
Few-Shot Histopathology Image Classification: Evaluating
State-of-the-Art Methods and Unveiling Performance Insights
Ardhendu Sekhar, Ravi Kant Gupta and Amit Sethi
Department of Electrical Engineering, Indian Institute of Technology Bombay, Mumbai, India
Keywords:
Deep Learning, Few-shot Classification, Medical Image.
Abstract:
This paper presents a study on few-shot classification in the context of histopathology images. While few-
shot learning has been studied for natural image classification, its application to histopathology is relatively
unexplored. Given the scarcity of labeled data in medical imaging and the inherent challenges posed by
diverse tissue types and data preparation techniques, this research evaluates the performance of state-of-the-
art few-shot learning methods for various scenarios on histology data. We have considered four histopathology
datasets for few-shot histopathology image classification and have evaluated 5-way 1-shot, 5-way 5-shot and 5-
way 10-shot scenarios with a set of state-of-the-art classification techniques. The best methods have surpassed
an accuracy of 70%, 80% and 85% in the cases of 5-way 1-shot, 5-way 5-shot and 5-way 10-shot cases,
respectively. We found that for histology images popular meta-learning approaches is at par with standard
fine-tuning and regularization methods. Our experiments underscore the challenges of working with images
from different domains and underscore the significance of unbiased and focused evaluations in advancing
computer vision techniques for specialized domains, such as histology images.
1 INTRODUCTION
Traditional deep learning models often require large
amounts of labeled data for training. These mod-
els learn representations and patterns from a sub-
stantial dataset to generalize well to unseen exam-
ples. The learning process involves adjusting nu-
merous parameters through backpropagation to mini-
mize the difference between predicted and actual out-
puts. Transfer learning is commonly used in tradi-
tional deep learning, where models pre-trained are
fine-tuned for specific tasks. The knowledge gained
from the pre-training on large dataset helps in solv-
ing related problems with smaller dataset. In medi-
cal imaging, traditional deep learning methods have
been successful for tasks such as image classifica-
tion and segmentation, but they often require exten-
sive labeled dataset. But traditional models might
struggle when faced with new tasks or limited data,
requiring substantial retraining or fine-tuning. This
is where few-shot learning can be useful. Few-shot
learning, as the name implies, is designed to make
accurate predictions when only a small number of
examples per class are available for training. This
can be achieved through various techniques such as
meta-learning, where the model is trained on a va-
riety of tasks and adapts quickly to new tasks with
minimal examples by leveraging knowledge gained
from previous tasks. One would think that meta learn-
ing would be particularly valuable in medical imaging
due to the scarcity and cost of labeling medical data.
However, such techniques have only been evaluated
on natural images. If successful on medical images,
this would enable demonstrate learning even when
only a handful of annotated medical images are avail-
able.
In this work, we have evaluated certain state-
of-the-art few-shot classification techniques on
histopathology medical datasets. A dataset prepared
by (Komura and Ishikawa, 2021) and FHIST (Shak-
eri et al., 2022) were considered for the experiments.
FHIST dataset comprises of many histology datasets.
These are : CRC-TP (Javed et al., 2020b), NCT-CRC-
HE-100K (Kather et al., 2019), LC25000 (Borkowski
et al., 2019) and BreakHis (Spanhol et al., 2016). For
our experiments, we have considered CRC-TP, NCT
and LC25000 datsets. CRC-TP is a colon cancer
dataset with six classes. NCT is also a colon cancer
dataset with nine classes. LC25000 consists of both
lung and colon cancer images with five classes. We
have also used a histology dataset proposed by (Ko-
mura and Ishikawa, 2021). It has around 1.6 million
244
Sekhar, A., Gupta, R. and Sethi, A.
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights.
DOI: 10.5220/0012568000003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 244-253
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

cancerous image patches of 32 different organs in the
body. The classes in this dataset are defined accord-
ing to the different organ sites. Dataset proposed by
(Komura and Ishikawa, 2021) is used to train the few-
shot classification models. The trained model is then
evaluated on various FHSIT dataset.
In our comprehensive exploration of few-shot
classification techniques, we have meticulously incor-
porated various state-of-the-art methodologies to en-
sure a thorough evaluation. The methods employed
in our experiments includes Prototypical Networks
(Snell et al., 2017), Model-Agnostic Meta-Learning
(MAML) (Finn et al., 2017), SimpleShot (Wang et al.,
2019), LaplacianShot (Ziko et al., 2020), and Deep-
EMD (Zhang et al., 2020). Prototypical Networks,
a benchmark technique for few-shot learning, lever-
age prototypes as representative embeddings for each
class. By minimizing the distance between query
examples and class prototypes, this model excels in
adapting quickly to new classes with limited labeled
data. MAML adopts a meta-learning paradigm, train-
ing a model to quickly adapt to new tasks with min-
imal examples. This approach proves invaluable in
scenarios where prompt adaptation to novel classes
is paramount. As its name implies, SimpleShot em-
phasizes simplicity in few-shot learning. This ap-
proach often relies on straightforward yet effective
techniques, showcasing the power of simplicity in
addressing complex classification tasks with limited
data. LaplacianShot introduces Laplacian regulariza-
tion to enhance few-shot learning performance. By
incorporating this regularization technique, the model
aims to improve generalization and robustness across
diverse classes. DeepEMD learns the image repre-
sentations by calculating the discrepancy between the
joint characteristics of embedded features and product
of the marginals.
2 RELATED WORK
In recent years, there has been a notable surge in
research endeavors for addressing the challenges of
few-shot learning within specific domains of medi-
cal imaging. This section outlines some contribu-
tions in the field, shedding light on how researchers
have applied various few-shot learning techniques to
tackle different problems in medical diagnostics. One
such study (Mahajan et al., 2020), focuses on skin-
disease identification. Here, the researchers applied
two prominent few-shot learning methods, Reptile
(Nichol and Schulman, 2018) and Prototypical Net-
works (Snell et al., 2017), showcasing their efficacy in
the task of distinguishing various skin diseases. The
task of transferring knowledge across different tissue
types was addressed in (Medela et al., 2019), where a
deep Siamese neural network was trained to transfer
knowledge from a dataset containing colon tissue to
another encompassing colon, lung, and breast tissue.
Meanwhile, (Teng et al., 2021) proposed a few-shot
learning algorithm based on Prototypical Networks,
specifically tailored for classifying lymphatic metas-
tasis of lung carcinoma from Whole Slide Images
(WSIs). In another work in (Chen et al., 2021), a two-
stage framework was adopted for the crucial task of
COVID-19 diagnosis from chest CT images. The ini-
tial stage involved capturing expressive feature repre-
sentations using an encoder trained on publicly avail-
able lung datasets through contrastive learning. Sub-
sequently, in the second stage, the pre-trained encoder
was employed within a few-shot learning paradigm,
leveraging the Prototypical Networks method. In
(Yang et al., 2022), the authors have incorporated
contrastive learning (CL) with latent augmentation
(LA) to build a few-shot classification model. Here,
contrastive learning learns important features without
needing labels, while Latent Augmentation moves se-
mantic variations from one dataset to another with-
out supervision. The model was trained on publicly
available colon cancer datasets and evaluated on PAIP
(Kim et al., 2021) liver cancer Whole Slide Images.
Shakeri et al. (Shakeri et al., 2022) introduced the
FHIST data coupled with assessments of certain few-
shot classification methodologies serves as a catalyst
for additional exploration in this domain.
The exploration of few-shot learning is not con-
fined to image classification alone. In (Ouyang et al.,
2020), a few-shot semantic segmentation framework
was introduced, aiming to alleviate the dependency
on labeled data during the training phase. Similarly,
(Yu et al., 2021) ventured into the domain of medi-
cal image segmentation, employing a prototype-based
method known as the location-sensitive local proto-
type network, which strategically incorporates spatial
priors to enhance segmentation accuracy.
3 METHODOLOGY
Few shot classification deals with the cases of lim-
ited data. For example, a medical dataset has five
rare classes and in each class there are five im-
ages. It is very difficult to train and test DL mod-
els with such small number of images. Training the
model with such small dataset my lead to over-fitting.
Since the classes are rare, there is a high probabil-
ity that the pre-trained models might not have seen
those images while being trained. Instead of tradi-
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights
245
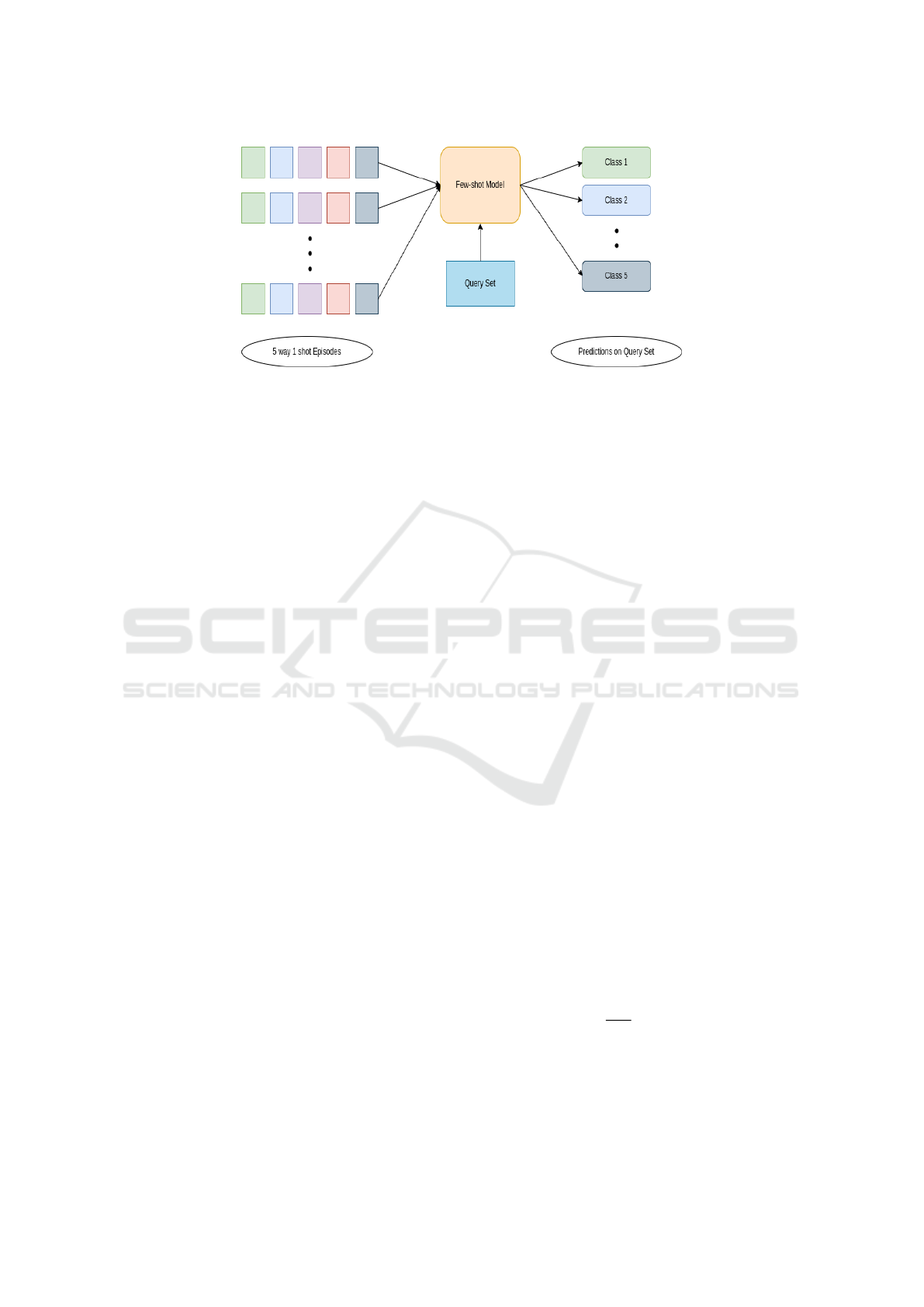
Figure 1: The diagram illustrates a few-shot learning model, showcasing its ability to effectively generalize and recognize
classes for unlabeled query set with only a limited number of support examples.The 5 different colors in support set represent
5 different classes(ways) having 1 sample(shot) each.
tional transfer-learning, the few-shot models follow
episodic-training paradigm.
In few-shot training, the train and test set are dis-
joint. The training set is denoted as D
Train
. It consists
of (X
Train
,Y
Train
)
M
i=1
. X
Train
are the images. Y
Train
are
the labels. M is the number of classes. In episodic
training, the huge labelled train dataset is broken into
many episodes. In each episode, there is a support
set(S) and a query set(Q). Each episode is defined by
K-way N-shot Q-query. It means, for each episode, K
classes are randomly selected from M classes. From
each K class, N images and Q images are selected.
N×K images form the support set and Q×K images
form the query set. The few shot model is trained in
such a way that it must learn from the support set to
predict the labels of the query set. Once the model is
trained, it is tested on the test set.
D
Train
= (X
Train
,Y
Train
) (1)
S := (X
i
,Y
i
)
K×N
i=1
(2)
Q := (X
i
,Y
i
)
K×Q
i=1
(3)
D
Test
= (X
Test
,Y
Test
) (4)
S := (X
i
,Y
i
)
K×N
i=1
(5)
Q := (X
i
) (6)
Y
Train
∩Y
Test
= Φ (7)
3.1 Prototypical Networks
The central idea of Prototypical Networks (Snell
et al., 2017) is rooted in the observation that data
points exhibiting proximity to a singular prototype
representation for each class contribute to a mean-
ingful embedding. To operationalize this concept, a
non-linear mapping transforms the input data into a
specialized embedded space. Within this embedded
space, the prototype representation for each class was
derived by calculating the mean of its respective sup-
port set. The process is initiated by the formation
of a non-linear mapping, effectively transforming the
original input data into a specialized embedded space.
This mapping is crucial for capturing intricate rela-
tionships and patterns within the data. Within the em-
bedded space, a class prototype was defined for each
category. The prototype representation for a specific
class was computed by determining the mean of its
corresponding support set. This means that the pro-
totype becomes a central point representing the class
in the embedded space. To perform classification on
a given query, the approach relies on identifying the
nearest distance between the embedded query and the
prototype representation of each class. The class as-
sociated with the closest prototype is then assigned to
the query, effectively determining its classification.
The small support set consists of K labeled exam-
ples which is represented by S = [(X
1
,Y
1
),...,(X
K
,Y
K
)].
X
i
is a D-dimensional feature vector for each image
and Y
i
represents the corresponding label of X
i
. There
are N classes with in a support set. The number of
examples with in a class N is represented by S
N
.
With the help of an embedding function, i.e. a
CNN, f
Φ
, prototypical networks estimate an M dimen-
sional representation C
N
of each class. C
N
is the mean
vector of the support points of each class. Φ are the
learnable parameters.
f
Φ
: R
D
− > R
M
(8)
C
N
∈ R
M
(9)
C
N
=
1
|S
N
|
∑
(X
i
,Y
i
)∈S
f
Φ
(X
i
) (10)
Prototypical networks generate a probability dis-
tribution across classes for a given query example X.
This distribution is computed using a softmax func-
BIOIMAGING 2024 - 11th International Conference on Bioimaging
246

tion applied to the distances between the query exam-
ple and other prototypes in the embedding space.
P
Φ
(Y = N|X) =
exp(−d( f
Φ
(X),C
N
))
∑
N
′
exp(−d( f
Φ
(X),C
N
′
))
(11)
The model learns by minimizing the negative of
the log probability of J(Φ) of the true class N through
Adam solver.
J(Φ) = −log(P
Φ
(Y = N|X)) (12)
3.2 Model Agnostic Meta
Learning(MAML)
MAML (Finn et al., 2017) introduces a meta-learning
objective that involves training a model on a distribu-
tion of tasks. The goal is to learn a good initialization
of the model’s parameters that facilitates quick adap-
tation to new, unseen tasks. The focus of MAML is
on few-shot learning scenarios, where the model is
required to generalize well from a small number of
examples per class. This is particularly important in
real-world applications where collecting extensive la-
beled dataset can be impractical. During the meta-
training phase, the model is exposed to a variety of
tasks. Each task consists of a support set (small la-
beled dataset) and a query set (unlabeled examples).
The model is trained to adapt quickly to new tasks
by updating its parameters through gradient descent.
MAML introduces a two-step gradient descent update
during meta-training. The first update involves com-
puting the gradients on the support set and using them
to update the model’s parameters. The second update
fine-tunes the model on the query set to improve its
performance on the specific task. The model’s ini-
tialization is learned in a way that allows it to adapt
quickly to new tasks. The meta-training process en-
courages the model to learn parameter initialization
that generalize well across different tasks. During
meta-testing, the model is fine-tuned quickly on new
tasks with limited examples. This is achieved by ap-
plying a small number of gradient descent steps using
the updated initialization. The goal is to improve the
model’s performance on the specific task.
The neural network is parameterized by a set of
weights θ. The weights are to be updated in such a
way that they can be rapidly adapted towards differ-
ent solutions. For instance, if there are 3 episodes E
1
,
E
2
, E
3
then through a few gradient steps the weights
should be able to move from a specific point θ to
another configuration of weights θ
∗
1
, θ
∗
2
and θ
∗
3
. It
should be done in such a way that the configuration of
weights are now well adapted on the task to be solved.
This can be achieved by optimizing three different
losses on three different support sets at the same time.
In this case, 2 losses will be optimized. The first loss
is across all the support to find θ. Another query set
specific loss will be optimized in θ
∗
1
, θ
∗
2
and θ
∗
3
direc-
tions.
S = [(X
1
,Y
1
), (X
2
,Y
2
)...(X
5
,Y
5
)] (13)
Q = [(X
1
,Y
1
] (14)
The two above equations represent a particular
episode. Suppose there are 3 different episode. In
each of of the support sets of these episodes, there
are five different images and five labels. And in each
query set, there is one image and one label. The ob-
jective is finding out the one particular class to which
the query image belongs.It is a standard few shot set-
ting. The below equations will depict how gradient
descent happens in one episodic training.
E
1
: f
θ
(S
1
) = V
1
(15)
θ
∗
1
= θ − α∆
θ
L (S
1
,V
1
) (16)
f
θ
∗
1
(Q
1
) = Z
1
(17)
θ = θ − β∆
θ
L (Q
1
, Z
1
) (18)
Considering episode 1, the model is parameterized
by f
θ
. The S
1
is passed through f
θ
, V
1
is obtained as
output.The θ is fine tuned towards a specific set of
parameters θ
1
. This θ
1
is used to do another forward
pass on the query set. This is the prediction on the
query image. The loss is calculated from the query set
which is used to do a backward pass on the θ. The task
is repeated for the rest other episodes E
2
and E
3
. Then
θ is updated by taking the gradient of accumulated
losses of all the query sets in all the tasks.
3.3 SimpleShot
SimpleShot (Wang et al., 2019) is a simple non-
episodic few-shot learning method. It uses the con-
cept of transfer-learning and nearest-neighbour rule.
A large scale deep neural network is trained on train-
ing classes. Then nearest-neighbour rule is carried out
on the images of the test episodes by using the trained
deep neural network as a feature encoder.
Training set is defined as D
Train
.
D
Train
= [(X
1
,Y
1
), (X
2
,Y
2
), ..., (X
N
, X
N
)] (19)
X are the images. Y are the corresponding labels.
The train set is trained on a CNN with cross entropy
loss.
argmin
θ
∑
(X,Y)∈D
T
rain
l(W
T
f
θ
(X),Y ) (20)
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights
247

f
θ
represents the CNN. W represents the weights
of the classification layer. l represents the cross en-
tropy loss function.
Classification on the test set is done using the near-
est neighbour rule. The features of an image are ob-
tained by passing it through the CNN. The features
are denoted by Z. The nearest neighbour classifica-
tion is carried out using the Euclidean distance.
Z = f
θ
(X) (21)
In one shot setting, support set S of test set has 1
labelled example from each of the N classes.
S = ((
ˆ
X
1
, 1), ..., (
ˆ
X
N
, N)) (22)
Using the Euclidean distance measure, the near-
est neighbour rule classifies the query image
ˆ
X to the
most similar support image.
Y (
ˆ
X) = argmin
N∈(1,2,..,N)
d(
ˆ
Z,
ˆ
Z
N
) (23)
ˆ
Z and
ˆ
Z
N
are the CNN features of the query and
support images respectively. In multi shot setting,
ˆ
Z
N
is the average of feature vector of each class in the
support set.
3.4 LalplacianShot
LaplacianShot (Ziko et al., 2020) introduces a new
approach for few-shot tasks - a transductive Lapla-
cian regularized interference. It minimises a quadratic
binary assignment function comprising of two essen-
tial terms. The first one is a unary term that allo-
cates query samples to their closest class prototype.
The second term is a pairwise-Laplacian term that
promotes consistent label assignments among nearby
query samples.
Similar to SimpleShot, train set is defined as
D
Train
. The train set is trained on a CNN with a simple
cross-entropy loss. It does not involve any episodic or
meta learning strategy.
argmin
θ
∑
(X,Y)∈D
T
rain
l(W
T
f
θ
(X),Y ) (24)
The regularization equations involved at the time
of few-shot test inference are described below.
E(Y ) = N(Y ) + λ
1
2
L(Y ) (25)
N(Y ) =
N
∑
q=1
C
∑
c=1
y
q,c
d(z
q
− m
c
) (26)
L(Y ) =
1
2
∑
q,p
w(z
q
, z
p
)||y
q
− y
p
||
2
2
(27)
In this objective, the first term N(Y) is minimized by
assigning each query point top the class of the near-
est prototype m
c
, from the support set, using a dis-
tance metric such as Euclidean distance. The second
term, L(Y), represents the Laplacian reguralizer and
is expressed as tr(Y
T
LY). L is a laplacian matrix that
corresponds to the affinity matrix W = w(z
q
, z
p
). It
measures the similarity between the feature vectors z
q
and z
p
with the help of a kernel function. z
q
and z
p
are feature vectors of the query images x
p
and x
q
.
3.5 DeepEMD
In DeepEMD (Zhang et al., 2020), Earth Mover’s Dis-
tance is used as a metric to calculate the structural dis-
tance dense image representations, determining im-
age similarity. The Earth Mover’s Distance produces
optimal matching flows between structural elements,
minimizing the matching cost. The minimized cost
is then utilized to signify the image distance for clas-
sification. To derive crucial weights for elements in
the EMD, a cross-reference mechanism is introduced
which mitigates the impact of clustered backgrounds
and intra-class variations. For K-shot classification,
a structured fully connected layer capable of directly
classifying dense image representations using EMD
is used.
The Earth Mover’s Distance serves as a distance
metric between the two sets of weighted objects or
distributions, leveraging the fundamental concept of
distance between individual objects. It adopts the
structure of the Transportation Problem in Linear
Programming. A set of supplier S = (S
i
|i=1,2....m)
must transport goods to a set of demanders D =
(d
j
|j=1,2....k). S
i
represents the supply unit of sup-
plier and d
j
indicates the demand of demander j.
The expense fro transporting one unit from sup-
plier i to demander j is represented by c
i j
. And
the quantity of units transported is represented by
x
i j
. The objective of the transportation problem is
to find the least-expensive flow of goods ˜x = ( ˜x
i j
|i=-
1,2,...,m,j=1,2....,k) from supplier i to demander j:
minimize
x
i j
m
∑
i=1
k
∑
j=1
c
i j
x
i j
(28)
In few shot classification, to calculate the similar-
ity between the support and query images , the images
are passed through a fully convolutional network to
generate image features S and Q.
S ∈ R
H×W ×C
(29)
Q ∈ R
H×W ×C
(30)
BIOIMAGING 2024 - 11th International Conference on Bioimaging
248

Each image feature is a collection of vectors i.e. S
= [s
1
,s
2
,....,s
HW
] and Q = [q
1
,q
2
,....,q
HW
]. The match-
ing cost between the 2 set of vectors is denoted as the
similarity of images. The cost between the 2 embed-
dings s
i
and q
j
is given by:
c
i j
= 1 −
s
T
i
q
j
∥
s
i
∥
q
j
, (31)
The optimal matching flow is represented by
˜
X ∈
R
HW X HW
. The distance between the support and the
query image is represented by:
d(S, Q) =
HW
∑
i=1
HW
∑
j=1
(1 − c
i j
) ˜x
i j
(32)
In a support set, when the shot is more than 1 then
the learnable embedding becomes a group of image
features for a class rather than one vector. Then the
mean of it is taken to give a single image feature. It
is similar to calculating the prototype of a class in a
support set. The fully connected network is a fea-
ture extractor and SGD optimizer is used to update
the weights by sampling few-shot episodes from the
dataset.
4 EXPERIMENTS AND RESULTS
4.1 Dataset
(Komura and Ishikawa, 2021) have created a dataset
of histology images from uniform tumor regions in
The Cancer Genome Atlas Program whole slide im-
ages(tcga, 2005). TCGA consist of tissue slides from
32 cancer types at different sites of a human body.
From this dataset, (Komura and Ishikawa, 2021) cre-
ated a set of 1,608,060 image patches with six dif-
ferent magnification levels i.e. 0.5µ/pixel, 0.6µ/pixel,
0.7µ/pixel, 0.8µ/pixel, 0.9µ/pixel, 1.0µ/pixel. As
these images are obtained from 32 different types of
cancer, they are categorized into 32 classes. NCT
(Kather et al., 2019) dataset comprises of 100,000
image patches of human colorectal cancer extratcted
from Hematoxylin and Eosin stained histological
images and normal tissue. The resolution of im-
ages are 224×224. These images are categorized
into seven classes which are Adipose(ADI), back-
ground(BACK), debris(DEB), lymphocytes(LYM),
mucus(MUC), smooth muscle(MUS), normal colon
mucosa (NORM), cancer-associated stroma (STR)
and colorectal adenocarcinoma epithelium (TUM).
LC25000 (Borkowski et al., 2019) or Lung and
Colon Histopathological dataset contains 25000 im-
age patches, As the name suggests, it comprises of
images from lung and colon cancer. The resolution of
the images is 768×768. they are categorized into five
classes. Three classes belong to lung cancer and the
remaining two classes belong to colon cancer. The
classes are benign colon tissues, colon adenocarci-
noma, lung squamous cell carcinoma and benign lung
tissues. CRC-TP (Javed et al., 2020b) is also a colon
cancer dataset. It consists of 280,000 image patches
categorized into six classes. They are tumor, stroma,
complex stroma muscle, debris, inflammatory and be-
nign. The images dimensions are 150×150. The
dataset prepared by Komura et al. is used as training
dataset. As the dataset is huge, a formidable few-shot
backbone network can be expected after training on it.
Rest other dataset are used as testing set. In few-shot
learning, the test and training set are disjoint.
4.2 Results and Discussion
All the experiments are conducted on an NVIDIA
A100 in pytorch. Two types of training regimes are
followed: Episodic training and standard training.
Standard training trains across an entire dataset with
abundant examples per class. In contrast, episodic
training, designed for few-shot learning, adapts a
model to quickly generalize from small episodes con-
taining very few examples of new classes. Models
such as Protonet (Snell et al., 2017), MAML (Finn
et al., 2017), DeepEMD (Zhang et al., 2020) follow
episodic training. SimpleShot (Wang et al., 2019) and
LaplacianShot (Ziko et al., 2020) follow the standard
training procedure.
In episodic training, the methods are trained for
120 epochs. In each epoch, 600 episodes are ran-
domly selected from the training set. The models are
trained on 5-way 1-shot, 5-way 5-shot and 5-way 10-
shot scenarios separately. In all these cases, the num-
ber of query images are set to 15. The initial learning
rate is set to 1e-3. γ is set to 0.1. The batch size is
set to 1 episode. During 5-way 1-shot, 5-way 5-shot
and 5-way 5-shot training, the number of images in
a batch are (5×1+5×15) 80, (5×5+5×15) 100 and
(5×10+5×15) 125 respectively. Few-shot methods
that follow standard CNN training have been trained
for 150 epochs. The batch size is set to 512. The ini-
tial learning rate, weight decay are initialized as 0.05
and 5e-4. Resnet18 is used as a backbone network in
both episodic and training procedure.
All the trained models follow meta-testing. In
this procedure, 5000 episodes are randomly sampled
from CRC-TP, NCT and LC25000 dataset. In all
K-shot testing scenarios, the number of query im-
ages are fixed as 15 in each episode. The results on
1-shot, 5-shot, 10-shot on different datasets are re-
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights
249
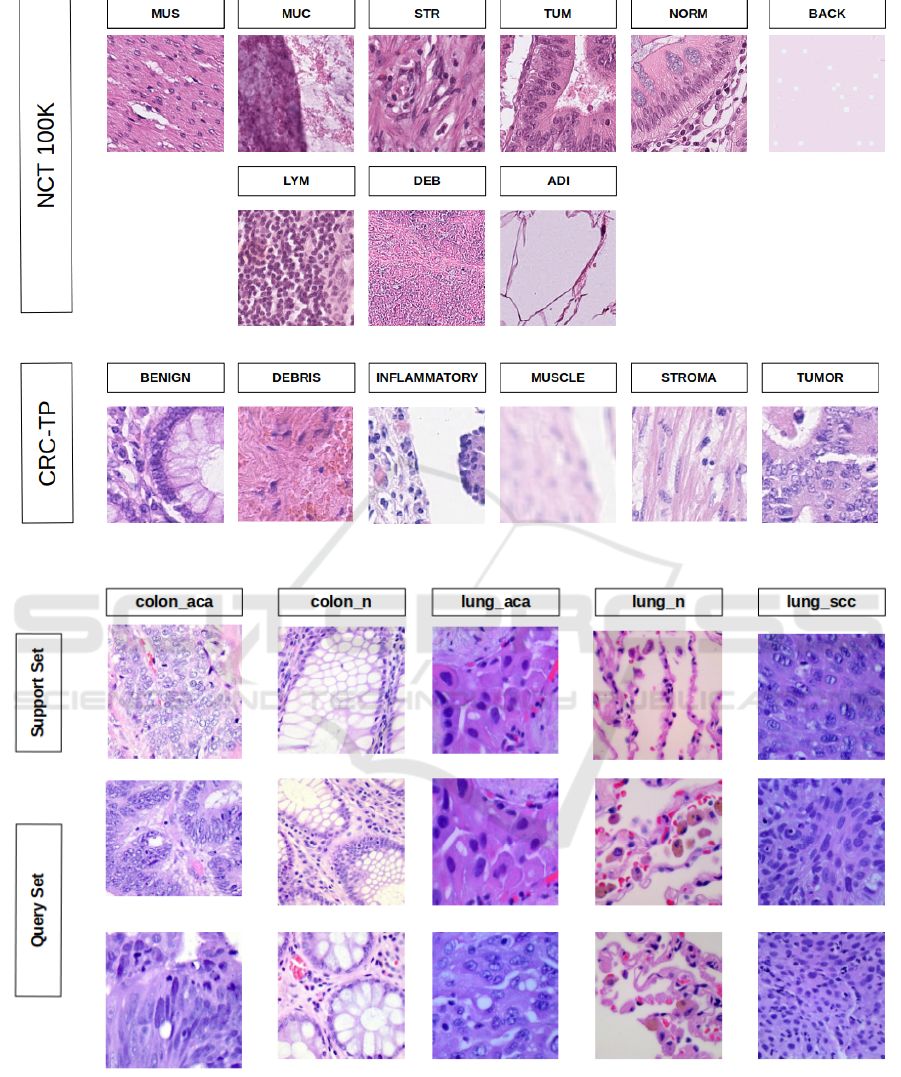
Figure 2: Snapshot of sample images of each class from NCT (top 2 rows) and CRC-TP (bottom row) of FHIST dataset.
Figure 3: An example of 5-way 1-shot 2-query episode. The first row represents the support set from 5 different classes of
LC25000 dataset. The last two rows represent the query set.
ported in the Table 1. The results we achieved by
employing few-shot methods on the histology dataset
align to a great extent with the outcomes obtained
from the datasets of natural images, but there were
a few surprises as well. As anticipated, few-shot
techniques trained using standard fine-tuning such
as SimpleShot and LaplacianShot yielded better re-
sults compared to approaches that adhered to episodic
BIOIMAGING 2024 - 11th International Conference on Bioimaging
250
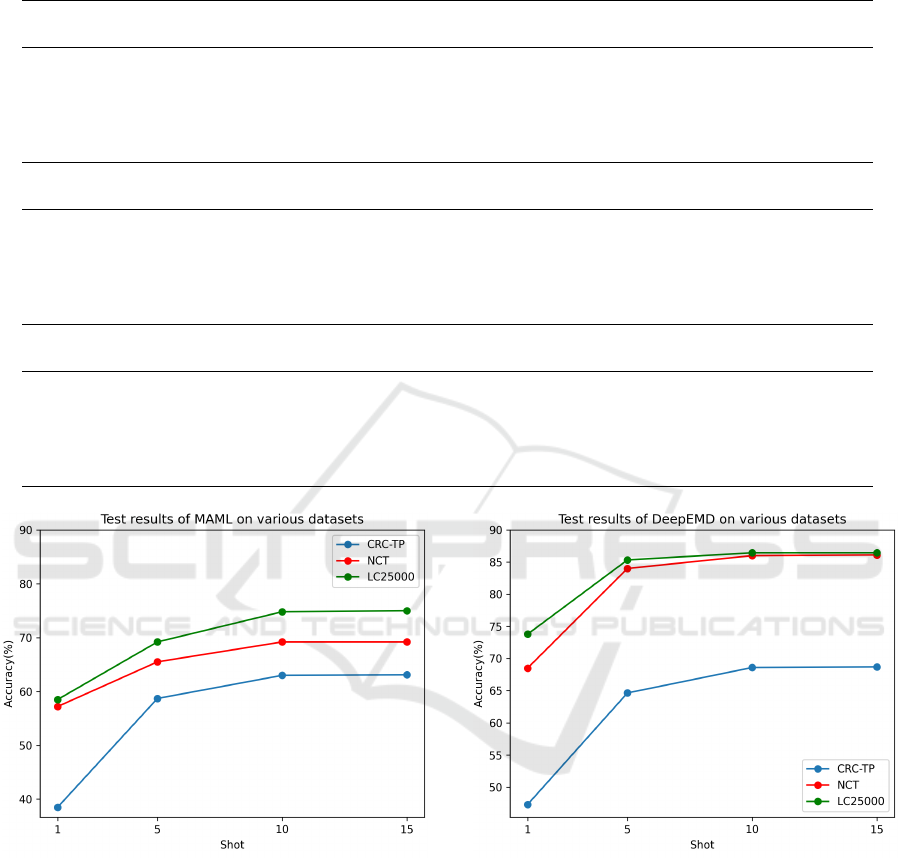
Table 1: Table 1: Accuracy(%) on three different datasets; CRC-TP (Javed et al., 2020b), NCT (Kather et al., 2019) and
LC25000 (Borkowski et al., 2019). The few-shot models are trained on the dataset proposed by (Komura and Ishikawa,
2021).
CRC-TP
Method Training Method 5-way 1-shot 5-way 5-shot 5-way 10-shot
MAML (Finn et al., 2017) Episodic 38.5 58.7 63.0
ProtoNet (Snell et al., 2017) Episodic 43.8 63.6 68.3
DeepEMD (Zhang et al., 2020) Episodic 47.3 64.6 68.6
SimpleShot (Wang et al., 2019) Standard 47.9 66.9 71.4
LaplacianShot (Ziko et al., 2020) Standard 48.5 68.0 72.8
NCT
Method Training Method 5-way 1-shot 5-way 5-shot 5-way 10-shot
MAML (Finn et al., 2017) Episodic 57.2 65.5 69.2
ProtoNet (Snell et al., 2017) Episodic 62.6 80.9 84.9
DeepEMD (Zhang et al., 2020) Episodic 68.5 84.0 86.0
SimpleShot (Wang et al., 2019) Standard 71.2 85.5 88.2
LaplacianShot (Ziko et al., 2020) Standard 71.8 86.9 89.5
LC25000
Method Training Method 5-way 1-shot 5way 5-shot 5-way 10-shot
MAML (Finn et al., 2017) Episodic 58.5 69.2 74.8
ProtoNet (Snell et al., 2017) Episodic 67.2 84.8 86.2
DeepEMD (Zhang et al., 2020) Episodic 73.8 85.3 86.4
SimpleShot (Wang et al., 2019) Standard 66.4 83.6 87.2
LaplacianShot (Ziko et al., 2020) Standard 67.5 84.2 87.9
Figure 4: Accuracy plot of MAML on different datasets.
training such as MAML, ProtoNet and DeepEMD.
The possible explanation for standard training pro-
cedures outperforming episodic training procedures
is due to training of these methods on the substan-
tial size of the dataset proposed by Komura et al.,
which includes cancer image patches from 32 diverse
classes or organs. For histopathology images, the re-
sults obtained from ProtoNet are comparable to those
achieved with DeepEMD. Considering that for natu-
ral images, DeepEMD exhibits significantly superior
performance compared to ProtoNet. MAML, a few-
shot learning-to-learn paradigm designed to learn and
Figure 5: Accuracy plot of DeepEMD on different datasets.
adapt quickly, does not appear to be well-suited for
histopathology images. As the number of shots are in-
creased in all few-shot methods, there is an observed
tendency for the results to reach a saturation point af-
ter 10 shots, as observed from the above plots. In
10-shot test scenarios, the accuracies of all methods,
except MAML, across all datasets, excluding CRC-
TP, fell within the range of 85% to 90%. In the con-
text of complete training, (Javed et al., 2020a) re-
ported an accuracy of 84.1% on the CRC-TP dataset.
Likewise, (Kather et al., 2019) achieved an accuracy
of 94.3% on the NCT dataset, and (Sarwinda et al.,
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights
251

Figure 6: Accuracy plot of Laplacianshot on different
datasets.
2020) reported an accuracy of 98.5% on the LC25000
dataset. The highest accuracy attained through the 10-
shot method is comparable to that achieved through
complete training.
5 CONCLUSION
In conclusion, this research has explored the appli-
cation of few-shot classification in the domain of
histopathology images. Despite the growing promi-
nence of few-shot learning in image classification, its
application in histopathology has remained relatively
uncharted. This study has specifically addressed the
challenges arising from the scarcity of labeled data
in medical imaging. Through the utilization of cer-
tain state-of-the-art few-shot learning methods, we
evaluated their performance across various scenarios
within the histology data domain. The consideration
of four histopathology datasets has contributed sig-
nificantly to our broader understanding of few-shot
histopathology image classification. The evaluation
process, scrutinizing 5-way 1-shot, 5-way 5-shot, and
5-way 10-shot scenarios, employing state-of-the-art
classification techniques, has yielded remarkable re-
sults. The selected methods demonstrated exceptional
capabilities, achieving accuracies close to 90% in the
5-way 10-shot scenarios. The top accuracy obtained
through the 10-shot method closely aligns with the
accuracy achieved through full training, emphasising
the effectiveness of Few-Shot Learning particularly
when ample pre-training data is not accessible. These
insights significantly contribute to the ongoing dis-
course surrounding the optimization of classification
techniques for medical imaging, particularly in sce-
narios where labeled data is limited.
Future work in the application of few-shot classifi-
cation in histopathology images could focus on multi-
modal image classification, transfer learning, and pre-
training, as well as the development of explainabil-
ity and interpretability methods. Few-shot learning
in multi-modal medical image classification involves
training models to accurately classify medical im-
ages with limited examples per pathology or disease,
leveraging information from various modalities like
visual (X-rays or MRIs), textual (medical reports or
descriptions), and potentially sensor data. In medi-
cal scenarios where obtaining a large labeled dataset
for every pathology is challenging, this approach be-
comes crucial. By combining information from dif-
ferent modalities and addressing the challenges posed
by limited examples, the model aims to make accurate
predictions in medical image classification tasks, par-
ticularly where traditional deep learning models may
struggle due to the scarcity of labeled medical data.
Additionally, researchers can explore innovative data
augmentation techniques and active learning strate-
gies. Improving robustness to image variability, fos-
tering collaboration with domain experts, establishing
benchmark datasets, and addressing real-time imple-
mentation challenges are also crucial for advancing
the field. Overall, these directions aim to enhance
diagnostic accuracy and efficiency in histopathology
through the effective use of limited labeled data and
the integration of machine learning into clinical work-
flows.
REFERENCES
Borkowski, A. A., Bui, M. M., Thomas, L. B., Wil-
son, C. P., DeLand, L. A., and Mastorides, S. M.
(2019). Lung and colon cancer histopathological im-
age dataset (lc25000).
Chen, X., Yao, L., Zhou, T., Dong, J., and Zhang, Y. (2021).
Momentum contrastive learning for few-shot covid-19
diagnosis from chest ct images. Pattern Recognition,
113:107826.
Finn, C., Abbeel, P., and Levine, S. (2017). Model-agnostic
meta-learning for fast adaptation of deep networks.
In International Conference on Machine Learning
(ICML).
Javed, S., Mahmood, A., Fraz, M. M., Koohbanani, N. A.,
Benes, K., Tsang, Y.-W., Hewitt, K., Epstein, D.,
Snead, D., and Rajpoot, N. (2020a). Cellular com-
munity detection for tissue phenotyping in colorec-
tal cancer histology images. Medical Image Analysis,
63:101696.
Javed, S., Mahmood, A., Werghi, N., Benes, K., and Ra-
jpoot, N. (2020b). Multiplex cellular communities in
multi-gigapixel colorectal cancer histology images for
tissue phenotyping. IEEE Transactions on Image Pro-
cessing, 29:9204–9219.
BIOIMAGING 2024 - 11th International Conference on Bioimaging
252

Kather, J. N., Krisam, J., Charoentong, P., Luedde, T., Her-
pel, E., Weis, C.-A., Gaiser, T., Marx, A., Valous,
N. A., Ferber, D., Jansen, L., Reyes-Aldasoro, C. C.,
Z
¨
ornig, I., J
¨
ager, D., Brenner, H., Chang-Claude, J.,
Hoffmeister, M., and Halama, N. (2019). Predicting
survival from colorectal cancer histology slides us-
ing deep learning: A retrospective multicenter study.
PLoS Med, 16(1):e1002730.
Kim, Y. J., Jang, H., Lee, K., Park, S., Min, S.-G., Hong,
C., Park, J. H., Lee, K., Kim, J., Hong, W., et al.
(2021). Paip 2019: Liver cancer segmentation chal-
lenge. Medical Image Analysis, 67:101854.
Komura, D. and Ishikawa, S. (2021). Histology images
from uniform tumor regions in tcga whole slide im-
ages. Cell Reports, 38(9):110424.
Mahajan, K., Sharma, M., and Vig, L. (2020). Meta-
dermdiagnosis: Few-shot skin disease identification
using meta-learning. In Proceedings of the IEEE/CVF
Conference on Computer Vision and Pattern Recogni-
tion Workshops, pages 730–731.
Medela, A., Picon, A., Saratxaga, C. L., Belar, O., Cabez
´
on,
V., Cicchi, R., Bilbao, R., and Glover, B. (2019). Few-
shot learning in histopathological images: reducing
the need of labeled data on biological datasets. In
2019 IEEE 16th International Symposium on Biomed-
ical Imaging (ISBI 2019), pages 1860–1864. IEEE.
Nichol, A. and Schulman, J. (2018). Reptile: A
scalable metalearning algorithm. arXiv preprint
arXiv:1803.02999.
Ouyang, C., Biffi, C., Chen, C., Kart, T., Qiu, H., and
Rueckert, D. (2020). Self-supervision with superpix-
els: Training few-shot medical image segmentation
without annotation. In European Conference on Com-
puter Vision, pages 762–780. Springer.
Sarwinda, D., Bustamam, A., Paradisa, R. H., Argyadiva,
T., and Mangunwardoyo, W. (2020). Analysis of deep
feature extraction for colorectal cancer detection. In
2020 4th International Conference on Informatics and
Computational Sciences (ICICoS), pages 1–5. IEEE.
Shakeri, F., Boudiaf, M., Mohammadi, S., Sheth, I., Havaei,
M., Ben Ayed, I., and Kahou, S. E. (2022). Fhist: A
benchmark for few-shot classification of histological
images. arXiv.
Snell, J., Swersky, K., and Zemel, R. (2017). Prototypical
networks for few-shot learning. In Advances in Neural
Information Processing Systems (NeurIPS).
Spanhol, F. A., Oliveira, L. S., Petitjean, C., and Heutte, L.
(2016). A dataset for breast cancer histopathological
image classification. IEEE Transactions on Biomedi-
cal Engineering, 63(7):1455–1462.
tcga (2005). The cancer genome atlas.
https://www.cancer.gov/ccg/research/genome-
sequencing/tcga. Accessed on: 2023-11-30.
Teng, H., Zhang, W., Wei, J., Lv, L., Tang, L., Fu, C.-
C., Cai, Y., Qin, G., Ye, M., and Fang, Qu, e. a.
(2021). Few-shot learning on the diagnosis of lym-
phatic metastasis of lung carcinoma. Research Square.
Wang, Y., Chao, W.-L., Weinberger, K. Q., and van der
Maaten, L. (2019). Simpleshot: Revisiting nearest-
neighbor classification for few-shot learning. arXiv
preprint arXiv:1911.04623.
Yang, J., Chen, H., Yan, J., Chen, X., and Yao, J.
(2022). Towards better understanding and better
generalization of few-shot classification in histology
images with contrastive learning. arXiv preprint
arXiv:2202.09059.
Yu, Q., Dang, K., Tajbakhsh, N., Terzopoulos, D., and Ding,
X. (2021). A location-sensitive local prototype net-
work for few-shot medical image segmentation. In
2021 IEEE 18th International Symposium on Biomed-
ical Imaging (ISBI), pages 262–266. IEEE.
Zhang, C., Cai, Y., Lin, G., and Shen, C. (2020). Deepemd:
Few-shot image classification with differentiable earth
mover’s distance and structured classifiers. In Con-
ference on Computer Vision and Pattern Recognition
(CVPR).
Ziko, I. M., Dolz, J., Granger, E., and Ben Ayed, I. (2020).
Laplacian regularized few-shot learning. In Interna-
tional Conference on Machine Learning (ICML).
Few-Shot Histopathology Image Classification: Evaluating State-of-the-Art Methods and Unveiling Performance Insights
253
