
Deep Learning in Digital Breast Pathology
Madison Rose
1
, Joseph Geradts
2 a
and Nic Herndon
1 b
1
Department of Computer Science, East Carolina University, Greenville, North Carolina, U.S.A.
2
Department of Pathology, Brody School of Medicine, East Carolina Univesity, Greenville, North Carolina, U.S.A.
Keywords:
Breast Cancer, Machine Learning, Deep Learning, Digital Pathology, Convolutional Neural Networks, Whole
Slide Imaging.
Abstract:
The development of scanners capable of whole slide imaging has transformed digital pathology. There have
been many benefits to being able to digitize a stained-glass slide from a tissue sample, but perhaps the most
impactful one has been the introduction of machine learning in digital pathology. This has the potential to
revolutionize the field through increased diagnostic accuracy as well as reduced workload on pathologists. In
the last few years, a wide range of machine learning techniques have been applied to various tasks in digital
pathology, with deep learning and convolutional neural networks being arguably the most popular choice.
Breast cancer, as one of the most common cancers among women worldwide, has been a topic of wide interest
since hematoxylin and eosin-stained (H&E)-stained slides can be used for breast cancer diagnosis. This paper
summarizes key advancements in digital breast pathology with a focus on whole slide image analysis and
provides insight into popular methods to overcome key challenges in the industry.
1 INTRODUCTION
Advancements in whole slide imaging (WSI) have
paved the way for digital pathology. This has driven
the increasing demand for more research into using
machine learning for whole slide image analysis. This
paper provides an overview of the main aspects of
deep learning in digital breast pathology. Background
information is included that can be used to gain an
understanding of the field. Key advancements, tools,
and insight into popular methods for overcoming key
challenges are discussed. While digital pathology is a
large field, the focus here will be on analysis of whole
slide images through deep learning techniques.
1.1 Whole Slide Imaging
Whole slide imaging shows many potential benefits
compared to its glass slide counterpart. Digitized
slides allow remote users to view slides for secondary
or even primary diagnosis. Digitization is also useful
for archiving and preserving samples, which is im-
portant since physical samples degrade over time. An
additional benefit of better archiving of tissue samples
a
https://orcid.org/0009-0002-3817-8499
b
https://orcid.org/0000-0001-9712-148X
is the preservation of rare specimens. Since digitized
slides can be accessed remotely, WSI also provides
the opportunity to make advancements in standardiz-
ing training for pathologists. It is important to con-
sider that while WSI can also be used for diagnosis,
there are still factors that can affect diagnostic accu-
racy from digitized images. While whole slide im-
ages are approved for diagnosis, some discrepancies
still make glass slide viewing the standard for diagno-
sis. These issues stem from poor image quality and
bad focus. Some specific microscopic details such as
mitotic figures that may be needed for analysis can
also be difficult to identify on the digitized images,
in some cases, due to faint scanning. However, it is
important to note that even glass to glass slide stud-
ies can show discrepancies due to observer variability,
among other factors (Pantanowitz et al., 2015).
1.2 Digital Pathology Tasks
In histopathology image analysis, three main tasks for
machine learning have emerged: classification, seg-
mentation, and object detection. Classification in-
volves analyzing an image and giving the image a la-
bel, sorting it into a class. There are two types of clas-
sification, binary and multi-class classification (Gupta
et al., 2022b). In binary classification, there are only
404
Rose, M., Geradts, J. and Herndon, N.
Deep Learning in Digital Breast Pathology.
DOI: 10.5220/0012576100003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 1, pages 404-414
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

two possible labels or classes for an image. In con-
trast, multi-class labeling has three or more possible
labels for a given image. A study by Araujo et al.
(2017) displayed both binary and multi-class classi-
fication. They used a convolutional neural network
for multi-class classification of breast biopsy images
into one of four categories: normal, benign, in situ
carcinoma, and invasive carcinoma. They also per-
formed binary classification into carcinoma and non-
carcinoma. More specific labelling as done in multi-
class classification is often very useful in medical di-
agnosis. However, due to using an increased num-
ber of classes, multi-class models often require more
complexity than binary models, which can impact
their accuracy. For example, in the study mentioned
above, the four-class model scored 65% accuracy on
test data compared to the binary class model achiev-
ing 77%.
Segmentation aims to separate parts of the images
– often cancerous cells vs. noncancerous cells. Object
detection focuses on finding landmarks in an image,
like individual cells or nuclei. In this paper, segmen-
tation and object detection will briefly be discussed,
while classification tasks will be the main focus.
2 BACKGROUND
2.1 Breast Cancer
In 2023, breast cancer accounted for 31% of all fe-
male cancers, making it one of the most common
cancers among women. Breast cancer occurrence
rates have been steadily increasing since the 2000s by
about 0.5% per year. Improvements in treatment have
seen the mortality rate for breast cancer decrease de-
spite the increase in incidence (Siegel et al., 2023). It
is well documented that early diagnosis and interven-
tion can greatly improve survival rates in breast can-
cer patients. Smaller tumors have notably better long-
term survival rates than larger tumors (Bhushan et al.,
2021). Many techniques are used to screen for and
diagnose breast cancer, including mammography and
ultrasonography (Watkins, 2019). However, while
these are helpful in screening and early detection of
breast cancer, a breast biopsy is the only definitive
method for diagnosing breast cancer (Nounou et al.,
2015). Tissue samples can provide information about
tumor type, grade, and biomarker status. A triple as-
sessment is often used to evaluate patients, consisting
of clinical evaluation and imaging in addition to a tis-
sue biopsy (Alkabban and Ferguson, 2020).
Once biopsied tissue is collected, it is fixed, pro-
cessed, sectioned, and stained to color different parts
of the cells in the tissue. Hematoxylin and eosin
(H&E) staining is considered the gold standard in
breast tissue biopsies and has been around for over
100 years (Huang et al., 2023). When H&E stain-
ing is performed, different cell parts will look distinct
based on which type of dye they have an affinity for.
Hematoxylin is a basic dye whereas eosin is an acidic
dye. Cell structures such as nuclei that have an affin-
ity for hematoxylin appear blue after staining. Struc-
tures such as cytoplasm that have an affinity for eosin
appear pink after staining. Structures with an affinity
for both basic and acidic dyes will appear purple after
staining (Chan, 2014; Bancroft and Layton, 2019).
2.2 Whole Slide Image Resolutions
Whole slide image scanners operate by capturing im-
ages of tissue sections tile by tile. The whole im-
age is reconstructed at the end. These scans can be
performed at multiple magnifications which increases
image detail. 20x magnification is a common scan ob-
jective and is adequate for typical viewing. However,
some types of slides require more detail and need
higher levels of magnification such as 40x (Zarella
et al., 2019).
2.3 Machine Learning Types
There are many learning types in machine learning.
The two main types are supervised and unsupervised
learning. These types differ based on the types of data
they receive.
Supervised learning supplies a machine learning
model with input data and its expected output. What
this will look like can vary depending on the task be-
ing performed. In cases of classification, the output is
typically a label. In segmentation tasks, often a mask
of pixels is used as the ground truth label (Khened
et al., 2021). In object detection, bounding boxes in
certain parts of the image are typically provided (Li
et al., 2019). The model will then try to predict the
desired output given only the input. The revolution-
ary idea behind deep learning models is that they can
compare their original predictions with the expected
output and internally modify their configurations to
see more accurate predictions during the next round
of training or testing. This is done through a method
called backpropagation (LeCun et al., 2015). Convo-
lutional neural networks are a popular type of super-
vised machine learning model.
In contrast to supervised learning, unsupervised
learning uses unlabeled data. Unsupervised learning
models will group data based on patterns and similar-
ities but are unable to provide a label (Gupta et al.,
Deep Learning in Digital Breast Pathology
405

Max-Pool Convolution Max-Pool Fully Connected
8@128x128
8@64x64
24@48x48
24@16x16
1x256
1x128
Figure 1: An example of a convolutional neural network architecture with convolutional layers, pooling layers, and fully
connected layers. As the input moves through the CNN, it continues to be reduced in size. Figure generated with NN-
SVG (LeNail, 2019).
2022b). This can be useful in histopathological im-
age analysis for detecting patterns in images that may
not be currently recognized by pathologists. Addi-
tionally, unsupervised learning can be used for fea-
ture extraction on histopathologic images (Sari and
Gunduz-Demir, 2019).
2.4 Deep Learning and CNNs
Deep learning is a subfield of machine learning that
focuses on using nodes to form a neural network
(Gupta et al., 2022a). These neural networks were
originally inspired by the human brain. The nodes in
neural networks are also referred to as neurons since
they mimic how neurons function in a human brain
(O’Shea and Nash, 2015). Deep learning has be-
come increasingly popular for several reasons. First,
these models are successful at a wide variety of tasks
such as natural language processing, speech and audio
processing, and digital image processing. The way
deep learning algorithms extract features decreases
the amount of domain knowledge and work needed
by researchers (Pouyanfar et al., 2018). Convolu-
tional neural networks (CNNs) are a type of neural
network and are particularly good at image recogni-
tion tasks. CNNs take in image pixel values as in-
put and pass these values through a series of layers
while performing various operations on the images.
Convolutional neural network architecture consists of
three main types of layers: convolutional, pooling,
and fully connected layers, which can be observed in
Figure 1. In convolutional layers, two dimensional
filters are applied to the image data to extract fea-
tures such as edges, objects, and colors. An exam-
ple of a convolution can be seen in Figure 2. These
features are used to create a feature map. Convo-
lutional layers are often paired with activation func-
tions, such as ReLU (rectified linear units), which
improve speed and performance by removing nega-
tive values after a convolution has been performed
(Krizhevsky et al., 2017; Zhang et al., 2021). The
pooling layer does downsampling which reduces the
number of parameters used while trying to maintain
the features (O’Shea and Nash, 2015; Zhang et al.,
2021). Lastly, fully connected layers take inputs re-
ceived by previous layers and connect them to activa-
tion units to produce output (Zhang et al., 2021).
2.5 CNN History
In 1989, Yann Lecun introduced LeNet-5, a convo-
lutional neural network originally designed to recog-
nize handwriting digits (LeCun et al., 1998). It be-
came one of the first widely recognized published
convolutional neural networks due to its performance
with an error rate of 0.95% on the test set of 16x16
pixel handwriting digit images (Zhang et al., 2021;
LeCun et al., 1998). LeNet-5 was also revolution-
ary for its use of backpropagation to reconfigure its
own internal weights using gradient descent (LeCun
et al., 1998). In 2012, convolutional neural networks
surged in popularity after AlexNet won the ImageNet
Large Scale Visual Recognition Challenge (ILSVRC)
(Krizhevsky et al., 2017). Since then, the devel-
opment of new CNN architectures rapidly expanded
with the emergence of VGG16/VGG19 (Simonyan
and Zisserman, 2015), Resnet (He et al., 2016) and
Inception (Szegedy et al., 2015). ImageNet is a pop-
ular dataset for training and benchmarking convolu-
tional neural networks and contains millions of an-
notated natural images (Deng et al., 2009). Classi-
fication and object detection are common computer
vision tasks and are included in popular challenges
such as ILSVRC (Russakovsky et al., 2015). Con-
volutional neural networks are particularly skilled at
computer vision tasks and in turn have been applied
to a wide range of medical imaging tasks.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
406
01010
01010
11110
01000
01000
010
010
010
313
312
311
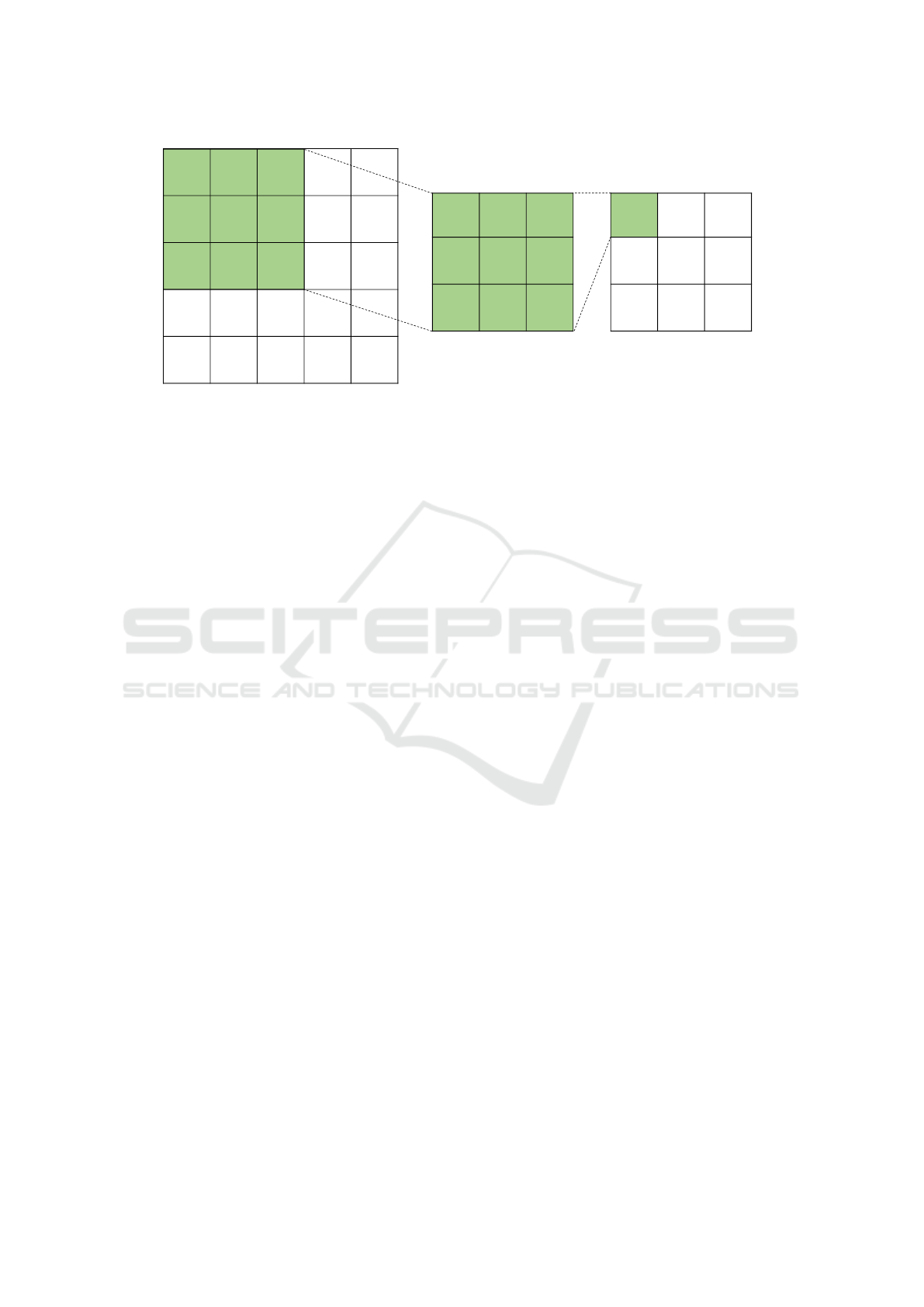
Figure 2: An example of the convolution operation. This convolution uses a 3x3 filter (center grid) on an image of size 5x5
(left grid) with a stride of 1 and padding of 0. The result of the convolution is the rightmost grid. The filter used here is an
example of a filter that detects vertical lines.
2.6 Modern CNNs
One of the most popular modern CNNs is VGG16.
The Visual Geometry Group (VGG) submitted
VGG16 to the ILSVRC in 2014. It proposed an in-
crease in CNN depth and was pretrained on the Ima-
geNet dataset. This model varied from its predeces-
sors by using stacks of smaller 3x3 receptive fields in-
stead of larger 11x11 or 7x7 receptive fields. This de-
creased the number of parameters throughout the net-
work. Small convolutions had previously been tried
but no other CNNs that used these smaller filters were
as deep as VGG16, which boasted sixteen layers as its
name suggests. The VGG were able to determine that
a larger depth increased the classification accuracy.
VGG19 was described in the same paper as VGG16
and follows a similar architecture but with nineteen
layers as opposed to sixteen (Simonyan and Zisser-
man, 2015).
Another popular modern CNN is Inception (also
known as GoogleLeNet). This neural network com-
peted in the ILSVRC 2014 challenge and acheived
high performance. Inception implements wider lay-
ers as opposed to making the entire network deeper
with more layers (Szegedy et al., 2015). This network
depth is why Inception is often referred to as a deep
convolutional neural network, while other CNNs like
VGG16 are considered shallow. This greatly reduces
the computational cost of the network in compari-
son to other modern CNNs such as VGG16 (Szegedy
et al., 2016). Like VGG16, Inception uses many
smaller filters in place of larger filters to reduce the
number of parameters needed. Inception was also
trained on the ImageNet dataset and achieved remark-
able error rates while cutting computational costs
(Szegedy et al., 2015).
Additional modern CNNs that will be discussed in
later sections include ResNet and MobileNet. ResNet
implements residual functions to reference layer in-
put, which was shown to make optimization eas-
ier. Additionally, this allowed for increased net-
work depth with lower complexity and increased ac-
curacy (He et al., 2016). MobileNet was built to be
a lightweight deep neural network by utilizing depth-
wise separable convolutions (Howard et al., 2017).
2.7 Whole Slide Image Annotations
In whole slide imaging, there are three main anno-
tation types. These types are patch level (sometimes
called pixel level), slide level, and patient level. These
three levels are illustrated in Figure 3. Each type of
annotation can be useful for different tasks. These
annotations can also be organized into a hierarchy of
specificity.
Patch level annotation is the most specific level of
annotation for whole slide images. Patch level anno-
tations guarantee that when taking patches from WSI,
every patch is fully annotated. Examples of patch
level annotations include instances where each patch
has its own classification label, segmentation mask,
or bounding boxes (Ciga and Martel, 2021). One
example of a segmentation mask would be when a
pathologist identifies cancerous regions within a tis-
sue sample and annotates all regions or pixels con-
taining cancerous cells (Khened et al., 2021). Patch
level annotations are extremely helpful for segmenta-
tion tasks as they provide the ability for high supervi-
sion. However, these annotations are much more time
consuming than slide level annotations and therefore
are less commonly available. Often, training will be
performed at a lower annotation level such as patch
level while expecting a final output at a higher level
such as slide level or patient level (Dimitriou et al.,
Deep Learning in Digital Breast Pathology
407

2019). Aggregation from lower to higher annotation
levels is discussed in Section 4.4.
The next level of annotation is at slide level.
Slide level annotations provide one label per whole
slide. For example, a whole slide image with a slide
level annotation may be labeled carcinoma vs non-
carcinoma (Araujo et al., 2017). Slide level annota-
tions are much less time consuming to do than pixel
level annotations and are therefore more abundant. If
looking at individual patches, as is common in WSI
analysis, it is possible for a patch to not match its slide
level annotation (Dimitriou et al., 2019; Hou et al.,
2016). This is why aggregation is needed when mov-
ing from one level during training to another at pre-
diction time.
The least specific level of annotations is patient
level. Patient level annotation is similar to slide level
annotations in that a single label/class is provided,
however, this label is provided to a patient rather than
a specific slide. Patients may have multiple images.
For example, in the CAMELYON17 dataset, each pa-
tient had 5 images (Litjens et al., 2018). Patient level
annotations mean that individual slides will not be
provided a label, but rather the patient, which means
the label applies to all slides associated with the pa-
tient. This is the least specific level of annotation be-
cause it is possible to incorrectly label a whole slide
(Dimitriou et al., 2019).
3 COMMON CHALLENGES
There are many unique challenges in pathology image
analysis when trying to apply deep learning. Solu-
tions to these challenges are the basis of many works
in the field.
3.1 Image Size
One major issue that must be addressed when trying
to apply any type of machine learning technique to
histopathology image analysis is the size of whole
slide images. Whole slide image scans are extremely
large, typically 100,000 x 100,000 pixels each (Dim-
itriou et al., 2019). With images this large, they are
not feasible for machine learning use without modi-
fications. For example, CNNs usually perform best
with smaller images around 224 x 224 pixels in size
(Ciga et al., 2021). Image compression would cer-
tainly be helpful but also has drawbacks including re-
duced image quality and distortion of important mark-
ers. It has been shown that there is a significant per-
formance decrease in benign vs. malignant breast
tissue classification once compression levels increase
past 32:1 (Krupinski et al., 2012). Even if extreme
downsampling were performed, the image would re-
main too large for use in a convolutional neural net-
work (Ciga et al., 2021). A common approach to ad-
dress this issue is to split the image into smaller im-
ages that would be more suitable for use by machine
learning models (Hou et al., 2016). These methods
are discussed in Section 4.3.
3.2 Data Availability
A lack of well-annotated and publicly available
training data is a well-known problem in digital
histopathology image analysis. Even when images
are available, domain knowledge is required to anno-
tate these images to make them suitable for analysis
via machine learning methods. Researchers have a
few options: use a publicly available dataset, or cre-
ate their own, using images provided to them by an
institution or pathologist. One of the most popular
datasets used in breast histopathology image analysis
is the CAMELYON dataset, which is a publicly avail-
able dataset of whole slide images along with their
associated pathologist annotations. This dataset was
collected from Dutch hospitals and contains 1,399
unique whole slide images totaling 2.95 terabytes.
Slides were scanned with three different scanners
based on which hospital they came from, with the ma-
jority of hospitals using the 3DHistech Pannoramic
Flash II 250 while the Hamamatsu NanoZoomer-
XR C12000-01 scanner and Philips Ultrafast Scanner
were both used by one hospital each. All 1,399 WSI
were annotated with a slide level label. Additionally,
399 slides from CAMELYON16 and 50 slides from
CAMELYON17 were also annotated at the patch level
(Litjens et al., 2018).
As of the 2018 publication about the dataset,
it had already been accessed by over 1000 users.
Along with the dataset came the CAMELYON16 and
CAMELYON17 challenges, which encouraged teams
to design models to classify breast cancer metastases
(Litjens et al., 2018). Although the main goal of
the CAMELYON challenges is breast cancer metas-
tases detection, the dataset is widely used by re-
searchers interested in a variety of breast histopathol-
ogy image tasks. As of December 2023, the CAME-
LYON17 challenge website boasts 205 submissions
to the leaderboard with 1,943 total participants. The
current top 10 submissions on the leaderboard all
boast Cohen-Kappa scores of greater than .90 when
evaluated by the CAMELYON team.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
408

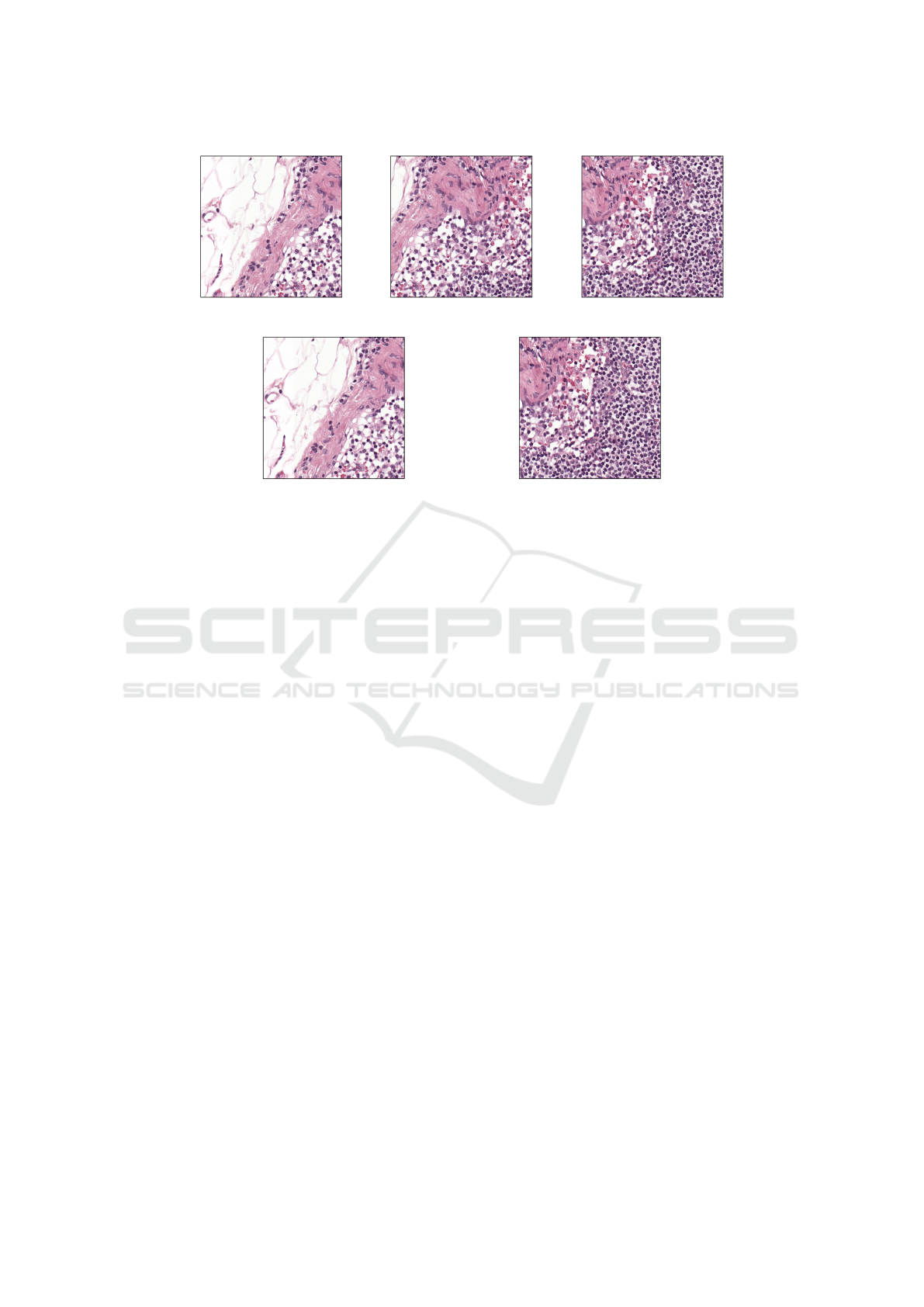
a) Image Patch b) Slide Image c) Collection of scans for a patient
Figure 3: Three main annotation types for whole slide images. (a) In patch level annotation, each image patch would have
its own classification label or would have a pixel annotation boundary. (b) In slide level annotation, there would be a single
classification label for the entire image. (c) In patient level annotation, there would be a single label associated with all five
images. Whole slide images come from the CAMELYON17 dataset (Litjens et al., 2018).
4 COMMON APPROACHES
4.1 Transfer Learning/Pretrained
Models
One downside to deep learning is the computational
complexity and the amount of well annotated data
needed. One technique that helps reduce model com-
plexity as well as the amount of domain specific an-
notated data needed is transfer learning (Wakili et al.,
2022). Transfer learning is another brain inspired
technique. It comes from the idea that knowledge in
one task can aid in performing a different, but some-
what related task. The use of transfer learning in con-
volutional neural networks is often used to pretrain
a CNN with large amounts of publicly available and
well annotated data, such as the ImageNet dataset.
Later, the CNN can be finetuned with domain spe-
cific data (Kim et al., 2022). Ultimately, this reduces
the amount of domain specific data needed since the
original weights will already be pretrained. Training
time is also reduced when using pretraining methods
since some portion of the training is already complete
(Gupta et al., 2022a). This is particularly useful in
fields such as digital pathology where there may be a
lack of widely available annotated data.
In a study comparing transfer learning methods in
medical imaging, Kim et al. (2022) defined four types
of transfer learning based on how the training is han-
dled after the original pretraining. The feature extrac-
tor method freezes the convolutional layers and only
retrains model weights in the fully connected layers.
The feature extractor hybrid also freezes the convo-
lutional layers but replaces the fully connected layers
with another machine learning model, such as a sup-
port vector machine (SVM). The fine-tuning method
unfreezes a few of the convolutional layers to be re-
trained. Finally, fine tuning from scratch completely
retrains the model on the new data. After analysis of
121 publications focused on using transfer learning on
convolutional neural networks with medical images,
they recommended the feature extractor approach and
then incrementally fine tuning the layers. Fine tun-
ing from scratch appeared to be a prevalent method
but did not show significant improvements in model
accuracy despite being much more computationally
expensive than other transfer learning methods (Kim
et al., 2022).
4.2 Common Models
There are many pretrained convolutional neural net-
works available for use. Some models such as Incep-
tion have become commonly used because of their
good performance. One review of medical imaging
using CNNs found that most works use multiple mod-
els. However, Inception was the leading model when
Deep Learning in Digital Breast Pathology
409

only one model was used (Kim et al., 2022).
While tumor detection is a common task in breast
digital pathology, it is not the only task that interests
researchers. One study attempted to predict early re-
currence from histopathological images. Early recur-
rence was defined as the return of a primary tumor
within three years of the original diagnosis. VGG16
pretrained on ImageNet was used in conjunction with
support vector machines (SVM). This approach ob-
served a 70.3% accuracy (67.7% sensitivity) using
within-patient validation (Shi et al., 2023). Another
study focused on predicting breast cancer recurrence
from whole slide images used six pretrained models
were used including VGG16, ResNet50, ResNet101,
Inception ResNet, EfficientB5 and Xception. Two
fully connected layers were added to help reduce the
computational load. Here, Xception was found to
have the highest accuracy on the training data (91%)
and was used for further testing where it achieved an
accuracy of 87% (Phan et al., 2021).
4.3 Image Patches
Due to the enormous size of whole slide images, one
common solution is to use patches of a whole slide
image rather than the entire image itself. However,
this adds another variable, what is the optimal patch
size? There isn’t a clear-cut answer and researchers
select different patch sizes based on their specific
needs. However, some patch sizes are more often
used and are selected as default values. When select-
ing patch size, it is important to consider several fac-
tors. Finding the optimal patch size is important be-
cause it plays a role in how long training takes and can
also impact model accuracy. Patch size often depends
on the overall goal of a work. For example, works
looking to perform slide level classification more of-
ten use larger patch sizes such as 512 x 512 and 1024
x 1024 pixels (Pinchaud, 2019; Khened et al., 2021;
Lee et al., 2021). This allows for more information to
be captured by each patch used in training and gives
a better overall view of the tissue elements and cell
architecture. However, other studies such as those fo-
cused on object detection and labeling individual cells
and nuclei, may decide to use smaller patch sizes.
Additionally, researchers need to decide whether they
will use overlapping or non-overlapping patches. An
example of the differences between overlapping and
non-overlapping patches can be found in Figure 4.
In the CAMELYON challenge there are a wide
range of approaches taken and this extends to selected
patch size. The top submission on the leaderboard for
CAMELYON17 uses a patch size of 704 x 704 pix-
els (Lee et al., 2021). However, the other submission
in the top 5 all use either 512 x 512 pixel patches,
1024 x 1024 pixel patches or some combination of
the two (Pinchaud, 2019; Khened et al., 2021; Lee
et al., 2021).
A study focusing on segmenting whole slide im-
ages from the CAMELYON dataset tried two en-
sembles with different patch sizes, 256 x 256 non-
overlapping patches and 1024 x 1024 overlapping
patches. The ensemble that used 1024 x 1024 over-
lapping patches performed slightly better than the en-
semble with 256 x 256 non-overlapping patches. This
study is currently a top 5 score on the CAMELYON17
leaderboard (Khened et al., 2021).
One work focused-on object detection of signet
ring cells. Images from 10 different organs were used,
with breast among them. Out of 127 images, each had
3 patches of size 2000 x 2000 selected for annotation
with a bounding box. A total of 12,381 signet ring
cells were annotated. However, due to overcrowd-
ing, some signet ring cells were not able to be anno-
tated. This work is also a top 5 scorer on the CAME-
LYON17 leaderboard (Li et al., 2019).
Another work used randomly selected 1000 x
1000 pixel sized patches for the task of tumor region
recognition. The patches had to be downsampled four
times to 224 x 224 to satisfy the requirements of their
selected model, MobileNetV2 (Huang et al., 2023).
There are many instances where a larger patch
size, such as 512 x 512, is initially chosen and then
cropped or resized to a smaller size like 128 x 128 to
make the image better match the selected model’s in-
put size (Phan et al., 2021). One work applied this,
originally selecting patch sizes of 512 x 512 before
randomly cropping to 448 x 448. The 448 x 448
pixel patches then went through dimension reduction
to achieve a size of 224 x 224 for training with the Ef-
ficientNet framework. This study found an improve-
ment in results in both slide level classification and
segmentation tasks with randomly cropped patches
(Ciga et al., 2021).
4.4 Annotation Aggregation
Training is often performed at a lower annotation
level such as patch level while expecting a final out-
put at a higher level such as slide level or patient level.
In these cases, aggregation is needed to combine the
results from many patches to achieve the output for
the higher level (Dimitriou et al., 2019). One study
converted from patch level to slide level predictions
for tumor and tumor bed detections. In this case, if
one or more patches were determined to be positive
or a tumor bed was detected, the entire WSI would
be labeled as tumor positive (Ciga et al., 2021). An-
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
410
(a)
(b)
Figure 4: An example of overlapping (a) and non-overlapping (b) patches. These patches cover the same image region, but
with overlapping, three patches are needed to cover the same area as two non-overlapping patches. The first and last patches
of (a) match the patches of (b), but the middle patch of (a) is a combination of the patches from (b). Note: these patches were
generated from whole slide images in the CAMELYON17 dataset (Litjens et al., 2018).
other study proposed a diffusion model for aggregat-
ing from patch level to slide level (Hou et al., 2016).
One study used patch level classification to ex-
tract features for slide level classification. 86.67% ac-
curacy was achieved using Inception for patch level
classification. An overall accuracy of 90.43% was
achieved for the slide level classification of normal,
benign, in situ carcinoma, and invasive carcinoma (Mi
et al., 2021).
4.5 Thresholding
When working with patches in an image, there are
hundreds of thousands of possible patches to be se-
lected depending on the patch size selected. Patch
selection methods vary between studies, with some
studies performing random patch selection while oth-
ers incorporate algorithms to select “best” patches
(Hou et al., 2016). However, one thing they all have
in common is avoiding irrelevant patches with no
cells and only background material. In most whole
slide images there are large background areas that
are irrelevant for image analysis (Veta et al., 2014).
With any patch selection technique, preprocessing
is typically performed to eliminate irrelevant back-
ground patches. Often, thresholding is used to sep-
arate the image background from the relevant mate-
rial. Thresholding is a technique that maps all im-
age pixels into one of two groups. This technique
is best used when there is high variance between an
image’s background and foreground. One popular
method of thresholding in whole slide imaging is the
Otsu thresholding technique. The Otsu threshold is
determined by finding the maximum inter-class vari-
ance (Otsu, 1979; Xu et al., 2011). While the Otsu
threshold is popular in whole slide image segmen-
tation of background and foreground, there are in-
stances where it is not as effective. For instance, in
Khened et al. (2021), the Otsu threshold could not
be used to segment the CAMELYON dataset due to
black regions within the WSI. Instead, the black pix-
els were changed to white first and then a median blur
filter of size 7x7 kernel was used prior to performing
the Otsu thresholding. (Khened et al., 2021).
While the Otsu threshold is popular, it is not the
only thresholding method used. One study used a cus-
tom threshold to segment areas without nuclei from
the image as regions that lack nuclei are not relevant
in tumor identification. Their thresholding removed
any regions that met the following criteria: hue be-
tween 0.5 and 0.65, saturation greater than 0.1, and
value between 0.5 and 0.9. These bounds were de-
rived from experimentation with whole slide images,
and patches with at least 25% foreground were in-
cluded in the study (Ciga et al., 2021). Neural net-
works have also been applied to the segmentation task
of tissue sample from its background with success
(Alomari et al., 2009).
Deep Learning in Digital Breast Pathology
411

4.6 Staining Techniques
Another common issue is variability in slide images.
Although hematoxylin and eosin (H&E) staining is
the most commonly used staining technique, it does
have some drawbacks. This staining technique does
not label the nuclei and cytoplasm in cells exclusively.
Sometimes other staining techniques are used such as
fluorescent staining, which is more common in tis-
sue morphology clinical research. Whole slide image
datasets using H&E stained slides that are publicly
available are already scarce, so these alternative stain-
ing methods have limited annotated data to be used
for machine learning. One study attempted to bridge
H&E stained images with fluorescent stained images.
Due to color variations, cross analysis can be diffi-
cult. Through methods involving color normaliza-
tion techniques for preprocessing and nuclei extrac-
tion, they were able to create a model that had 89.6%
accuracy in identifying tumor regions in H&E images
and 80.5% accuracy in identifying those same regions
in fluorescent stained slides. Further work into cross
analysis between staining methods will increase the
amount of available data for all types of stained whole
slide image analysis (Huang et al., 2023).
4.7 Tools for Whole Slide Image
Analysis
Several approaches have produced free and open-
source software to aid others conducting research in
this area. One available tool is DigiPathAI. This
is a generalized deep learning-based framework for
histopathology tissue analysis. When creating Digi-
PathAI, four main problems were addressed – the
large size of WSI images, minimal training sam-
ples, stain variability and extraction of clinically rel-
evant features. Four datasets were used in train-
ing the model including CAMELYON16 and CAME-
LYON17 along with DigestPath (colon) and PAIP
(liver). DigiPathAI used an ensemble of 3 fully con-
volutional networks – Deeplabv3, Inception-ResNet
and DenseNet. A divide and conquer approach was
taken for the WSI image size problem. Patches of the
image were selected and segmented. Once all patches
were segmented, they stitched together the segments
to generate the whole slide image segmentation. The
researchers used data augmentation to combat a lower
number of training samples as well as to generalize
across different staining and scanning protocols. This
included horizontal/vertical flip, rotations, and Gaus-
sian blurring and color augmentation (Khened et al.,
2021).
MIA (Microscopic Image Analyzer) is another
open-source tool developed for deep learning on mi-
croscopic images. MIA provides a graphical user
interface for using deep learning tools for classifi-
cation, segmentation, and object detection of micro-
scopic images. By providing the graphical user in-
terface, programming skills are not required to work
with MIA. MIA simply requires training data, al-
though the user needs to be able to select a model and
hyperparameters. MIA also provides image labeling
tools for annotating datasets (K
¨
orber, 2023).
The creators of the CAMELYON dataset also have
created an open-source tool for visualizing and in-
teracting with the CAMELYON dataset. This tool
is called ASAP (Automated Slide Analysis Platform)
and works on Linux and Windows operating systems.
ASAP offers tools for both viewing and annotation
(Litjens et al., 2018).
4.8 Comparing Machine Learning
Approaches to Pathologist Analysis
In 2017, a study put pathologists and coding teams up
to the CAMELYON16 challenge. They split pathol-
ogists into two groups and provided them with the
same WSI images for two tasks – metastases identi-
fication through pixel level annotation and slide level
labeling of metastases. 129 WSI were provided for
annotation. The first group of pathologists was given
a time constraint of two hours while the second group
had no time constraint. The group without time con-
straint took approximately 30 hours to assess all 129
images. The challenge was open to coding teams and
32 total algorithms were submitted across 23 teams.
Of the 32 algorithms, 25 were based on deep con-
volutional nueral networks, showing their popularity
for whole slide imaging tasks. GoogleLeNet team
scored 0.994 AUC on the image classification task. In
comparison, the median AUC for pathologists with-
out time constraint was 0.966 and 0.81 for patholo-
gists with time constraint. This showed that the model
outperformed pathologists with time constraint. This
is more realistic since pathologists have many cases
to analyze and a limited amount of time. The al-
gorithm was comparable to the results achieved by
human pathologists with unlimited time to view and
classify whole slide imaging (Bejnordi et al., ). Im-
portantly, while not perfect, CNNs can be used to pre-
dict the phenotype of breast cancers, potentially re-
ducing the need for expensive biomarker assays (Cou-
ture et al., 2018; Su et al., 2023). Deep learning algo-
rithms also have the potential to predict patient out-
come, which is hard to achieve with pathologic evalu-
ation of a breast cancer tissue sample (Shi et al., 2023;
Fernandez et al., 2022).
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
412

5 CONCLUSION
Since the advent of whole slide imaging, research in
digital pathology has surged. Computer-aided diag-
nosis and medical image analysis have become a fo-
cus for researchers, especially in digital pathology.
While there are still many challenges when work-
ing with whole slide images, current research shows
promise for finding the solutions to overcome these
challenges. Deep learning in digital pathology has the
potential to become a powerful tool for pathologists
and assist them with the high demand of the field,
which could ultimately lead to better care for breast
cancer patients. This paper provides background in-
formation about breast cancer, whole slide images,
and deep learning along with key challenges and the
techniques employed by researchers in the field to
overcome these challenges. The implementation of
deep learning shows potential for incredible benefits
that can both propel digital pathology forward as well
as help patients.
REFERENCES
Camelyon17 - grand challenge. https://camelyon17.
grand-challenge.org/Home/. Accessed: 2023-11-27.
Alkabban, F. M. and Ferguson, T. (2020). Breast cancer. In
StatPearls. Treasure Island (FL):Stat Pearls Publish-
ing.
Alomari, R. S., Allen, R., Sabata, B., and Chaudhary, V.
(2009). Localization of tissues in high-resolution dig-
ital anatomic pathology images. In Medical Imaging
2009: Computer-Aided Diagnosis, volume 7260.
Araujo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P.,
Eloy, C., ..., and Campilho, A. (2017). Classification
of breast cancer histology images using convolutional
neural networks. PLoS ONE, 12(6).
Bancroft, J. D. and Layton, C. (2019). 10 - the hematoxylins
and eosin. In Suvarna, S. K., Layton, C., and Bancroft,
J. D., editors, Bancroft’s Theory and Practice of His-
tological Techniques, volume 1, pages 126–138. Else-
vier, eighth edition edition.
Bejnordi, B. E., Paul, M. V., Diest, J. V., Ginneken, B. V.,
Karssemeijer, N., Litjens, G., ..., and Ven
ˆ
ancio, R.
Bhushan, A., Gonsalves, A., and Menon, J. U. (2021). Cur-
rent state of breast cancer diagnosis, treatment, and
theranostics. Pharmaceutics, 13(5).
Chan, J. K. (2014). The wonderful colors of the
hematoxylin-eosin stain in diagnostic surgical pathol-
ogy. International Journal of Surgical Pathology,
22(1):12–32.
Ciga, O. and Martel, A. L. (2021). Learning to segment im-
ages with classification labels. Medical Image Analy-
sis, 68.
Ciga, O., Xu, T., Nofech-Mozes, S., Noy, S., Lu, F. I., and
Martel, A. L. (2021). Overcoming the limitations of
patch-based learning to detect cancer in whole slide
images. Scientific reports, 11.
Couture, H. D., Williams, L. A., Geradts, J., Nyante, S. J.,
Butler, E. N., Marron, J. S., ..., and Niethammer, M.
(2018). Image analysis with deep learning to predict
breast cancer grade, er status, histologic subtype, and
intrinsic subtype. npj Breast Cancer, 4.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). Imagenet: A large-scale hierarchical
image database. In IEEE Conference on Computer
Vision and Pattern Recognition.
Dimitriou, N., Arandjelovi
´
c, O., and Caie, P. D. (2019).
Deep learning for whole slide image analysis: An
overview. Frontiers in Medicine, 6.
Fernandez, G., Prastawa, M., Madduri, A. S., Scott, R.,
Marami, B., Shpalensky, N., ..., and Donovan, M. J.
(2022). Development and validation of an ai-enabled
digital breast cancer assay to predict early-stage breast
cancer recurrence within 6 years. Breast Cancer Re-
search, 24.
Gupta, J., Pathak, S., and Kumar, G. (2022a). Deep learn-
ing (cnn) and transfer learning: A review. Journal of
Physics: Conference Series, 2273(1):012029.
Gupta, V., Mishra, V. K., Singhal, P., and Kumar, A.
(2022b). An overview of supervised machine learning
algorithm. In 2022 11th International Conference on
System Modeling & Advancement in Research Trends
(SMART).
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In IEEE Confer-
ence on Computer Vision and Pattern Recognition.
Hou, L., Samaras, D., Kurc, T. M., Gao, Y., Davis, J. E.,
and Saltz, J. H. (2016). Patch-based convolutional
neural network for whole slide tissue image classifi-
cation. In IEEE Conference on Computer Vision and
Pattern Recognition.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., ..., and Adam, H. (2017). Mo-
bilenets: Efficient convolutional neural networks for
mobile vision applications. CoRR, abs/1704.04861.
Huang, P. W., Ouyang, H., Hsu, B. Y., Chang, Y. R.,
Lin, Y. C., ..., Y. A. C., and Pai, T. W. (2023).
Deep-learning based breast cancer detection for cross-
staining histopathology images. Heliyon, 9(2).
Khened, M., Kori, A., Rajkumar, H., Krishnamurthi, G.,
and Srinivasan, B. (2021). A generalized deep learn-
ing framework for whole-slide image segmentation
and analysis. Scientific Reports, 11.
Kim, H. E., Cosa-Linan, A., Santhanam, N., Jannesari, M.,
Maros, M. E., and Ganslandt, T. (2022). Transfer
learning for medical image classification: a literature
review. BMC Medical Imaging, 22(1).
K
¨
orber, N. (2023). Mia is an open-source standalone deep
learning application for microscopic image analysis.
Cell Reports Methods, 3(7).
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2017). Im-
agenet classification with deep convolutional neural
networks. Communications of the ACM, 60(6).
Krupinski, E. A., Johnson, J. P., Jaw, S., Graham, A. R.,
and Weinstein, R. S. (2012). Compressing pathol-
Deep Learning in Digital Breast Pathology
413

ogy whole-slide images using a human and model ob-
server evaluation. Journal of Pathology Informatics,
3:17.
LeCun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
LeCun, Y., Hinton, G., and Bengio, Y. (2015). Deep learn-
ing. Nature, 521:436–444.
Lee, S., Cho, J., and Kim, S. W. (2021). Automatic classifi-
cation on patient-level breast cancer metastases.
LeNail, A. (2019). Nn-svg: Publication-ready neural net-
work architecture schematics. Journal of Open Source
Software, 4(33):747.
Li, J., Yang, S., Huang, X., Da, Q., Yang, X., ..., Z. H.,
and Li, H. (2019). Signet ring cell detection with a
semi-supervised learning framework. In Information
Processing in Medical Imaging, volume 11492, pages
842–854.
Litjens, G., Bandi, P., Bejnordi, B. E., Geessink, O., Balken-
hol, M., Bult, P., ..., and van der Laak, J. (2018). 1399
h&e-stained sentinel lymph node sections of breast
cancer patients: The camelyon dataset. GigaScience,
7(6).
Mi, W., Li, J., Guo, Y., Ren, X., Liang, Z., Zhang, T., and
Zou, H. (2021). Deep learning-based multi-class clas-
sification of breast digital pathology images. Cancer
Management and Research, 13.
Nounou, M. I., ElAmrawy, F., Ahmed, N., Abdelraouf, K.,
Goda, S., and Syed-Sha-Qhattal, H. (2015). Breast
cancer: Conventional diagnosis and treatment modal-
ities and recent patents and technologies. Breast Can-
cer: Basic and Clinical Research, 9s2.
O’Shea, K. and Nash, R. (2015). An introduction to convo-
lutional neural networks. ArXiv e-prints, 10.
Otsu, N. (1979). Threshold selection method from gray-
level histograms. IEEE Transactions on Systems,
Man, and Cybernetics, 9(1):62–66.
Pantanowitz, L., Farahani, N., and Parwani, A. (2015).
Whole slide imaging in pathology: advantages, lim-
itations, and emerging perspectives. Pathology and
Laboratory Medicine International, 7:23–33.
Phan, N. N., Hsu, C. Y., Huang, C. C., Tseng, L. M., and
Chuang, E. Y. (2021). Prediction of breast cancer re-
currence using a deep convolutional neural network
without region-of-interest labeling. Frontiers in On-
cology, 11.
Pinchaud, N. (2019). Camelyon17 challenge.
Pouyanfar, S., Sadiq, S., Yan, Y., Tian, H., Tao, Y., Reyes,
M. P., ..., and Iyengar, S. S. (2018). A survey on
deep learning: Algorithms, techniques, and applica-
tions. ACM Computing Surveys, 51(5):1–36.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh,
S., Ma, S., ..., and Fei-Fei, L. (2015). Imagenet large
scale visual recognition challenge. International Jour-
nal of Computer Vision, 115:211–252.
Sari, C. T. and Gunduz-Demir, C. (2019). Unsupervised
feature extraction via deep learning for histopatholog-
ical classification of colon tissue images. IEEE Trans-
actions on Medical Imaging, 38(5):1139–1149.
Shi, Y., Olsson, L. T., Hoadley, K. A., Calhoun, B. C.,
Marron, J. S., Geradts, J., ..., and Troester, M. A.
(2023). Predicting early breast cancer recurrence from
histopathological images in the carolina breast cancer
study. npj Breast Cancer, 9:92.
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A.
(2023). Cancer statistics, 2023. CA: A Cancer Journal
for Clinicians, 73(1):17–48.
Simonyan, K. and Zisserman, A. (2015). Very deep convo-
lutional networks for large-scale image recognition. In
ICLR.
Su, Z., Niazi, M. K. K., Tavolara, T. E., Niu, S., Tozbikian,
G. H., Wesolowski, R., and Gurcan, M. N. (2023).
Bcr-net: A deep learning framework to predict breast
cancer recurrence from histopathology images. PLoS
ONE, 18(4).
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., ..., and Rabinovich, A. (2015). Go-
ing deeper with convolutions. In IEEE Conference on
Computer Vision and Pattern Recognition.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna,
Z. (2016). Rethinking the inception architecture for
computer vision. In IEEE Conference on Computer
Vision and Pattern Recognition, pages 2818–2826.
Veta, M., Pluim, J. P., Diest, P. J. V., and Viergever, M. A.
(2014). Breast cancer histopathology image analysis:
A review. IEEE Transactions on Biomedical Engi-
neering, 61(5):1400–1411.
Wakili, M. A., Shehu, H. A., Sharif, M. H., Sharif, M.
H. U., Umar, A., Kusetogullari, H., ..., and Uyaver,
S. (2022). Classification of breast cancer histopatho-
logical images using densenet and transfer learning.
Computational Intelligence and Neuroscience, 2022.
Watkins, E. J. (2019). Overview of breast cancer. Jour-
nal of the American Academy of Physician Assistants,
32(10):13–17.
Xu, X., Xu, S., Jin, L., and Song, E. (2011). Characteristic
analysis of otsu threshold and its applications. Pattern
Recognition Letters, 32(7).
Zarella, M. D., Bowman, D., Aeffner, F., Farahani, N.,
Xthona, A., Absar, S. F., ..., and Hartman, D. J. (2019).
A practical guide to whole slide imaging a white pa-
per from the digital pathology association. Archives of
Pathology & Laboratory Medicine, 143:222–234.
Zhang, A., Lipton, Z. C., Li, M., and Smola, A. J. (2021).
Dive into Deep Learning. Cambridge University
Press. https://D2L.ai.
BIOINFORMATICS 2024 - 15th International Conference on Bioinformatics Models, Methods and Algorithms
414
