoma becomes paramount. This
not only expedites the overall process of determining
the MGMT promoter methylation status in
glioblastoma patients but also reduces the need for
multiple surgeries such patients would have to
undergo. Therefore, developing non-invasive
methods for quantifying the MGMT promoter
methylation status has been already researched in the
literature, e.g., using texture features extracted from
T2-weighted MR images and Support Vector
Machines (Korfiatis et al., 2016). It was also
demonstrated that the use of radiomic features
together with machine learning algorithms can enable
non-invasive prediction of the MGMT promoter
methylation status (Hajianfar et al., 2019) – here, a
pipeline of the radiomic feature extraction, feature
selection, and classification were employed for each
patient. Also, there are deep learning-powered
approaches, e.g., exploiting various network
architectures (Korfiatis et al., 2017). In their recent
work, Saeed et al. 2023 performed an extensive
evaluation study of an array of deep learning models
for estimating MGMT methylation status from MRI
data, and showed that the reliability of the deep
learning approaches should be verified using external
cohorts before exploiting them in clinical
applications. Here, capturing large, heterogeneous
and representative datasets that would allow for
training large-capacity learners is a practical
challenging which may ultimately hamper
generalization capabilities of deep learning models.
In this work, we tackle the problem of quantifying
the MGMT methylation status based on MRI data,
and introduced a classic machine learning algorithm
for this task. We hypothesize that the features
extracted from the whole brain region scanned using
the T2 Fluid Attenuation Inversion Recovery (T2-
FLAIR) MR sequence, as such sequences have been
designed to suppress the signal from cerebrospinal
fluid, providing improved visualization of lesions
near cerebrospinal fluid spaces, may be utilized in
differentiating the MGMT methylation status (Alpar,
2023). Here, since the lesion segmentation step is
skipped in our processing chain, we may not only
accelerate the computation, as a single MR sequence
is processed, but we can also rely on the widely-
established brain extraction (skull stripping)
algorithms (Isensee et al., 2019) for removing the
skull that are known to be generalizing well over the
unseen MR scans. Once the T2-FLAIR sequence is
skull-stripped, we extract nearly 120 radiomic-based
features that are fed (with or without additional
dimensionality reduction) to the classification engine.
The generalization capabilities of the proposed
technique for quantifying the MGMT methylation
status were verified over the RSNA-MICCAI Brain
Tumor Radiogenomic Classification benchmark
dataset (Baid et al. 2021; Bakas et al., 2017a; Bakas
et al., 2017b; Bakas et al., 2017c; Menze et al., 2015).
In this study, we frame the problem of assessing the
MGMT methylation status as the classification task,
with the patients being assigned to unmethylated and
methylated classes.
The remainder of the paper is structured as
follows. In Section 2, we present the RSNA-MICCAI
Brain Tumor Radiogenomic Classification
benchmark dataset, and introduce our machine
learning pipeline for assessing the MGMT
methylation status based on the radiomic-based
features extracted from T2-FLAIR MR sequences. In
Section 3, we report and discuss the experimental
study performed to investigate the generalization
capabilities of the algorithms, as well as to verify the
impact of various dimensionality reduction
techniques on its capabilities (both classic and deep
learning-powered, with the latter benefiting from
autoencoder architectures). Finally, Section 4
summarized the findings and sheds more light on the
Predicting the MGMT Promoter Methylation Status in T2-FLAIR Magnetic Resonance Imaging Scans Using Machine Learning
873
most promising research directions that may emerge
from the results obtained in this article.
a) The axial plane
b
) The sagittal plane
c
)
The coronal
p
lane
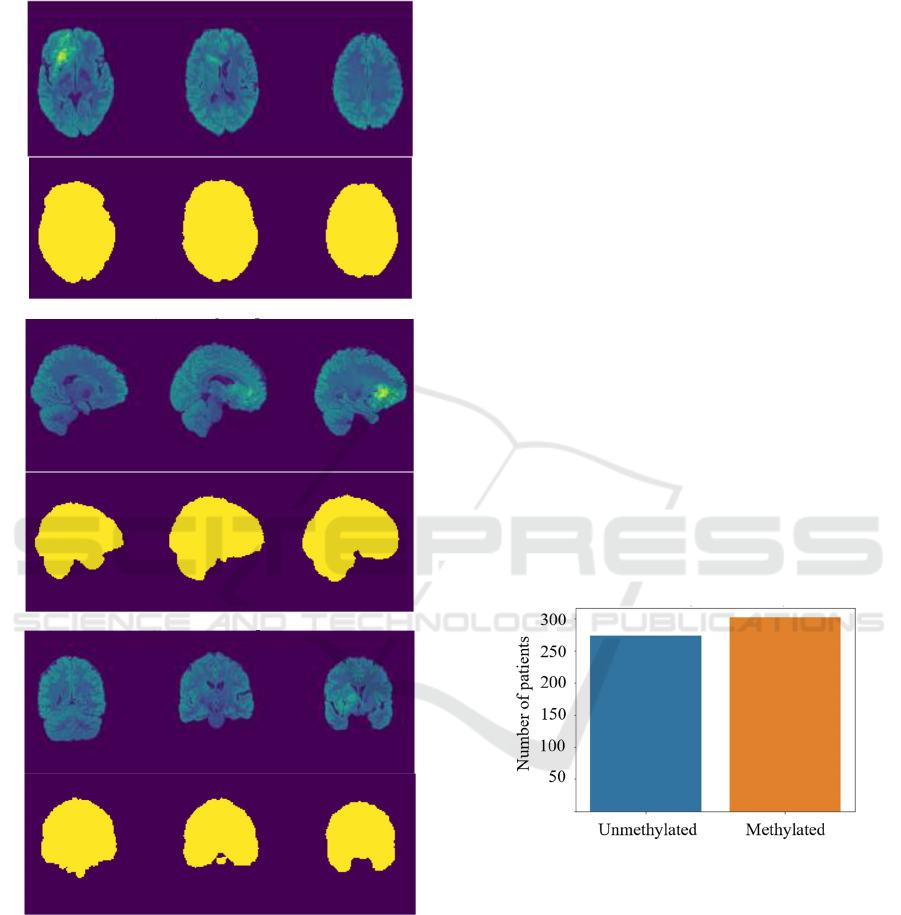
Figure 1: An example of a skull-stripped T2-FLAIR MR
frames (visualized in the false-color scheme), together with
the corresponding brain regions in the a) axial, b) sagittal,
and c) coronal planes.
2 MATERIALS AND METHODS
In this section, we discuss the dataset used in our
study (Section 2.1). In Section 2.2, we present the
most important steps of our processing chain for
classifying the patients into the unmethylated and
methylated classes, based on the radiomic features
extracted from T2-FLAIR MR sequences.
2.1 The RSNA-ASNR-MICCAI Brain
Tumor Segmentation Dataset
In this study, we build upon the RSNA-ASNR-
MICCAI Brain Tumor Segmentation (BraTS)
benchmark dataset (the 2021 edition, for which the
clinical information related to the MGMT promoter
methylation status was obtained as well) (Baid et al.
2021; Bakas et al., 2017a; Bakas et al., 2017b; Bakas
et al., 2017c; Menze et al., 2015). This dataset
contains multi-modal MRI scans captured with
different protocols and scanners from multiple
institutions, and the BraTS dataset is commonly
considered the state-of-the-art benchmark dataset for
confronting the brain tumor segmentation algorithms,
thanks to its size and heterogeneity. The MRI scans
contained within the dataset were interpolated to the
same shape (the size of an MRI scan is 240 × 240 ×
155, therefore there are 155 images of 240 × 240 MR
images, with the voxel size of 1 mm
3
). All of the
available images are skull-stripped – a set of example
T2-FLAIR frames (obtained for a single patient) with
the corresponding brain ground-truth segmentation
masks are rendered in Figure 1.
Figure 2: Distribution of the unmethylated and methylated
patients in the dataset used in this study.
The MGMT promoter methylation status data was
defined as a binary label, corresponding to the
unmethylated and methylated patients. The
distribution of the methylated and unmethylated
patients within the training set of BraTS 2021 (for
which the ground-truth labels are known, as they were
revealed by the organizers of the challenge) is
visualized in Figure 2. Out of all 585 patients, we
removed nine patients due to an incorrect registration
of their brain segmentation masks and corresponding
image data. Therefore, the final dataset included 576
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
874

patients with the MRI scans and the corresponding
MGMT promoter methylation status. We can observe
that the dataset is balanced, and includes a similar
number of unmethylated and methylated patients.
2.2 Predicting the MGMT Promoter
Methylation Status Using Machine
Learning and Radiomic Features
In this work, we introduce an end-to-end processing
chain benefiting from classic machine learning
classification models (trained in a supervised way)
operating over the radiomic features extracted from
T2-FLAIR sequences of brain MRI (Figure 3). The
feature extraction may be followed by an optional
dimensionality reduction step which can play a
pivotal role if a very large number of radiomic
features are extracted, as it may easily lead to
overfitting the model to the training data (Kotowski
et al., 2023). Of note, our approach for determining
the MGMT promoter methylation status offers a high
level of flexibility, and the specific algorithms may be
easily updated at each processing step – this
flexibility will be further proven in the experimental
section of this article.
Figure 3: A high-level flowchart presenting the proposed
processing chain. The optional step is rendered as a dashed
block, whereas the input and output steps are presented as
white and orange ones.
The input T2-FLAIR images undergo brain
extraction, which might be performed using an array
of thoroughly-evaluated state-of-the-art techniques,
such as the HD-BET algorithm (Isensee et al., 2019)
(note that the scans included in BraTS are already
skull-stripped, hence this step was omitted in our
study). Afterwards, we extract the following radiomic
features (as suggested by van Griethuysen et al., 2017
and by Ponikiewski et al., 2022) from the 3D brain
region of the T2-FLAIR scan:
• First Order Statistics (18 features),
• Shape-based (3D) features (14 features),
• Gray Level Co-occurrence Matrix (24 features)
• Gray Level Run Length Matrix (16 features),
• Gray Level Size Zone Matrix (16 features),
• Neighboring Gray Tone Difference Matrix (5
features),
• Gray Level Dependence Matrix (14 features).
The majority of the features are in compliance
with the feature definitions as suggested by the
Imaging Biomarker Standardization Initiative
(Zwanenburg et al., 2020). Overall, we extract 119
features (which were scaled to the unit variance).
Since the number of features is large, especially
when confronted with a relatively small number of
patients, exploiting all of them while training
supervised learners may easily lead to overfitting
them to the training set, hence memorizing it – it
would render them impossible to generalize over the
unseen test patients (Ying et al., 2019). To deal with
this issue, we exploit the additional (yet optional)
dimensionality reduction step, and employ the
following techniques for this task (although we are
aware that the hyperparameters of the following data
dimensionality methods are tunable, we present them
here, rather than in the experimental section in order
to make this section self-contained):
• Principal component analysis (PCA), for which
the number of principal components (PCs) was
selected to explain 98% of the data variance (21
PC were exploited). In Figure 4, we can observe
that exploiting just two PCs would make the
classification process (i.e., distinguishing the
methylated and unmethylated patients) virtually
impossible due to heavy overlaps across these
two classes in the PC space for 2 PCs.
Figure 4: The first two PCs show that discriminating
unmethylated (black dots) and methylated (orange dots)
patients would be virtually impossible using only two PCs.
In this study, we selected 21 PCs to explain 98% variance
within the dataset.
• Autoencoder (AE) with a fully-connected
architecture with the scaled exponential linear
unit activations, containing two encoding and
decoding layers (with 50 and 30 neurons), and
Predicting the MGMT Promoter Methylation Status in T2-FLAIR Magnetic Resonance Imaging Scans Using Machine Learning
875

elaborating the latent representation of 21
features (to ensure consistency with the number
of PCs elaborated by PCA).
• Feature selection (FS), where we selected 21
features with the largest variance (as previously,
we ensured consistency with the number of PCs).
Such variance-based feature selection might be
useful to ensure interpretability of the extracted
features (this is not necessarily the case for the
radiomic features, as they may be fundamentally
challenging to interpret by human readers).
In Figure 5, we render the distributions of the
selected features for all dimensionality reduction
techniques – these features (extracted by each
dimensionality reduction approach) are later fed into
the supervised learner for elaborating the predicted
class label (i.e., methylated or unmethylated patient).
There are numerous established supervised
classification models that could be exploited in our
processing pipeline. In this study, we investigated the
following machine learning models which have
proven their classification capabilities in a range of
real-world applications: logistic regression (LR),
support vector machines (SVMs), random forests
(RFs), k-nearest neighbor classifiers (k-NN), extreme
gradient boosting classifiers (XGBoost), and artificial
neural networks (ANNs) with a single hidden layer
containing 10 neurons. As for the feature extraction
and dimensionality reduction techniques, other
machine learning models (also deep learning
techniques) can be easily exploited in our approach.
3 EXPERIMENTAL STUDY
In this section, we discuss the results obtained in our
experimental study. To quantify the generalization
capabilities of the classification engine, we follow the
5-fold cross-validation procedure, where each fold is
stratified according to the ratio of unmethylated and
methylated patients within the full dataset. The
performance of the models was assessed using classic
metrics, including precision (Pr), recall (Re), F1 score
and the Matthews's correlation coefficient (MCC).
All metrics should be maximized, where one
indicates the perfect classification (additionally, we
tracked accuracy during the ANN training to verify if
it started overfitting). The hyperparameters of all
investigated machine learning models were fine-
tuned using an internal cross-validation procedure
performed over the corresponding training set (the
test set in the k-fold cross-validation approach was
never used here).
a) Principal component analysis
b) Autoencoder
c) Feature selection
Figure 5: Distribution of the features selected using a)
principal component analysis, b) a fully-connected
autoencoder, and c) variance-based feature selection.
Finally, to make sure that the processing chain is
straightforward to reproduce (the full approach was
implemented in Python 3.6), we exploited a well-
established pyradiomics package to extract radiomic
features from the brain areas, and the scikit-learn
package for the classification models.
In Figure 6, we gather the experimental results
(quantified as all above-mentioned quality metrics)
obtained for all investigated machine learning models
and dimensionality reduction techniques, averaged
across all five test folds. We can appreciate that
various dimensionality reduction gave consistently
similar results for virtually all classification models
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
876
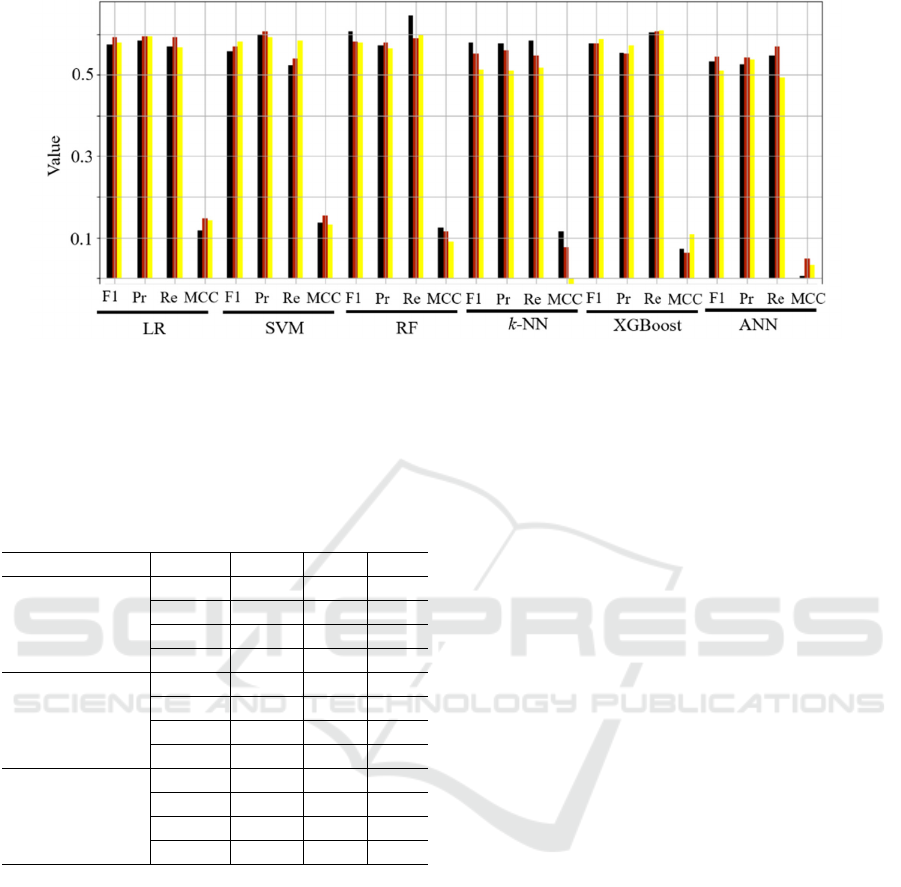
Figure 6: Classification results (averaged across all five test folds) obtained for all investigated machine
learning classification models and dimensionality reduction techniques (the black color corresponds to
principal component analysis, red to the autoencoder, and yellow to variance-based feature selection).
Table 1: The results obtained using the ANN model without
and with regularization techniques applied (averaged across
all test sets in the five-fold cross-validation scenario). The
best metrics for each dimensionality reduction approach are
boldfaced.
Regularization Metric PCA AE FS
None F1 0.53 0.54 0.51
P
r
0.53 0.54 0.54
Re 0.55 0.57 0.49
MCC 0.01 0.05 0.03
Dropout F1 0.55 0.49 0.57
P
r
0.54 0.56 0.58
Re 0.56 0.43 0.56
MCC 0.04 0.06 0.12
Dropout and
early stopping
F1 0.56 0.56 0.56
P
r
0.59 0.59 0.59
Re 0.52 0.54 0.54
MCC 0.13 0.12 0.12
(the smallest differences between different
dimensionality reduction routines were captured for
the LR classifier), with PCA outperforming the other
methods for RF. Here, this model resulted in the
highest recall values which is of paramount clinical
significance, as identifying methylated patients may
lead to designing their more effective treatment
pathways. Of note, it was observable that the ANN
model started overfitting the training set – as an
example for the PCA dimensionality reduction, the
accuracy over the training folds exceeded 0.9, with
the corresponding accuracy over the validation set
reaching approx. 0.6. This phenomenon was,
however, observed for all dimensionality reduction
approaches, indicating that the training sample may
be too small to elaborate a well-generalizing
classifiers. To verify if applying additional
regularization techniques could help improve the
abilities of the ANN model, we investigated two
additional (yet well-established in the field)
regularization approaches, being the dropout within
the ANN, together with an early stopping routine. The
results gathered in Table 1 indeed confirm that
applying additional regularization techniques help
improve the generalization capabilities of the ANN
models.
4 CONCLUSIONS
Glioblastoma is the most common form of brain
cancer, and the detailed profiling of patients suffering
from this disease is of pivotal importance. We
approached this issue, and proposed a machine
learning pipeline to predict the MGMT promoter
methylation from T2-FLAIR, as it is an important
biomarker for the patient prognosis. The experiments
indicated that radiomic features extracted from
whole-brain scans allow to elaborate classifiers that
identify the methylated patients. The generalization
of models, thus their clinical utility might be
improved by gathering more heterogeneous and
representative training sets, as we observed that the
models started overfitting, and by explicitly tackling
the problem of the dataset imbalance. This issue may
be also tackled using model-level regularization
which was shown effective in this study.
Predicting the MGMT Promoter Methylation Status in T2-FLAIR Magnetic Resonance Imaging Scans Using Machine Learning
877

ACKNOWLEDGEMENTS
This work was supported by the Silesian University
of Technology funds through the grant for
maintaining and developing research potential, and
by the Silesian University of Technology funds
through the Excellence Initiative—Research
University program (Grant 02/080/SDU/10-21-01).
REFERENCES
Alpar, O. (2023). A mathematical fuzzy fusion framework
for whole tumor segmentation in multimodal MRI
using Nakagami imaging. Expert Systems with
Applications, 216. https://doi.org/10.1016/j.eswa.
2022.119462.
Baid, U., Ghodasara, S., Bilello, M., Mohan, S., Calabrese,
E., Colak, E., Farahani, K., Kalpathy-Cramer, J.,
Kitamura, F.C., Pati, S., Prevedello, L.M., Rudie, J.D.,
Sako, C., Shinohara, R.T., Bergquist, T., Chai, R.,
Eddy, J.A., Elliott, J., Reade, W.C., Schaffter, T., Yu,
T., Zheng, J., Annotators, B., Davatzikos, C., Mongan,
J.T., Hess, C.P., Cha, S., Villanueva-Meyer, J.E.,
Freymann, J.B., Kirby, J.S., Wiestler, B., Crivellaro,
P.S., R.Colen, R., Kotrotsou, A., Marcus, D.,
Milchenko, M., Nazeri, A., Fathallah-Shaykh, H.M.,
Wiest, R., Jakab, A., Weber, M., Mahajan, A., Menze,
B.H., Flanders, A.E., & Bakas, S. (2021). The RSNA-
ASNR-MICCAI BraTS 2021 Benchmark on Brain
Tumor Segmentation and Radiogenomic Classification.
ArXiv, abs/2107.02314.
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M.,
Kirby, J. S., Freymann, J. B., Farahani, K., &
Davatzikos, C. (2017). Advancing The Cancer Genome
Atlas glioma MRI collections with expert segmentation
labels and radiomic features. Scientific data, 4, 170117.
https://doi.org/10.1038/sdata.2017.117.
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby
J, Freymann J, Farahani K, Davatzikos C. (2017).
Segmentation Labels for the Pre-operative Scans of the
TCGA-GBM collection [Data set]. The Cancer Imaging
Archive. DOI: 10.7937/K9/TCIA.2017.KLXWJJ1Q.
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby
J, Freymann J, Farahani K, Davatzikos C. (2017)
Advancing The Cancer Genome Atlas glioma MRI
collections with expert segmentation labels and
radiomic features Nature Scientific Data, 4:170117
DOI: 10.1038/sdata.2017.117.
Beyer, T., Bidaut, L., Dickson, J., Kachelriess, M.,
Kiessling, F., Leitgeb, R., Ma, J., Shiyam Sundar, L. K.,
Theek, B., & Mawlawi, O. (2020). What scans we will
read: imaging instrumentation trends in clinical
oncology. Cancer imaging : the official publication of
the International Cancer Imaging Society, 20(1), 38.
https://doi.org/10.1186/s40644-020-00312-3
Bukhari, S.T., Mohy-ud-Din, H. (2022). E1D3 U-Net for
Brain Tumor Segmentation: Submission to the RSNA-
ASNR-MICCAI BraTS 2021 challenge. In: Crimi, A.,
Bakas, S. (eds) Brainlesion: Glioma, Multiple
Sclerosis, Stroke and Traumatic Brain Injuries.
BrainLes 2021. Lecture Notes in Computer Science, vol
12963. Springer, Cham. https://doi.org/10.1007/978-3-
031-09002-8_25
van Griethuysen, J. J. M., Fedorov, A., Parmar, C., Hosny,
A., Aucoin, N., Narayan, V., Beets-Tan, R. G. H.,
Fillion-Robin, J. C., Pieper, S., & Aerts, H. J. W. L.
(2017). Computational Radiomics System to Decode
the Radiographic Phenotype. Cancer research, 77(21),
e104–e107. https://doi.org/10.1158/0008-5472.CAN-
17-0339
Hajianfar, G., Shiri, I., Maleki, H., Oveisi, N., Haghparast,
A., Abdollahi, H., Oveisi, M. (2019). Noninvasive O6
Methylguanine-DNA Methyltransferase Status
Prediction in Glioblastoma Multiforme Cancer Using
Magnetic Resonance Imaging Radiomics Features:
Univariate and Multivariate Radiogenomics Analysis.
World Neurosurgery. DOI: 10.1016/j.wneu.2019.
08.232.
Hu, X., Luo, W., Hu, J., Guo, S., Huang, W., Scott, M. R.,
Wiest, R., Dahlweid, M., & Reyes, M. (2020). Brain
SegNet: 3D local refinement network for brain lesion
segmentation. BMC medical imaging, 20(1), 17.
https://doi.org/10.1186/s12880-020-0409-2.
Korfiatis, P., Kline, T. L., Coufalova, L., Lachance, D. H.,
Parney, I. F., Carter, R. E., Buckner, J. C., & Erickson,
B. J. (2016). MRI texture features as biomarkers to
predict MGMT methylation status in glioblastomas.
Medical physics, 43(6), 2835–2844. https://doi.org/
10.1118/1.4948668.
Korfiatis, P., Kline, T. L., Lachance, D. H., Parney, I. F.,
Buckner, J. C., & Erickson, B. J. (2017). Residual Deep
Convolutional Neural Network Predicts MGMT
Methylation Status. Journal of digital imaging, 30(5),
622–628. https://doi.org/10.1007/s10278-017-0009-z.
Kotowski, K., Kucharski, D., Machura, B., Adamski, S.,
Gutierrez Becker, B., Krason, A., Zarudzki, L., Tessier,
J., & Nalepa, J. (2023). Detecting liver cirrhosis in
computed tomography scans using clinically-inspired
and radiomic features. Computers in biology and
medicine, 152, 106378. https://doi.org/10.1016/j.
compbiomed.2022.106378.
Isensee, F., Schell, M., Pflueger, I., Brugnara, G.,
Bonekamp, D., Neuberger, U., Wick, A., Schlemmer,
H. P., Heiland, S., Wick, W., Bendszus, M., Maier-
Hein, K. H., & Kickingereder, P. (2019). Automated
brain extraction of multisequence MRI using artificial
neural networks. Human brain mapping, 40(17), 4952–
4964. https://doi.org/10.1002/hbm.24750.
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J.,
Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom,
J., Wiest, R., Lanczi, L., Gerstner, E., Weber, M. A.,
Arbel, T., Avants, B. B., Ayache, N., Buendia, P.,
Collins, D. L., Cordier, N., Corso, J. J., … Van
Leemput, K. (2015). The Multimodal Brain Tumor
Image Segmentation Benchmark (BRATS). IEEE
transactions on medical imaging, 34(10), 1993–2024.
https://doi.org/10.1109/TMI.2014.2377694.
ICPRAM 2024 - 13th International Conference on Pattern Recognition Applications and Methods
878

Peiris, H., Chen, Z., Egan, G., Harandi, M. (2022).
Reciprocal Adversarial Learning for Brain Tumor
Segmentation: A Solution to BraTS Challenge 2021
Segmentation Task. In: Crimi, A., Bakas, S. (eds)
Brainlesion: Glioma, Multiple Sclerosis, Stroke and
Traumatic Brain Injuries. BrainLes 2021. Lecture
Notes in Computer Science, vol 12962. Springer,
Cham. https://doi.org/10.1007/978-3-031-08999-2_13.
Ponikiewski, W., Nalepa, J. (2022). Deep Learning Meets
Radiomics For End-To-End Brain Tumor MRI
Analysis. 1301-1305. 10.1109/ICIP46576.2022.
9897847.
Puttagunta, M., & Ravi, S. (2021). Medical image analysis
based on deep learning approach. Multimedia tools and
applications, 80(16), 24365–24398. https://doi.org/10.
1007/s11042-021-10707-4.
Qi, D., Li, J., Quarles, C. C., Fonkem, E., & Wu, E. (2023).
Assessment and prediction of glioblastoma therapy
response: challenges and opportunities. Brain : a
journal of neurology, 146(4), 1281–1298.
https://doi.org/10.1093/brain/awac450
Saeed, N., Ridzuan, M., Alasmawi, H., Sobirov, I., Yaqub,
M. (2023). MGMT promoter methylation status
prediction using MRI scans? An extensive
experimental evaluation of deep learning models.
Medical Image Analysis. 90. 102989.
10.1016/j.media.2023.102989.
Shi, Y., Micklisch, C., Mushtaq, E., Avestimehr, S., Yan,
Y., Zhang, X. (2022). An Ensemble Approach to
Automatic Brain Tumor Segmentation. In: Crimi, A.,
Bakas, S. (eds) Brainlesion: Glioma, Multiple
Sclerosis, Stroke and Traumatic Brain Injuries.
BrainLes 2021. Lecture Notes in Computer Science, vol
12963. Springer, Cham. https://doi.org/10.1007/978-3-
031-09002-8_13.
Vadmal, V., Junno, G., Badve, C., Huang, W., Waite, K.
A., & Barnholtz-Sloan, J. S. (2020). MRI image
analysis methods and applications: an algorithmic
perspective using brain tumors as an exemplar. Neuro-
oncology advances, 2(1), vdaa049. https://doi.org/10.
1093/noajnl/vdaa049.
Weller, M., Stupp, R., Reifenberger, G., Brandes, A. A.,
van den Bent, M. J., Wick, W., & Hegi, M. E. (2010).
MGMT promoter methylation in malignant gliomas:
ready for personalized medicine?. Nature reviews.
Neurology, 6(1), 39–51. https://doi.org/10.1038/
nrneurol.2009.197.
Xing, F., Liu, X., Kuo, C. J., Fakhri, G. E., & Woo, J.
(2022). Brain MR Atlas Construction Using Symmetric
Deep Neural Inpainting. IEEE journal of biomedical
and health informatics, 26(7), 3185–3196.
https://doi.org/10.1109/JBHI.2022.3149754.
Xuan, K., Xiang, L., Huang, X., Zhang, L., Liao, S., Shen,
D., & Wang, Q. (2022). Multimodal MRI
Reconstruction Assisted With Spatial Alignment Network.
IEEE transactions on medical imaging, 41(9), 2499–
2509. https://doi.org/10.1109/TMI.2022.3164050.
Yan, B., Wei, Y., Jagtap, J., Moassefi, M., Vera Gracia,
DV., & Singh, Y., Vahdati, S., Faghani, S., Erickson,
B., Conte, G. (2022). MRI Brain Tumor Segmentation
Using Deep Encoder-Decoder Convolutional Neural
Networks. In: Crimi, A., Bakas, S. (eds) Brainlesion:
Glioma, Multiple Sclerosis, Stroke and Traumatic
Brain Injuries. BrainLes 2021. Lecture Notes in
Computer Science, vol 12963. Springer, Cham.
https://doi.org/10.1007/978-3-031-09002-8_7.
Ying, X. (2019). An Overview of Overfitting and its
Solutions. Journal of Physics: Conference Series. 1168.
022022. 10.1088/1742-6596/1168/2/022022.
Zwanenburg, A., Vallières, M., Abdalah, M. A., Aerts, H.
J. W. L., Andrearczyk, V., Apte, A., Ashrafinia, S.,
Bakas, S., Beukinga, R. J., Boellaard, R., Bogowicz,
M., Boldrini, L., Buvat, I., Cook, G. J. R., Davatzikos,
C., Depeursinge, A., Desseroit, M. C., Dinapoli, N.,
Dinh, C. V., Echegaray, S., … Löck, S. (2020). The
Image Biomarker Standardization Initiative:
Standardized Quantitative Radiomics for High-
Throughput Image-based Phenotyping. Radiology,
295(2), 328–338. https://doi.org/10.1148/radiol.
2020191145.
Predicting the MGMT Promoter Methylation Status in T2-FLAIR Magnetic Resonance Imaging Scans Using Machine Learning
879
