
Improving Explainability of the Attention Branch Network with CAM
Fostering Techniques in the Context of Histological Images
Pedro Lucas Miguel
1
, Alessandra Lumini
2 a
, Giuliano Cardozo Medalha
3
, Guilherme F. Roberto
4 b
,
Guilherme Botazzo Rozendo
1
, Adriano Mauro Cansian
1 c
, Tha
´
ına A. A. Tosta
5 d
,
Marcelo Z. do Nascimento
6 e
and Leandro A. Neves
1 f
1
Department of Computer Science and Statistics, S
˜
ao Paulo State University, S
˜
ao Jos
´
e do Rio Preto-SP, Brazil
2
Department of Computer Science and Engineering, University of Bologna, Italy
3
WZTECH NETWORKS, Avenida Romeu Strazzi (room 503-B), 325, 15084-010, S
˜
ao Jos
´
e do Rio Preto-SP, Brazil
4
Faculty of Engineering, University of Porto, Porto, Portugal
5
Institute of Science and Technology, Federal University of S
˜
ao Paulo, S
˜
ao Jos
´
e dos Campos-SP, Brazil
6
Faculty of Computer Science, Federal University of Uberl
ˆ
andia, Uberl
ˆ
andia-MG, Brazil
Keywords:
Attention Branches, CAM Fostering, Convolutional Neural Networks, Grad-CAM, Histological Images.
Abstract:
Convolutional neural networks have presented significant results in histological image classification. Despite
their high accuracy, their limited interpretability hinders widespread adoption. Therefore, this work proposes
an improvement to the attention branch network (ABN) in order to improve its explanatory power through
the gradient-weighted class activation map technique. The proposed model creates attention maps and applies
the CAM fostering strategy to them, making the network focus on the most important areas of the image.
Two experiments were performed to compare the proposed model with the ABN approach, considering five
datasets of histological images. The evaluation process was defined via quantitative metrics such as coherency,
complexity, confidence drop, and the harmonic average of those metrics (ADCC). Among the results, the
proposed model through the ResNet-50 was able to provide an improvement of 4.16% in the average ADCC
metric and 3.88% in the coherence metric when compared to the respective ABN model. Considering the
DesneNet-201 network as the explored backbone, the proposed model achieved an improvement of 14.87% in
the average ADCC metric and 9.77% in the coherence metric compared to the corresponding ABN model. The
contributions of this work are important to make the results via computer-aided diagnosis more comprehensible
for clinical practice.
1 INTRODUCTION
Computational systems based on Convolutional Neu-
ral Networks (CNN) have shown great results in dif-
ferent image classification and pattern recognition
problems (H
¨
ohn et al., 2021; Shihabuddin and K.,
2023; Majumdar et al., 2023). However, despite the
very high levels of accuracy presented by some archi-
tectures, the adoption of this type of system is still re-
a
https://orcid.org/0000-0003-0290-7354
b
https://orcid.org/0000-0001-5883-2983
c
https://orcid.org/0000-0003-4494-1454
d
https://orcid.org/0000-0002-9291-8892
e
https://orcid.org/0000-0003-3537-0178
f
https://orcid.org/0000-0001-8580-7054
stricted in several critical fields of society, especially
in medical images. (Miotto et al., 2017). This fact oc-
curs due to the difficulty in interpreting how the clas-
sification process is carried out internally by CNNs,
leading to a lack of confidence in the way these mod-
els operate (Xu et al., 2019).
To enhance the reliability of those approaches, dif-
ferent techniques have been developed to make CNN
more explainable, particularly techniques that provide
visual solutions. For instance, the gradient-weighted
class activation mapping (Grad-CAM) technique cal-
culates the gradient of the network to obtain activation
maps that show the most important regions for the fi-
nal classification of the image (Selvaraju et al., 2019).
This type of technique allows human operators to see
more clearly which regions of the image are most im-
456
Miguel, P., Lumini, A., Medalha, G., Roberto, G., Rozendo, G., Cansian, A., Tosta, T., Z. do Nascimento, M. and Neves, L.
Improving Explainability of the Attention Branch Network with CAM Fostering Techniques in the Context of Histological Images.
DOI: 10.5220/0012595700003690
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 26th International Conference on Enterprise Information Systems (ICEIS 2024) - Volume 1, pages 456-464
ISBN: 978-989-758-692-7; ISSN: 2184-4992
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

portant for the final classification of the model, mak-
ing this approach particularly interesting for clinical
practice, especially in the context of histological im-
ages.
The analysis of histological samples is one of the
stages widely used in medicine to define diagnostics
and prognostics for different diseases. The images
are obtained through a series of steps, such as: col-
lecting a small tissue sample; fixation of the tissue;
processing; embedding; sectioning; staining, and mi-
croscopy analysis (Gurina and Simms, 2023). Con-
sidering the steps required to analyze a tissue sample,
the staining process is particularly important. Among
the different approaches to staining, the most popular
is the use of hematoxylin and eosin (H&E). Hema-
toxylin stains the nucleic acids of tissues in a deep
blue-purple color. Eosin stains proteins in a pink color
(Fischer et al., 2008). This process allows specialists
in the field to investigate more clearly the regions that
may point to the presence of diseases or other clin-
ical conditions. Considering the methodology used
in this stage, computer-aided diagnosis (CAD) can be
developed to support specialists in the process of an-
alyzing stained tissue samples. In this context, CNN-
based models that make use of so-called class acti-
vation maps (CAM) are especially interesting, as they
visually show which are the most important regions of
a given image that led to its final classification (Poppi
et al., 2021).
In order to improve the explainability power of
CNN, different approaches have been developed to
obtain increasingly precise and easy-to-understand
explanations. The study presented by (Fukui et al.,
2019) makes use of so-called attention branches
to improve explanations. The proposed Attention
Branch Network (ABN) architecture is made up of
three main blocks: the feature extractor; the attention
branch and the perception branch. The feature extrac-
tor is responsible for extracting the features from the
input images into feature maps. These maps are then
supplied to the attention branch to provide a label for
the input data and also create an attention map indi-
cating the most important regions of the images. The
attention map is then combined with the attributes ex-
tracted by the feature extractor, and the result of this
operation is supplied as input to the perception branch
to obtain a second label from the new data. Finally,
the loss values related to the attention branch and per-
ception branch classifications are combined, and by
doing so, all the weights of the model are updated
from this single loss value. This process, in addition
to improving the network’s accuracy, allows it to be-
come more attentive to the most important regions of
the image for the final classification.
In this context, the study presented by (Sch
¨
ottl,
2022) takes a different approach to obtaining better
explanations. This approach is called CAM foster-
ing and consists of an activation map created by any
CNN during the training process. Thus, it is possible
to calculate the entropy of this activation map, and af-
ter calculating the entropy, a weight is associated with
this value. This operation results in a value that can
be added to the training loss so that the weights are
later adjusted with this new loss value. This approach
presented relevant results in terms of the quality of
the explanations, although there was a slight accuracy
drop in the tested models.
Although the techniques presented (Fukui et al.,
2019; Sch
¨
ottl, 2022) show interesting conclusions
in terms of explainability, the methodology used to
evaluate the quality of the explanations involves only
qualitative methods, which do not indicate the impact
of the results for clinical practice. Thus, the study
presented by (Poppi et al., 2021) proposes the use
of a series of quantitative metrics such as coherency,
complexity and confidence drop. These metrics can
be represented through a single metric called average
DCC (ADCC), thus allowing a quantitative evaluation
of the explanations obtained by the models, which is
one of the motivations for the development of this
study.
Thus, to enhance the explanatory power of convo-
lutional neural networks in the context of histologi-
cal images, this work proposes an improvement to the
ABN model in association with the CAM fostering
strategy. The proposal explores the attention maps
generated by the attention branch, where the CAM
fostering strategy can be applied by calculating the
entropy of these maps. The explanations generated af-
ter this modification are then evaluated using a series
of quantitative metrics, allowing for a more complete
analysis of the impact of the proposed technique. The
main contributions presented here are:
• A improvement over the ABN model to provide
better explanations considering the context of his-
tological images;
• The use of quantitative metrics to evaluate the ex-
planatory power of convolutional neural network
models;
• The development of a pipeline for evaluating ex-
planations that can be used with other models;
2 METHODOLOGY
The proposed methodology was divided into three
steps. The first step was the process of splitting up five
Improving Explainability of the Attention Branch Network with CAM Fostering Techniques in the Context of Histological Images
457
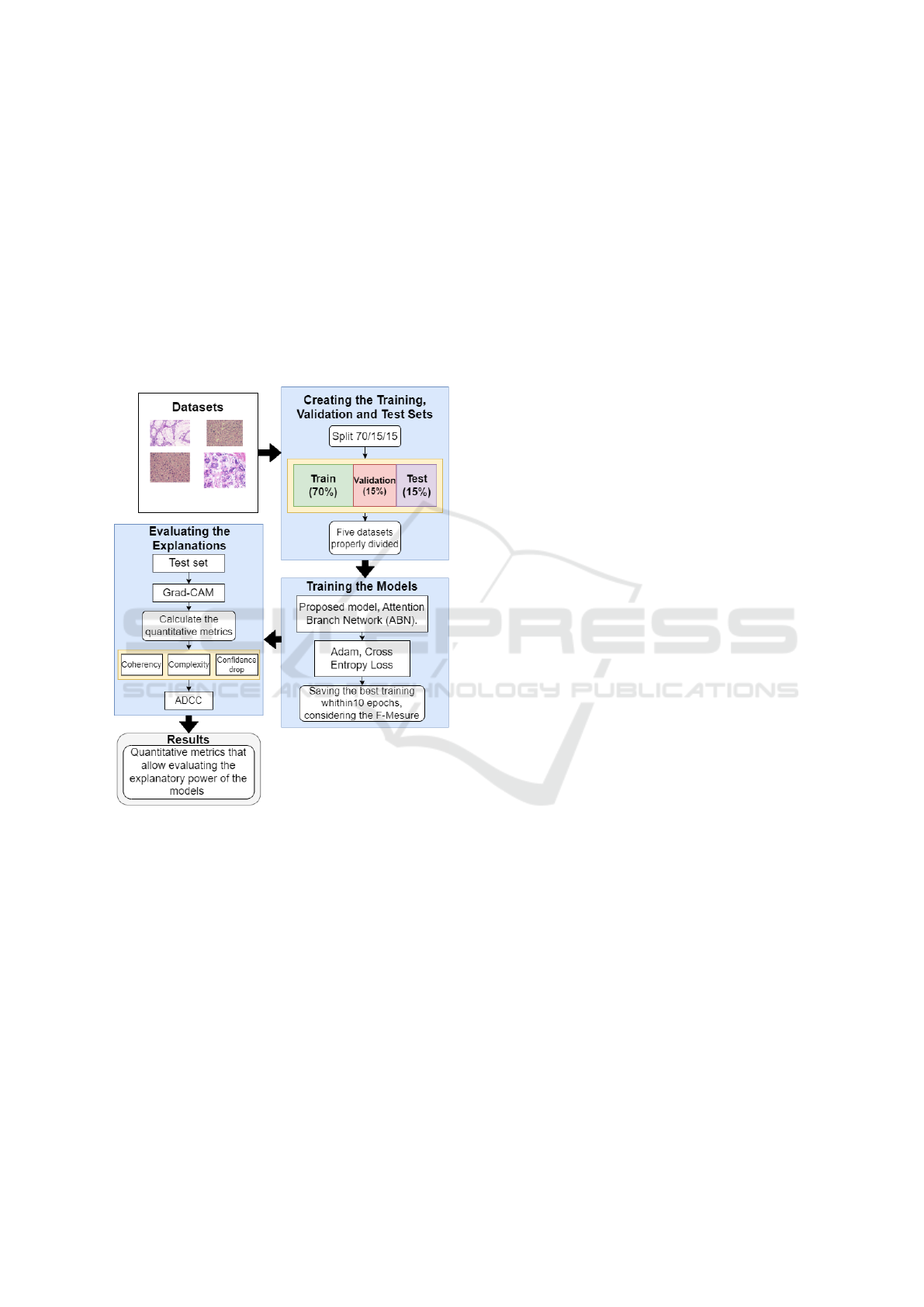
datasets of histological images, through the hold-out
strategy (Comet, 2023), considering a 70/15/15 split.
The second step consisted of training the ABN model
and the proposed model on each of the previously di-
vided datasets, using the F-measure as the metric for
selecting the best training among the epochs. Finally,
the models trained in the previous step were used to
obtain the activation maps of the images present in
the test split of each dataset, using the Grad-CAM
technique, so that all the quantitative metrics were
calculated to assess the explanatory capacity of each
model. An overview of the proposal is shown in Fig-
ure 1.
Figure 1: Proposed methodology to evaluate the explana-
tory capacity of the proposed architecture.
2.1 Proposed Architecture
To develop the architecture proposed in this work,
the approaches presented by (Fukui et al., 2019) and
(Sch
¨
ottl, 2022) were considered. The proposed model
used two backbones, the ResNet-50 (He et al., 2016)
and DenseNet-201(Huang et al., 2017) networks.
These architectures were chosen because of the rele-
vant results presented in (Fukui et al., 2019). The pro-
posed architecture considered three modules (feature
extractor, attention branch and perception branch).
In addition, the proposed architecture considers two
mechanisms (the attention mechanism and CAM fos-
tering) aimed at improving explanations. Figure 2
gives an overview of the proposed architecture, which
also shows the proposed modifications made to the
ABN model.
The proposed model considered a feature extrac-
tor module based on all residual (ResNet-50) or dense
blocks (DenseNet-201), excluding the last block in
both cases. It is important to note that the last block
was not considered in this case, as it is used to com-
pose the attention branch and the perception branch.
The main purpose was to extract feature maps g(X
i
)
from the input image X
i
, where these maps were then
provided as input to the attention branch and attention
mechanism.
The attention branch module received the feature
maps g(X
i
) obtained by the feature extractor so that
these maps were then processed by a series of con-
volutional layers. The composition of these convolu-
tional layers is the same as those present in the last
residual (ResNet-50) or dense block (DenseNet-201)
from the backbone model. The output provided by
these convolutional layers is presented in the format
K × w × h, where K is the number of feature maps, w
is the width and h is the height of each map. This data
was then processed by a block composed of a batch
normalization layer, a 1 ×1 × 1 convolution layer and
a ReLU activation. This configuration allowed all K
maps to be aggregated into a single map. The map
was then normalized by a block composed of a batch
normalization layer, a 1 ×1 × 1 convolution layer and
a sigmoid activation. Finally, an M(X
i
) attention map
was created in order to be used in the attention mech-
anism and CAM fostering mechanism. It is important
to note that, unlike the original ABN model, our at-
tention branch does not have the classification module
for the attention map. This modification was made in
order to use the CAM fostering strategy when training
the model.
The use of attention mechanisms has become
an increasingly common practice in different com-
puter vision systems, especially for sequential mod-
els (Yang et al., 2016; You et al., 2016; Vaswani et al.,
2017). For the proposed model, the attention mecha-
nism follows the indications of (Fukui et al., 2019).
From a set of feature maps g(X
i
) and an attention
map M(X
i
), it was possible to use the attention map to
create new feature maps g
′
(X
i
), whose important ar-
eas for the model’s final classification are reinforced.
Equation 1 indicates the association of the attention
map with the feature maps.
g
′
(X
i
) = (g(X
i
) × M(X
i
)) + g(X
i
) (1)
The perception branch was the module responsi-
ble for providing the final classification (label). This
module received the g
′
(X
i
) feature maps as input and
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
458
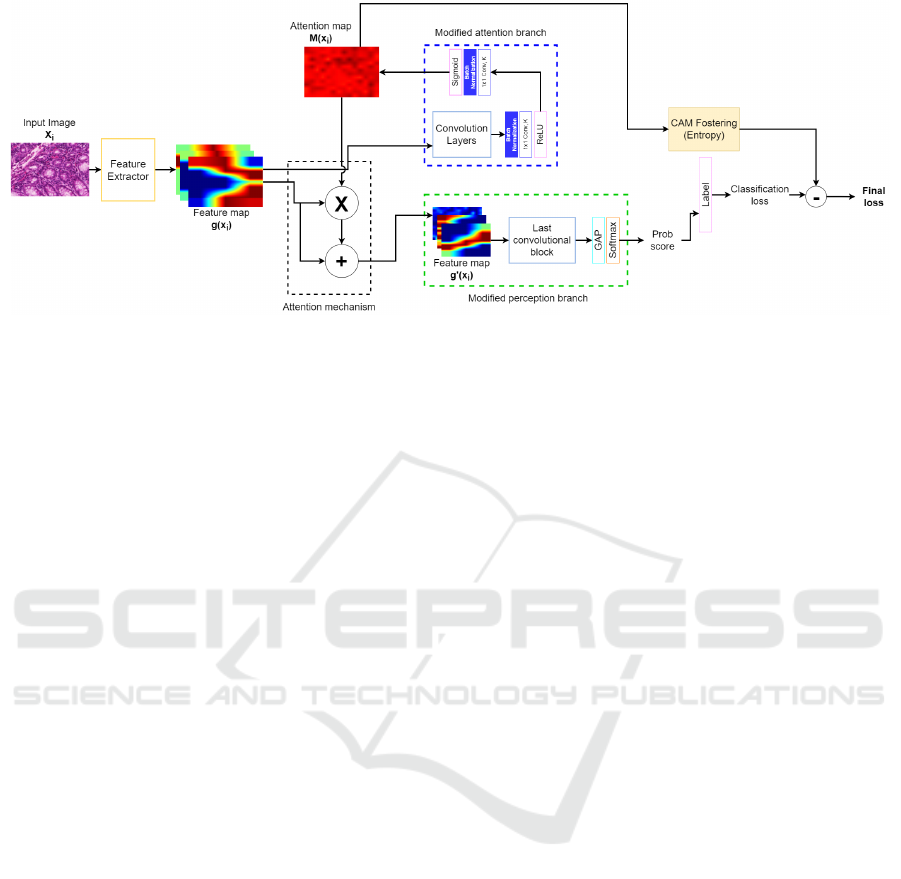
Figure 2: Overview of the proposed architecture, where the blue segmented rectangle shows the attention branch after the
proposed modifications, while the green segmented rectangle shows the perception branch after the modifications.
from the attention mechanism. The perception branch
was made up of convolutional layers that correspond
to the configuration of the last residual (ResNet-50) or
dense block (ResNet-50) from the backbone model,
as well as a global average pooling layer (GAP) used
to provide the model’s final classification in associ-
ation with a softmax function. The choice to use the
GAP layer instead of the fully connected layer present
in the original ABN model followed the indications
presented by (Zhou et al., 2016). By using the GAP
layer to perform the model’s final classification, the
convolutional layers’ ability to locate objects in the
image is preserved, consequently improving the ar-
chitecture’s explanatory power.
The CAM fostering mechanism follows the de-
scription presented by (Sch
¨
ottl, 2022), in which it is
possible to calculate the entropy value ce of an ac-
tivation map. The entropy computed from an activa-
tion map measures the variability of activations across
different regions or pixels in the map. A uniform map
with consistent activations yields low entropy, while a
varied map with diverse activations shows higher en-
tropy. Adding entropy as a term in the loss function
can serve as regularization, encouraging the model to
generate more diverse and information-rich activation
maps.
Thus, for the proposed model, the entropy factor
ce was calculated from the attention map M(X
i
) and
weighed by a regularization factor γ
e
equal to 10. The
chosen value for γ
e
followed the indications given by
(Sch
¨
ottl, 2022). The weighed ce value was then sub-
tracted from the classification loss l
n
measured by a
cross-entropy loss function (Mao et al., 2023), giving
the new loss value l
′
n
. Equation 2 shows how the ce
value is calculated, while Equation 3 shows how the
new loss value was calculated considering the CAM
fostering strategy.
ce(M(X
i
)) = −
∑
M(X
i
)
i j
− ln M(X
i
)
i j
(2)
l
′
n
= l
n
− γ
e
∗ ce(M(X
i
)), (3)
where i j represents the pixel’s index from the at-
tention map.
2.2 Dataset
This study used five datasets representing four differ-
ent types of histological tissues. For all five datasets,
the tissue samples were stained with hematoxylin and
eosin (H&E). The first dataset (UCSB) is composed
of breast cancer images provided by the University
of California, Santa Barbara (Drelie Gelasca et al.,
2008). This dataset consists of 58 samples divided
into two classes: benign (32) and malignant (26).
The second dataset (CR) is composed of images of
colorectal tissues (Sirinukunwattana et al., 2017), to-
taling 165 samples divided between two classes: be-
nign (74) and malignant (91). To acquire the images,
histological areas were digitally photographed using
a Zeiss MIRAX MIDI Slide Scanner with a resolu-
tion scale of 0.620 µm, which is equivalent to a 20x
magnification.
The third dataset (NHL) was published by the Na-
tional Cancer Institute and the National Institute on
Ageing (Shamir et al., 2008), and consists of 173
samples of non-Hodgkin lymphoma divided into three
classes: MCL — mantle cell lymphoma (99); FL —
follicular lymphoma (62); and CLL— chronic lym-
phocyte leukemia (12). To obtain the images, a light
microscope Zeiss Axioscope with a 20x objective and
an AXio Cam MR5 digital camera were used. The
images obtained by this process were stored without
compression with a resolution of 1388 × 1040 pixels,
a 24-bit quantization ratio and the RGB color model;
Improving Explainability of the Attention Branch Network with CAM Fostering Techniques in the Context of Histological Images
459
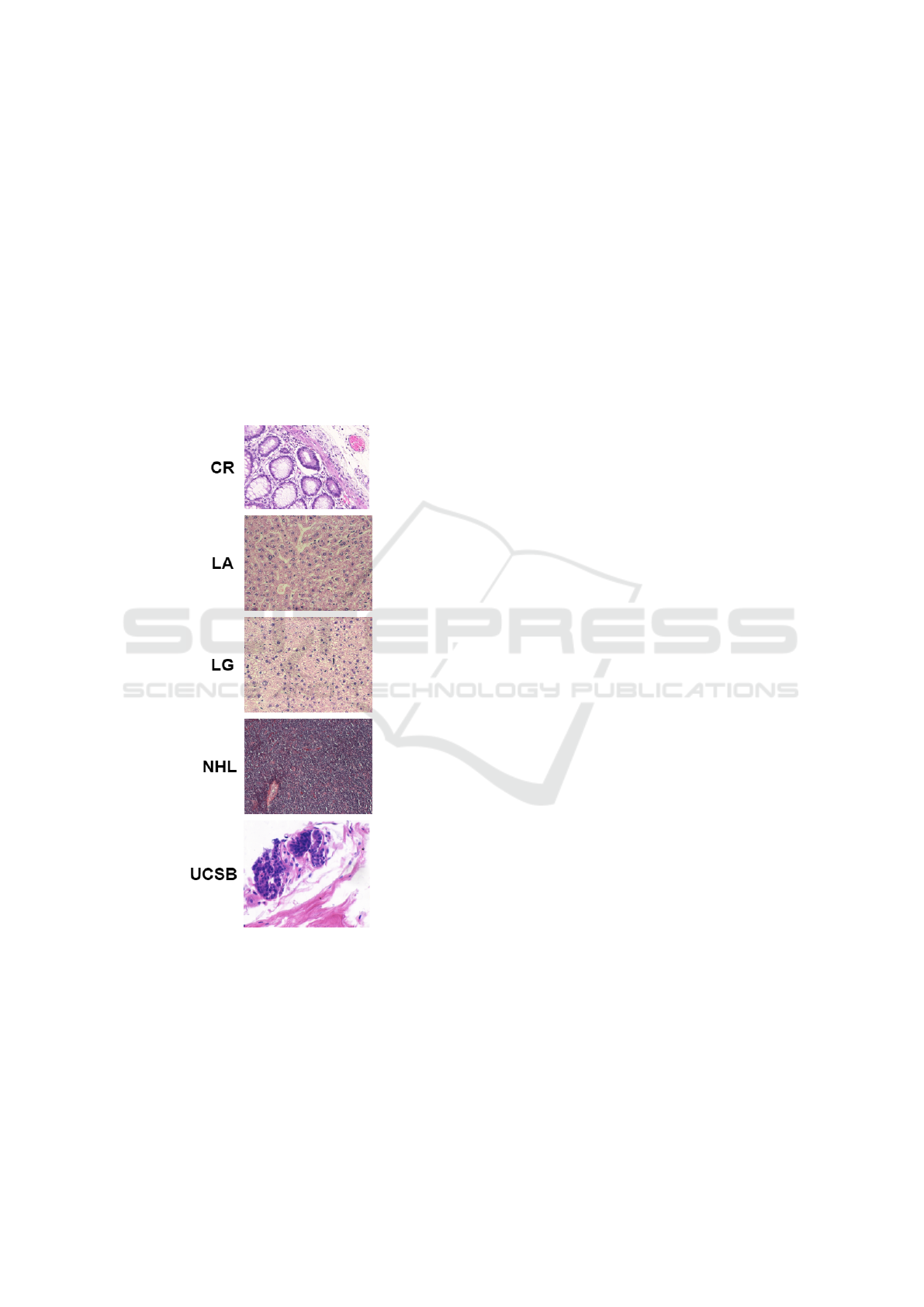
Finally, the fourth and fifth datasets were provided
by the Atlas of Gene Expression in Mouse Ageing
Project (AGEMAP) and are composed of liver tissue
images obtained from mice (AGEMAP, 2020). The
images were acquired by a Carl Zeiss Axiovert 200
microscope and 40x objective. The fourth dataset
(LG) consists of 265 liver tissue samples obtained
from male (150) and female (115) mice on a caloric
restriction diet. The fifth dataset (LA) is composed
of 529 images divided into four classes, where each
class represents different age groups of female mice
on an ad libitum diet, the classes being: one (100);
six (115); 16 (162) and 24 (152) months old.
Figure 3 shows a sample from each dataset, while
Table 1 displays an overview of all datasets.
Figure 3: Examples of histological images from each
dataset: UCSB (Drelie Gelasca et al., 2008); CR (Sir-
inukunwattana et al., 2017); NHL (Shamir et al., 2008); LA
and LG (AGEMAP, 2020).
2.3 Step 1: Creating the Training,
Validation and Test Sets
To ensure consistent results in terms of the explana-
tory power of each compared model, the hold-out
strategy was applied to each dataset individually
(Comet, 2023). A 70/15/15 split was applied to each
dataset, whereby: 70% of the dataset was dedicated to
the training process, 15% to the validation stage, and
15% to the tests. It is worth noting that the images be-
longing to the split were randomly selected from the
original sets.
2.4 Step 2: Training the Models
For this step, the ABN models and the proposed
here were trained. In both cases, the ResNet-50 and
DenseNet-201 architectures were used as backbones,
so that a fair comparison could be made of the impact
of the modifications proposed by our methodology in
both cases. To speed up the training process and avoid
problems such as overfitting due to the small num-
ber of samples in some datasets, the transfer learning
strategy was applied (Zhuang et al., 2019). Therefore,
all models were pre-trained on the ImageNet image
database (Deng et al., 2009), so that it was possible to
fine-tune the weights considering a small number of
epochs.
For training, a total of 10 epochs were chosen,
considering a learning rate of 0.0001 and a batch size
of 16. To update the weights, the Adam optimizer was
chosen given its rapid convergence, considering a re-
duced number of epochs (Kingma and Ba, 2014). It
is worth mentioning that all the weights in the model
were updated during the training step. The loss func-
tion chosen for training was the cross-entropy loss
(Mao et al., 2023). The loss value obtained loss value
was then used together with the CAM fostering strat-
egy to obtain a new loss value, which was used to
update the network weights during this step.
2.5 Step 3: Evaluating the Explanations
In this step, the quantitative metrics relating to the
quality of the Grad-CAM of each of the models
trained in step 2 were calculated. Thus, the set of
images belonging to the test split of each dataset pre-
viously defined in step 1 was used to obtain the acti-
vation maps. The metrics used to assess the explana-
tory power of each model were proposed by (Poppi
et al., 2021) such as coherency, complexity, confi-
dence drop and average DCC. The coherency metric
indicates that, given an image x referring to a class of
interest c, the activation map obtained by the image x
should not be altered when the activation map itself
is provided to the network. This property is presented
as
CAM
c
(x ⊙CAM
c
(x)) equal to CAM
c
(x). (4)
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
460

Table 1: Overview of all five datasets.
Dataset Tissue type Classes Samples Resolution
UCSB Breast tumours 2 58 896 × 768
CR Colorectal tumours 2 165 From 567 × 430 to 775 × 522
NHL Non-Hodgkin lymphoma 3 173 From 86 × 65 to 1388 × 1040
LG Liver tissue 2 265 417 × 312
LA Liver Tissue 4 529 417 × 312
Therefore, to measure the extent to which an acti-
vation map respects this property, the Pearson Corre-
lation Coefficient was calculated between two CAMs
considering Equation 5.
Coherency(x) =
Cov(CAM
c
(x ⊙CAM
c
(x)),CAM
c
(x))
σCAM
c
(x ⊙CAM
c
(x))σCAM
c
(x)
,
(5)
where Cov is the covariance between two maps
and σ indicates the standard deviation. It is worth
noting that as the Pearson Correlation Coefficient is
defined in the [-1, 1] interval, therefore, the values
obtained were subsequently normalized in the [0, 1]
interval to maintain the same scale as the other met-
rics. This metric takes on values closer to one when
the method is invariant to the input image.
The complexity metric was responsible for calcu-
lating the quantity of information presented in an acti-
vation map, since the more pixels an explanation has,
the more complex it is, making this explanation not so
significant. Thus, adopting the L
1
norm as a proxy, it
was possible to calculate complexity using Equation
6.
Complexity(x) = ∥CAM
c
(x)∥
1
(6)
Therefore, the lower the number of pixels as-
signed to a given explanation, the lower the complex-
ity value, this value being limited to the interval [0,
1].
The confidence drop was a metric that indicated
the loss of confidence in a model when only the acti-
vation map was provided as input instead of the full
image. This metric was defined by Equation 7
Drop(x) = max(0,y
c
− o
c
)/y
c
, (7)
Where y
c
is the class score considering the com-
plete image, and o
c
is the class score considering the
activation map of the complete image. This metric
was defined in the interval [0, 1], where the closer it
is to zero, the lower the model’s loss of confidence.
Finally, considering all the metrics described
above, the Average DCC (ADCC) was calculated as
the harmonic mean between all the metrics, as defined
by Equation 8.
ADCC(x) = 3
1
Coherency(x)
+
1
1 −Complexity(x)
+
1
1 − Drop(x)
−1
(8)
In this way, it was possible to assess the overall
quality of the explanations generated by the models
tested in this study using a single metric.
3 RESULTS
The methodology developed in this work was applied
to evaluate the explanatory power of the proposed
model concerning other models in the literature, con-
sidering the context of histological images. Thus, the
evaluation process was defined via the quantitative
metrics coherency (COH), complexity (COM), confi-
dence drop (CD) and Average DCC (ADCC). Tables 2
and 3 show the results obtained in the first experiment,
considering the proposed model using the ResNet-50
and DenseNet-201 networks as backbones, respec-
tively.
Table 2: Percentage values for coherency (COH), com-
plexity (COM), confidence drop (CD) and ADCC for the
proposed model (ResNet-50, ABN, CAM fostering, GAP),
considering all datasets.
Dataset COH↑ COM↓ CD↓ ADCC↑
LG 31.66 0.24 35.01 47.35
CR 31.82 0.13 12.00 54.32
NHL 25.13 0.07 62.50 35.32
UCSB 32.47 0.11 11.11 55.24
LA 20.82 0.24 77.22 26.35
Mean 28.38 0.16 39.57 43.72
Considering the results presented in Table 2, the
proposed model via the ResNet-50 network as a back-
bone showed the best results among all the experi-
ments. For the CR and UCSB datasets, the proposed
approach was able to achieve an ADCC index equal to
54.32% and 55.24% respectively, contributing to the
average of 43.72% achieved in this metric. This result
indicates that the model has an important explanatory
capability in the context of histological images. It is
Improving Explainability of the Attention Branch Network with CAM Fostering Techniques in the Context of Histological Images
461

Table 3: Percentage values for coherency (COH), complex-
ity (COM), confidence drop (CD) and ADCC for the pro-
posed model (DenseNet-201, ABN, CAM fostering, GAP),
considering all datasets.
Dataset COH↑ COM↓ CD↓ ADCC↑
LG 28.44 0.23 52.40 24.96
CR 33.96 0.13 14.94 57.58
NHL 38.70 0.07 60.07 37.81
UCSB 32.92 0.11 15.95 56.02
LA 38.17 0.24 71.07 27.17
Mean 34.44 0.16 42.89 40.71
also relevant to highlight the coherence indices ob-
tained by this model, where it was observed that the
proposed approach was able to reach a value higher
than 30% in three of the five datasets, totaling an av-
erage of 28.38%. This indicates that this model has a
better ability to generate more concise explanations.
For the results obtained by the proposed model
using the DenseNet-201 network as a backbone, an
average ADCC metric of 40.71% was observed, es-
pecially for the CR dataset, where the configuration
used in this test was able to achieve an ADCC value
of 57.58% which is the highest metric obtained in this
work. It is also important to note that this configu-
ration had the highest average coherence index of all
the models tested, with an average coherence value of
34.44%.
Taking into account the second experiment, the at-
tention branch network model (ABN) was evaluated
in the same way using the ResNet-50 and DenseNet-
201 architectures. Tables 4 and 5 show the results
obtained for each backbone, respectively.
Table 4: Percentage values for coherency (COH), complex-
ity (COM), confidence drop (CD) and ADCC for the ABN
model (ResNet-50), considering all datasets.
Dataset COH↑ COM↓ CD↓ ADCC↑
LG 30.05 0.24 19.72 52.55
CR 26.36 0.13 4.75 50.13
NHL 23.57 0.07 54.84 27.84
UCSB 26.70 0.11 8.39 50.37
LA 15.82 0.24 75.34 16.94
Mean 24.50 0.16 32.60 39.56
Table 5: Percentage values for coherency (COH), complex-
ity (COM), confidence drop (CD) and ADCC for the ABN
Model (DenseNet-201), considering all datasets.
Dataset COH↑ COM↓ CD↓ ADCC↑
LG 21.40 0.23 47.74 7.71
CR 32.66 0.13 29.84 54.08
NHL 24.43 0.07 60.51 19.10
UCSB 13.65 0.06 11.46 25.08
LA 31.21 0.24 74.48 23.24
Mean 24.67 0.15 44.81 25.84
For the results obtained by the ABN model us-
ing the ResNet-50 as a backbone, it was observed
that in three of the five datasets the ADCC metric
was greater than 50%, totaling an average ADCC of
39.56%. This result is 4.16% lower in relation to the
proposed model using the same backbone. It is also
important to highlight the confidence drop index ob-
tained by the architecture in the CR dataset, where
a total of 4.75% was observed, the lowest value ob-
tained in all the experiments in this study. Finally,
it is worth noting the value of the coherence index
obtained by the ABN model, in which an average in-
dex of 24.50% was observed, this result being 3.88%
lower compared to the proposed model. These data
indicate that the model was unable to generate expla-
nations that showed more restricted areas in the im-
age.
As for the results presented by the ABN model
using the DesNet-201 network as a backbone, low
ADCC indices were observed for each of the five
datasets, totaling an average ADCC of 25.84%. This
result is 14.87% lower than the proposed model us-
ing the same backbone. It is also worth noting that
the average coherence value obtained by this model
was 24.67%, which is 9.77% lower than the model
proposed in this work. These results prove the effec-
tiveness of the modifications proposed in this study
in terms of increasing the explanatory power of the
models.
Finally, Figures 4 and 5 show some samples of
explanations obtained by the proposed model and the
ABN model, using the ResNet-50 and DenseNet-201
networks as backbones, respectively. These images
show the impact of the coherence metric on the ex-
planations generated by each model, where the ex-
planations are expected to have smaller areas that are
important for the final classification.
From the samples of explanations shown in fig-
ures 4 and 5, it could be seen that the proposed model
was able to provide more concise explanations com-
pared to the ABN model using the same backbones,
a fact that is directly related to the coherence metrics
obtained by each model. Therefore, it is possible to
observe that the application of the proposed method-
ology was able to generate better explanations in the
context of histological image analysis.
4 CONCLUSIONS
In this work, a proposed model was defined to
improve the attention branch network architecture
through the use of the CAM fostering strategy. The
proposal was tested to increase the explanatory power
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
462
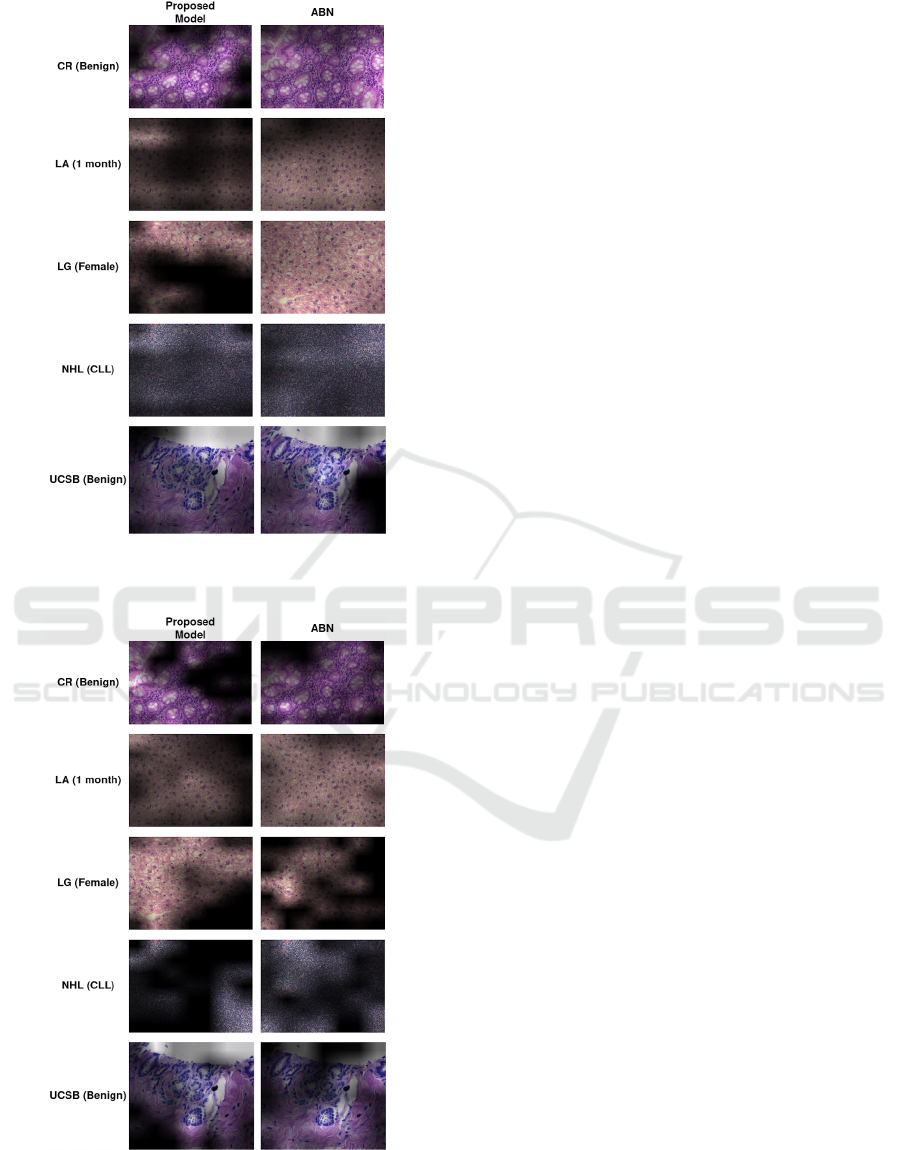
Figure 4: Explanations obtained with the Grad-CAM tech-
nique, considering the proposed model and the ABN model,
using the ResNet-50 as the backbone.
Figure 5: Explanations obtained with the Grad-CAM tech-
nique, considering the proposed model and the ABN model,
using the DenseNet-201 as the backbone.
of the model in the context of histological images.
Thus, a pipeline was developed to train the models,
as well as allow fair comparisons between the expla-
nations generated by the proposed approach and the
ABN model. In this pipeline, quantitative metrics
were used to assess the quality of the explanations
generated by each model.
The proposed model using the ResNet-50 network
as a backbone obtained the highest average ADCC
when compared to the other configurations tested
(43.3%), indicating an improvement of 4.16% when
compared to the ABN model using the same back-
bone. The model proposed using the DenseNet-201
network as a backbone showed a significant improve-
ment in its explanatory power, reaching an average
ADCC of 40.71%. This shows that the proposed
model explanatory power was increased by 14.87%
when compared to the ABN model using the same
backbone. In addition, this configuration provided the
best coherence metric in the study totaling an average
of 34.44%, which indicates that this model is capable
of creating explanations that emphasize only the most
important regions of the image. These results prove
that the modifications proposed in this work were able
to improve the explanatory power of the models, and
are important contributions to the development of re-
liable CAD systems.
For future work, we intend to investigate the im-
pact of the modifications using other models as back-
bones, as well as adapting the explanation evaluation
pipeline to support explanations generated by vision
transformer models.
ACKNOWLEDGEMENTS
This research was funded in part by the: Coordenac¸
˜
ao
de Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
- Brasil (CAPES) – Finance Code 001; National
Council for Scientific and Technological Develop-
ment CNPq (#313643/2021-0 and #311404/2021-9);
the State of Minas Gerais Research Foundation -
FAPEMIG (Grant #APQ-00578-18); S
˜
ao Paulo Re-
search Foundation - FAPESP (Grant #2022/03020-1);
WZTECH NETWORKS, S
˜
ao Jos
´
e do Rio Preto, S
˜
ao
Paulo.
REFERENCES
AGEMAP, N. I. o. A. (2020). The atlas of gene expression
in mouse aging project. AGEMAP. https://ome.grc.ni
a.nih.gov/iicbu2008/agemap/index.html.
Comet (2023). Understanding hold-out methods for training
Improving Explainability of the Attention Branch Network with CAM Fostering Techniques in the Context of Histological Images
463

machine learning models. Comet. https://www.comet.
com/site/blog/understanding-hold-out-methods-for-t
raining-machine-learning-models.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). Imagenet: A large-scale hierarchical
image database. In 2009 IEEE conference on com-
puter vision and pattern recognition, pages 248–255.
Ieee.
Drelie Gelasca, E., Byun, J., Obara, B., and Manjunath, B.
(2008). Evaluation and benchmark for biological im-
age segmentation. In 2008 15th IEEE International
Conference on Image Processing, pages 1816–1819.
Fischer, A. H., Jacobson, K. A., Rose, J., and Zeller, R.
(2008). Hematoxylin and eosin staining of tissue and
cell sections. CSH Protoc, 2008:db.prot4986.
Fukui, H., Hirakawa, T., Yamashita, T., and Fujiyoshi, H.
(2019). Attention branch network: Learning of at-
tention mechanism for visual explanation. In 2019
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition (CVPR), pages 10697–10706.
Gurina, T. S. and Simms, L. (2023). Histology, Staining.
StatPearls Publishing.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In 2016 IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR), pages 770–778.
Huang, G., Liu, Z., Maaten, L. V. D., and Weinberger, K. Q.
(2017). Densely connected convolutional networks.
In 2017 IEEE Conference on Computer Vision and
Pattern Recognition (CVPR), pages 2261–2269, Los
Alamitos, CA, USA. IEEE Computer Society.
H
¨
ohn, J., Krieghoff-Henning, E., Jutzi, T. B., von Kalle,
C., Utikal, J. S., Meier, F., Gellrich, F. F., Hobels-
berger, S., Hauschild, A., Schlager, J. G., French, L.,
Heinzerling, L., Schlaak, M., Ghoreschi, K., Hilke,
F. J., Poch, G., Kutzner, H., Heppt, M. V., Haferkamp,
S., Sondermann, W., Schadendorf, D., Schilling, B.,
Goebeler, M., Hekler, A., Fr
¨
ohling, S., Lipka, D. B.,
Kather, J. N., Krahl, D., Ferrara, G., Haggenm
¨
uller,
S., and Brinker, T. J. (2021). Combining cnn-based
histologic whole slide image analysis and patient data
to improve skin cancer classification. European Jour-
nal of Cancer, 149:94–101.
Kingma, D. and Ba, J. (2014). Adam: A method for
stochastic optimization. International Conference on
Learning Representations.
Majumdar, S., Pramanik, P., and Sarkar, R. (2023). Gamma
function based ensemble of cnn models for breast can-
cer detection in histopathology images. Expert Sys-
tems with Applications, 213:119022.
Mao, A., Mohri, M., and Zhong, Y. (2023). Cross-entropy
loss functions: theoretical analysis and applications.
In Proceedings of the 40th International Conference
on Machine Learning, ICML’23. JMLR.org.
Miotto, R., Wang, F., Wang, S., Jiang, X., and Dudley, J. T.
(2017). Deep learning for healthcare: review, oppor-
tunities and challenges. Briefings in Bioinformatics,
19(6):1236–1246.
Poppi, S., Cornia, M., Baraldi, L., and Cucchiara, R. (2021).
Revisiting the evaluation of class activation mapping
for explainability: A novel metric and experimental
analysis. In 2021 IEEE/CVF Conference on Computer
Vision and Pattern Recognition Workshops (CVPRW),
pages 2299–2304.
Sch
¨
ottl, A. (2022). Improving the interpretability of grad-
cams in deep classification networks. Procedia Com-
puter Science, 200:620–628. 3rd International Con-
ference on Industry 4.0 and Smart Manufacturing.
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., and Batra, D. (2019). Grad-CAM: Visual
explanations from deep networks via gradient-based
localization. International Journal of Computer Vi-
sion, 128(2):336–359.
Shamir, L., Orlov, N., Mark Eckley, D., Macura, T. J., and
Goldberg, I. G. (2008). Iicbu 2008: a proposed bench-
mark suite for biological image analysis. Medical
& Biological Engineering & Computing, 46(9):943–
947.
Shihabuddin, A. R. and K., S. B. (2023). Multi cnn
based automatic detection of mitotic nuclei in breast
histopathological images. Computers in Biology and
Medicine, 158:106815.
Sirinukunwattana, K., Pluim, J. P., Chen, H., Qi, X., Heng,
P.-A., Guo, Y. B., Wang, L. Y., Matuszewski, B. J.,
Bruni, E., Sanchez, U., B
¨
ohm, A., Ronneberger, O.,
Cheikh, B. B., Racoceanu, D., Kainz, P., Pfeiffer,
M., Urschler, M., Snead, D. R., and Rajpoot, N. M.
(2017). Gland segmentation in colon histology im-
ages: The glas challenge contest. Medical Image
Analysis, 35:489–502.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, L. u., and Polosukhin,
I. (2017). Attention is all you need. In Guyon,
I., Luxburg, U. V., Bengio, S., Wallach, H., Fer-
gus, R., Vishwanathan, S., and Garnett, R., editors,
Advances in Neural Information Processing Systems,
volume 30. Curran Associates, Inc.
Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., and Zhu,
J. (2019). Explainable ai: A brief survey on history,
research areas, approaches and challenges. In Tang,
J., Kan, M.-Y., Zhao, D., Li, S., and Zan, H., edi-
tors, Natural Language Processing and Chinese Com-
puting, pages 563–574, Cham. Springer International
Publishing.
Yang, Z., He, X., Gao, J., Deng, L., and Smola, A. (2016).
Stacked attention networks for image question an-
swering. In 2016 IEEE Conference on Computer Vi-
sion and Pattern Recognition (CVPR), pages 21–29.
You, Q., Jin, H., Wang, Z., Fang, C., and Luo, J. (2016). Im-
age captioning with semantic attention. In 2016 IEEE
Conference on Computer Vision and Pattern Recog-
nition (CVPR), pages 4651–4659, Los Alamitos, CA,
USA. IEEE Computer Society.
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., and Tor-
ralba, A. (2016). Learning deep features for discrimi-
native localization. In 2016 IEEE Conference on Com-
puter Vision and Pattern Recognition (CVPR), pages
2921–2929, Los Alamitos, CA, USA. IEEE Computer
Society.
Zhuang, F., Qi, Z., Duan, K., Xi, D., Zhu, Y., Zhu, H.,
Xiong, H., and He, Q. (2019). A comprehensive sur-
vey on transfer learning. CoRR, abs/1911.02685.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
464
