
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with
Metric Access Method
Larissa R. Teixeira
1 a
, Igor A. R. Eleut
´
erio
1 b
, Mirela T. Cazzolato
1,2 c
, Marco A. Gutierrez
2 d
,
Agma J. M. Traina
1 e
and Caetano Traina-Jr.
1 f
1
Institute of Mathematics and Computer Science (ICMC), University of S
˜
ao Paulo (USP), S
˜
ao Carlos, Brazil
2
The Heart Institute (InCor), University of S
˜
ao Paulo (USP), S
˜
ao Paulo, Brazil
Keywords:
Dimensional Data, k-medoids, Clustering, Indexing, Metric Access Method.
Abstract:
Clustering algorithms are powerful data mining techniques, responsible for identifying patterns and extracting
information from datasets. Scalable algorithms have become crucial to enable data mining techniques on large
datasets. In literature, k-medoid-based clustering algorithms stand out as one of the most used approaches.
However, these methods face scalability challenges when applied to massive datasets and high dimensional
vector spaces, mainly due to the high computational cost in the swap step. In this paper, we propose the KluSIM
method to improve the computational efficiency of the swap step in the k-medoids clustering process. KluSIM
leverages Metric Access Methods (MAMs) to prune the search space, speeding up the swap step. Additionally,
KluSIM eliminates the need of maintaining a distance matrix in memory, successfully overcoming memory
limitations in existing methodologies. Experiments over real and synthetic data show that KluSIM outperforms
the baseline FasterPAM, with a speed up of up to 881 times, requiring up to 3,500 times fewer distance
calculations, and maintaining a comparable clustering quality. KluSIM is well-suited for big data analysis,
being effective and scalable for clustering large datasets.
1 INTRODUCTION
Clustering is a data mining task applied across differ-
ent applications, such as bio-informatics, image pro-
cessing, pattern recognition, financial risk analysis,
and more (Qaddoura et al., 2020). Clustering is the
process of identifying patterns in a dataset through
grouping into clusters. Objects within the same clus-
ter have higher similarity to each other compared to
objects in different clusters. Among several clus-
tering algorithms in the literature, the most popular
are k-means and k-medoids, whereas the most em-
ployed variant of the latter is the Partitioning Around
Medoids (PAM) (Kaufman, 1990). The k-means al-
gorithm is based on defining a centroid, which is the
arithmetic mean of all objects within a cluster. On the
a
https://orcid.org/0009-0007-9917-4404
b
https://orcid.org/0009-0007-3987-8880
c
https://orcid.org/0000-0002-4364-010X
d
https://orcid.org/0000-0003-0964-6222
e
https://orcid.org/0000-0003-4929-7258
f
https://orcid.org/0000-0002-6625-6047
other hand, k-medoids are algorithms that seek for a
medoid, an actual object within a cluster that have the
smallest distance sum to all other objects.
One of the advantages of k-medoids over k-means
algorithm is the results interpretability (Kenger et al.,
2023). By assigning actual objects as cluster rep-
resentatives, k-medoids results can provide insights
into the dataset based on the characteristics of the
medoids.
Medoid-based approaches have shown high clus-
tering quality. However, these methods show many
difficulties when running over large amounts of
data and high dimensional vector spaces. The k-
medoid-based algorithms rely on accurate initializa-
tion heuristics to preselect initial medoids as seed be-
fore the swap step. Good seeds can accelerate the
convergence of the algorithm and avoid distance cal-
culations during the swap step. The swap step iterates
over the entire dataset to find the best combination of
non-medoid and medoid that minimizes a objective
function. This step is computationally costly and is
the bottleneck of the algorithm. Several approaches
maintain a in-memory distance matrix to speed up
Teixeira, L., Eleutério, I., Cazzolato, M., Gutierrez, M., Traina, A. and Traina-Jr., C.
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method.
DOI: 10.5220/0012599900003690
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 26th International Conference on Enterprise Information Systems (ICEIS 2024) - Volume 1, pages 73-84
ISBN: 978-989-758-692-7; ISSN: 2184-4992
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
73

swap and avoid recomputing the distance between the
same objects. However, this option is unfeasible for
large datasets, due to limited memory.
We focus on clustering dimensional data over
large datasets. We propose KluSIM, a method
that speeds up the swap step of medoid-based ap-
proaches by taking advantage of Metric Access Meth-
ods (MAMs) to index and prune the search space,
without trading the semantic meaning of generated
clusters for performance. Thus, KluSIM avoids the
need for maintaining a distance matrix and reduces
the number of distance calculations. The key contri-
butions of our work are:
• MAM-Based Space Pruning. We strategically
take advantage of MAMs for search space prun-
ing during swap, drastically reducing the num-
ber of distance calculations. We also eliminate
the need for precomputing and keeping a in-
memory distance matrix. The MAM allows per-
forming a range query over the candidates of
medoid objects, even over large high-dimensional
datasets. The range query operation associates
each non-medoid object to the cluster with its
nearest medoid. KluSIM computes the minimum
distance, for each medoid, among other medoids,
and uses it to prune the search space.
• Centroid-Guided Candidate Selection. KluSIM
computes the centroid of each cluster and se-
lects its nearest objects through a k-NN query
to the centroid. The answers are the candi-
dates to be tested as medoid objects during swap.
Thus, unlike existing methods, KluSIM reduces
the medoids candidates to be evaluated at each it-
eration, also reducing the number of distance cal-
culations needed in the overall clustering process.
• KluSIM is Fast and Effective. Our method is up
to 881 faster than FasterPAM, the medoid-based
clustering approach baseline, while maintaining
comparable cluster quality.
KluSIM combines both k-NN and range queries
during clustering. Our experimental evaluation shows
a significant improvement in the execution time, with
comparable quality. KluSIM improves efficiency,
captures spatial relationships, and enhances the over-
all clustering process.
Paper Outline. Section 2 gives the background.
Section 3 reviews the related work. Section 4 in-
troduces the proposed method KluSIM. Section 5
presents the experimental evaluation and discussion.
Finally, Section 6 concludes this work.
2 BACKGROUND
We present the relevant background on clustering al-
gorithms, k-medoids approaches, and metric access
methods. Table 1 shows the symbols and acronyms.
2.1 Clustering Algorithms
Clustering, as a data mining technique, partitions
a dataset into k groups, where objects within the
same group have high similarity among themselves,
while maintaining dissimilarity with objects from
other groups (Han et al., 2011). The literature
presents several clustering algorithms, mainly cate-
gorized into hierarchical and partitioning methods,
sometimes also including density-based approaches
(Ran et al., 2023). Hierarchical clustering builds a
cluster tree structure, also called dendrogram. The
objects are organized into a hierarchy of clusters by
iteratively merging (agglomerative approach) or di-
viding (divisive approach) them based on a distance
measure (Ran et al., 2023). Despite the dendrograms
being effective for small datasets, they do not provide
a flat partitioning of the data. Therefore, simpler alter-
natives are generally preferred by users, such as par-
titioning methods (Schubert and Rousseeuw, 2021).
Partitioning methods assign each object to one of
a predefined number of clusters (Han et al., 2011).
The goal is to optimize a certain objective function,
such as minimizing the within-cluster variance or
maximizing inter-cluster distances. Examples include
k-means and k-medoids (Kaufman, 1990). Given
the better results interpretability of clusters achieved
by k-medoid-based algorithms compared to k-means,
this paper focuses on the former.
2.1.1 k-medoids Algorithms
The Partitioning Around Medoids (PAM) (Kaufman,
1990) was one of the first k-medoid-based algorithms
introduced in the literature. PAM has two main steps:
• build: Select k objects as initial medoids with a
greedy search.
• swap: Based on the initial medoids, PAM repeat-
edly performs the swap step to improve the clus-
tering quality until convergence. At each iteration,
a non-medoid object is evaluated to improve the
clustering quality by replacing one of the current
medoids with the non-medoid object.
2.1.2 Cluster Quality Measure
As part of the cluster quality assessment, many tech-
niques have been proposed in the literature. The clus-
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
74

Table 1: Summary of symbols and definitions.
Symbol Description
k Number of clusters
n Number of objects in the dataset
S Set of objects
s
i
, s
j
, s
k
, s
q
Objects ∈ S
δ Dissimilarity (distance) measure
M Set of medoids
m
i
A medoid object ∈ M
C Set of k clusters
C A cluster
x
C
An object ∈ C
p Number of nearest neighbors
S
p
Set of objects returned from kNNq
TD The total deviation of cluster quality
SS Silhouette score
DBI Davies Bouldin Index
ξ Range query radius
tering quality of k-medoids is evaluated using the ab-
solute error criterion denoted as total deviation (TD),
defined as:
TD =
k
∑
i=1
∑
x
c
∈C
i
δ(x
c
,m
i
) (1)
which is the sum of distances from each object x
c
∈
C
i
to the medoid m
i
of its cluster (Schubert and
Rousseeuw, 2021).
The Silhouette Score (SS) is another popular mea-
sure to evaluate clustering quality (Lenssen and Schu-
bert, 2024). For each object, the Silhouette Score con-
siders two key metrics: i. The average distance of a
object i to other objects in the same cluster. ii. The
average distance of a object i to other objects in the
nearest neighbor cluster. The overall SS is the av-
erage of individual SS across all objects. The score
ranges from −1 to 1, where a higher score means
better-defined and well-separated clusters.
Another cluster quality metric is the Davies-
Bouldin Index (DBI) (Davies and Bouldin, 1979).
The index considers both intra-cluster similarity and
inter-cluster dissimilarity to calculate the average
similarity of each cluster with all others. The DBI
index is obtained as the sum of average similarity val-
ues of all clusters, divided by the number of clusters.
Lower DBI indicates a better clustering result (mini-
mum index is zero).
2.2 Metric Access Methods
Metric Access Methods (MAMs) are built on top of
indexing structures designed to optimize the search
for nearest neighbors within a dataset. These meth-
ods take advantage of the inherent properties of met-
ric spaces, such as the triangular inequality, to prune
subsets of the search space, thus speeding up query
processing. Examples of MAMs include the VP-Tree
(Yianilos, 1993), Ball-Tree (Omohundro, 1989) and
KD-Tree (Bentley, 1975).
A metric space is represented as a domain of
objects and a distance function δ between two ob-
jects s
i
and s
j
from the domain that satisfy the fol-
lowing properties of a metric space (Zezula et al.,
2006): i. non-negativity: s
i
̸= s
j
→ δ(s
i
,s
j
) > 0;
ii. identity: δ(s
i
,s
j
) = 0 ⇔ s
i
= s
j
; iii. symme-
try: δ(s
i
,s
j
) = δ(s
j
,s
i
); iv. triangle inequality:
δ(s
i
,s
k
) ≤ δ(s
i
,s
j
) + δ(s
j
,s
k
).
There are two main elementary similarity queries
in a metric space (Zezula et al., 2006):
i. Range Query (Rq): given a query object s
q
, a
distance function δ and a radius ξ, the operation
retrieves all objects that differ from s
q
by at most
the distance ξ.
ii. k-Nearest Neighbors (kNNq): retrieves the k ob-
jects closest to the query object s
q
∈ S considering
the distance function δ.
2.3 Vantage Point Tree
One notable MAM is the Vantage Point Tree (VP-
Tree) introduced by (Yianilos, 1993). Using a ball
partitioning approach, VP-Tree selects a vantage point
(vp) to partition a dataset S into two subsets S
1
⊂ S
and S
2
⊂ S. This partitioning is based on the me-
dian distance ˜x between the chosen vp to all other
objects. The objects are distributed to S
1
or S
2
as:
S
1
← {s
i
|δ(vp,s
i
) ≤ ˜x} and S
2
← {s
i
|δ(vp,s
i
) > ˜x}
where vp ∈ S. After this, the same process is recur-
sively applied to each subset, creating a hierarchy.
3 RELATED WORK
In this work, we focus on improving the efficiency of
k-medoids. Table 2 summarizes related studies. Here
we compare our proposal KluSIM with related meth-
ods considering the following aspects:
• Memory Optimization: most algorithms require
precomputing a distance matrix, which is a bot-
tleneck for large datasets. This aspect informs
whether the algorithm can perform without an in-
memory matrix, showing a significant advantage
for the specific approach.
• Large Datasets: this aspect informs whether the
algorithm runs over the entire dataset within a fea-
sible time frame.
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method
75

Table 2: KluSIM optimizes the usage of memory when performing clustering through a space-pruning technique.Comparison
of state-of-art algorithms with our proposed.
Works
Memory
optimization
Executes with
whole dataset
Main memory
Space-pruning
SWAP
optimization
PAM-SLIM Ë é é é
SFKM é Ë Ë é
FastPAM1 é Ë Ë é
FasterPAM é Ë Ë é
(Proposal) KluSIM Ë Ë Ë Ë
• Main Memory: this aspect informs whether the
algorithm runs in main memory, which is faster
than algorithms operating in secondary memory.
• Space-Pruning swap Optimization: whether
the algorithm takes advantage of space-pruning
heuristics, which can significantly enhance the ef-
ficiency of the swap step in k-medoids algorithms
by avoiding many distance calculations.
The discussion of the related works is organized
into two parts, each addressing key aspects of en-
hancing the efficiency of k-medoid algorithms. Sub-
section 3.1 explores alternative approaches for k-
medoids initialization. Subsection 3.2 discusses tech-
niques related to k-medoids swap optimization.
This organization provides a comprehensive com-
parison of the proposed KluSIM method with existing
methodologies.
3.1 k-medoids Initialization Methods
PAM’s build step is well known for being state-of-
the-art heuristic for initializing k-medoids. However,
alternative approaches for initialization have been ex-
plored in the literature, such as the random selection
of initial medoids (Schubert and Rousseeuw, 2021).
In (Arthur and Vassilvitskii, 2007) the authors
propose a seeding strategy to improve the k-means
initialization, denoted as k-means++. The algorithm
randomly selects the first centroid from the dataset.
Then, k-means++ selects the next centroids with the
probability proportional to their squared distance to
the nearest centroid. New centroid are likely to be far
from the previously chosen ones.
In (Barioni et al., 2008) the authors present a vari-
ation of PAM called PAM-SLIM. The algorithm uses
the Slim-Tree MAM (Traina et al., 2002), stored in
secondary memory, to assist the selection of initial
medoids. The algorithm leverages the tree structure to
choose the initial medoids, selecting objects located
at the middle level of the tree as possible medoids.
Unfortunately, as this method does not consider the
entire dataset when selecting the initial medoids, it
may affect the clustering quality. Additionally, PAM-
SLIM does not take advantage of the benefits of
MAM to optimize the k-medoids swap step. It only
leverages the advantage of how the tree is organized,
focusing to select a subset of the dataset rather than
considering the whole dataset.
In (Park and Jun, 2009) the authors propose the
Simple and Fast k-medoids (SFKM), an initializa-
tion method for k-medoids that operates like k-means.
SFKM computes a distance matrix between all ob-
jects, aiming at finding new medoids in each iteration.
The algorithm selects the k-medoids with the small-
est normalized sum of distances within the dataset.
SFKM significantly reduces the computation time and
shows performance comparable to PAM, but it is not
effective in improving cluster quality. Also, SFKM
tends to perform worse than randomly selecting ini-
tial medoids, as reported in (Schubert and Rousseeuw,
2021). SFKM lacks memory optimization as it re-
quires the creation of a distance matrix in memory for
the entire dataset.
3.2 k-medoids Swap Optimization
One of the main challenges of k-medoids for large
datasets is the high computational cost, leading to
long execution times and memory limitations. The
swap step contributes the most to the computational
cost of PAM, since it seeks for the best pair of medoid
and non-medoid objects, among all possible k × (n −
k) pairs, which can improve the clustering quality.
Our proposal does not rely on existing methods.
Many methods include techniques for parallel cluster-
ing algorithms, such as (Vandanov et al., 2023). Sta-
tistical estimation techniques have also improved the
performance of k-medoids, such as BanditPAM (Ti-
wari et al., 2020), an algorithm inspired by multi-
armed bandits. Despite the potential advantages of
existing approaches, our proposal concentrates on op-
timizing the clustering algorithm in its original form
without changing the overall problem-solving strat-
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
76

egy. Parallel clustering algorithms and statistical esti-
mation techniques have proven beneficial in overcom-
ing computational challenges. However, they often
introduce complexities or modifications to the meth-
ods. Our goal is to enhance the efficiency and perfor-
mance of the clustering algorithm while preserving its
intrinsic structure and characteristics.
Additionally, other techniques have been intro-
duced to enhance existing approaches, such as faster
k-medoids (Schubert and Rousseeuw, 2021). The
work proposes FastPAM1 and FasterPAM algorithms,
both bringing an enhancement to the PAM algorithm.
FastPAM1 ensures the same output as PAM, while
FasterPAM does not, although maintains equivalent
quality. Both approaches improve the k-medoids
complexity in a factor of O(k). However, when the
number of objects in the dataset n is much larger than
the number of medoids k, the improvement may not
be advantageous, as also observed by (Tiwari et al.,
2020). Based on experimental results, FasterPAM is
faster than FastPAM1. Both approaches lack memory
optimization, as require a distance matrix as an input,
and lack space pruning optimization.
3.3 Open Issues
There is a gap in the existing literature concerning the
k-medoids strategy to employ MAMs in main mem-
ory. Such a strategy should not require a distance ma-
trix to enhance the swap step of the algorithm, which
would be beneficial for large datasets. Our proposed
KluSIM method fills this gap. The baseline competi-
tor is FasterPAM, which has a specific focus on im-
proving the swap step. In contrast, other related stud-
ies have mainly focused on developing new strategies
for selecting initial medoids.
We propose a method to improve the swap of
PAM, incorporating a space pruning optimization ap-
proach. We aim to optimize memory usage enabling
working with large datasets, exchanging the distance
matrix with a much smaller distance indexing MAM.
We detail our proposal in the next section.
4 THE KluSIM METHOD
In this work, we propose the k-medoids clustering
Swap Improvement with Metric Access Method
(KluSIM) method, designed to enhance the efficiency
of the swap step of k-medoids clustering. KluSIM
clusters objects in a vector space, satisfies the metric
space constraints (see Section 2.2), and takes advan-
tage of MAMs for space pruning.
Algorithm 1 presents the pseudocode of KluSIM.
It takes as input: the set of objects; the number of
clusters; the number of nearest neighbors of medoid
candidates during the swap step; and the initialization
method. The outputs are the set of k medoids, and the
produced clusters.
Roman numbers in parentheses indicate the steps
of the algorithm. In line 2, the algorithm starts in-
stantiating a MAM with the objects in S, and then
selects the initial set of k medoids (line 3). Line 4
performs the ASq operation, which computes overall
cluster quality TD and the current cluster assignment
Cusing the initial set of medoids M (line 4). Then,
KluSIM repeats the following steps for each cluster
(lines 6-17): Computes cluster centroids (line 7); Per-
forms k-NN search (line 8); Executes ASq operation
for each object o
j
within the set S
p
of nearest objects
of the centroid (line 12); Measures cluster quality im-
provement (line 13). The loop repeats while TD de-
creases, i.e. the cluster quality improves.
Algorithm 1: KluSIM (S,k, p, initMethod).
input : S: Set of objects
input : k: Number of clusters
input : p: Number of nearest neighbors of
medoid candidate
input : initMethod: Initialization method (e.g.,
build, k-means++)
output : M: Set of k medoids
output : C: Set of clusters
1 begin
2 mam ← CreateMAM(S) ; (i)
3 M ← InitializeMedoids(k, initMethod) ; (ii)
4 TD, C← mam.ASq(M) ; (iii)
5 while TD decreases do
6 for cluster C
i
∈ C do
7 µ
i
← ComputeCentroid(C
i
) ; (iv)
8 S
p
← mam.kNNq(µ
i
, p) ; (v)
9 for o
j
∈ S
p
do
/* Check if object o
j
improves
cluster quality */
10 M
′
← M ;
11 m
′
i
← o
j
; m
′
i
∈ M
′
12 TD
′
,C
′
← mam.ASq(M
′
) ; (iii)
13 if TD
′
< TD then (vi)
14 TD ← TD
′
;
15 C ← C
′
;
16 swap m
i
with o
j
;
Figure 1 illustrates the workflow of KluSIM. It di-
vides the algorithm into six steps, as detailed next.
The step numbering is the same as indicated in Algo-
rithm 1.
The initial step (i) of KluSIM instantiates a MAM
with all objects in the dataset. In step (ii), the algo-
rithm initializes by selecting k initial medoids using
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method
77
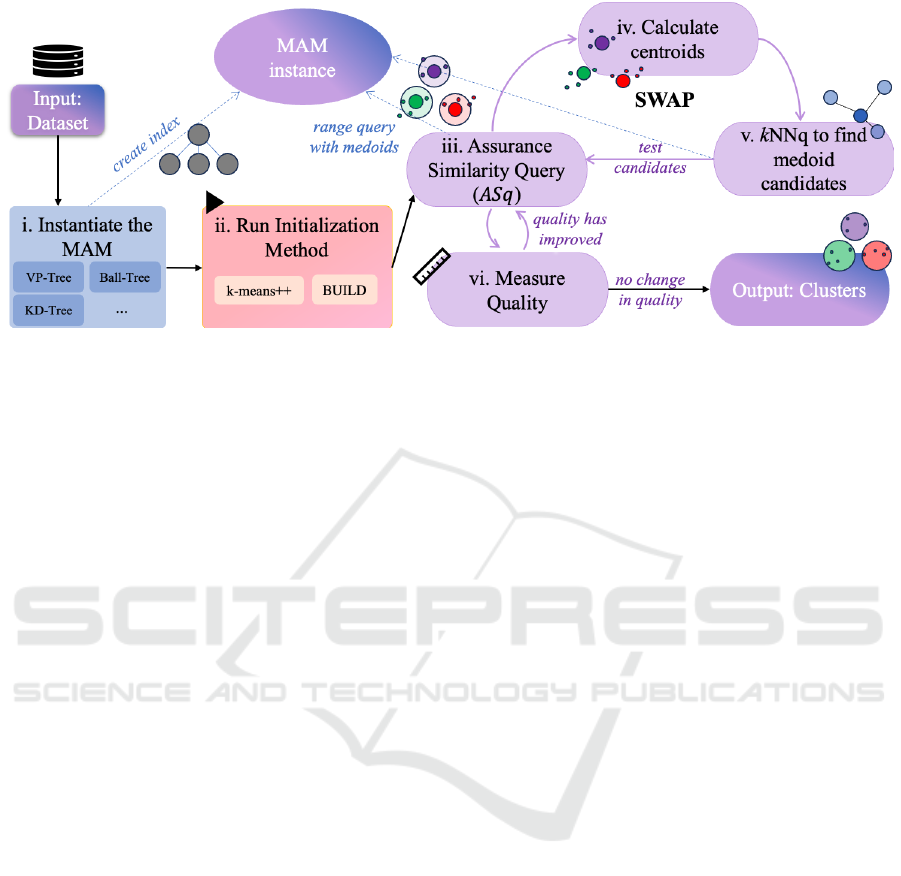
Figure 1: The KluSIM algorithm. i. Create a MAM with input objects. ii. Select k initial medoids using a given initialization
method. iii. Perform the Assurance Similarity Query (ASq). iv. Calculate the centroid for each cluster. v. Find the p nearest
objects from each centroid. vi. Calculate TD. Repeat the process while TD decreases.
an initialization method such as build or k-means++.
Step (iii) executes a search operation, referred to as
Assurance Similarity Query (ASq), using the MAM
instance. Then, following steps are executed itera-
tively until convergence:
• Compute Cluster Centroids (step iv): For each
cluster, compute the centroid object.
• Perform a k-NN Search to Find Medoid Can-
didates (step v): Based on the centroid of each
cluster, select the p nearest objects to the centroid
employing a MAM. In this step, parameter p is
the number of nearest neighbors. Taking advan-
tage of MAMs data organization allows to effi-
ciently prune the search space when performing
query operations. During the kNNq, the MAM
discards regions, as they are unlikely to contain
nearest neighbors, avoiding unnecessary distance
calculations.
• Execute the assurance similarity query opera-
tion ASq (step iii): Analyze each object o
j
∈
S
p
returned from the kNNq as a potential new
medoid within a cluster C
i
∈ C (line 9). For each
candidate, we replace the current medoid m
i
to o
j
(lines 10-11). Function ASq returns the clustering
quality TD and k clusters (line 12).
• Measure Cluster Quality Improvement (step
6): If the non-medoid object o
j
decreases TD,
then execute a swap operation between m
i
and o
j
.
In this work, we evaluate our algorithm using the
VP-Tree MAM. However, other MAMs that rely on
using representative objects to partition the data space
can be employed.
4.1 Assurance Similarity Query (ASq)
This operation is designed to easy creating k groups,
based on medoid objects. Function ASq gets as in-
put the set of medoids. It assures that each object is
assigned to the cluster with the most similar medoid.
The operation involves three steps: (1) Definition of
coverage radius; (2) Assignment of nearest objects;
(3) Association of remaining objects.
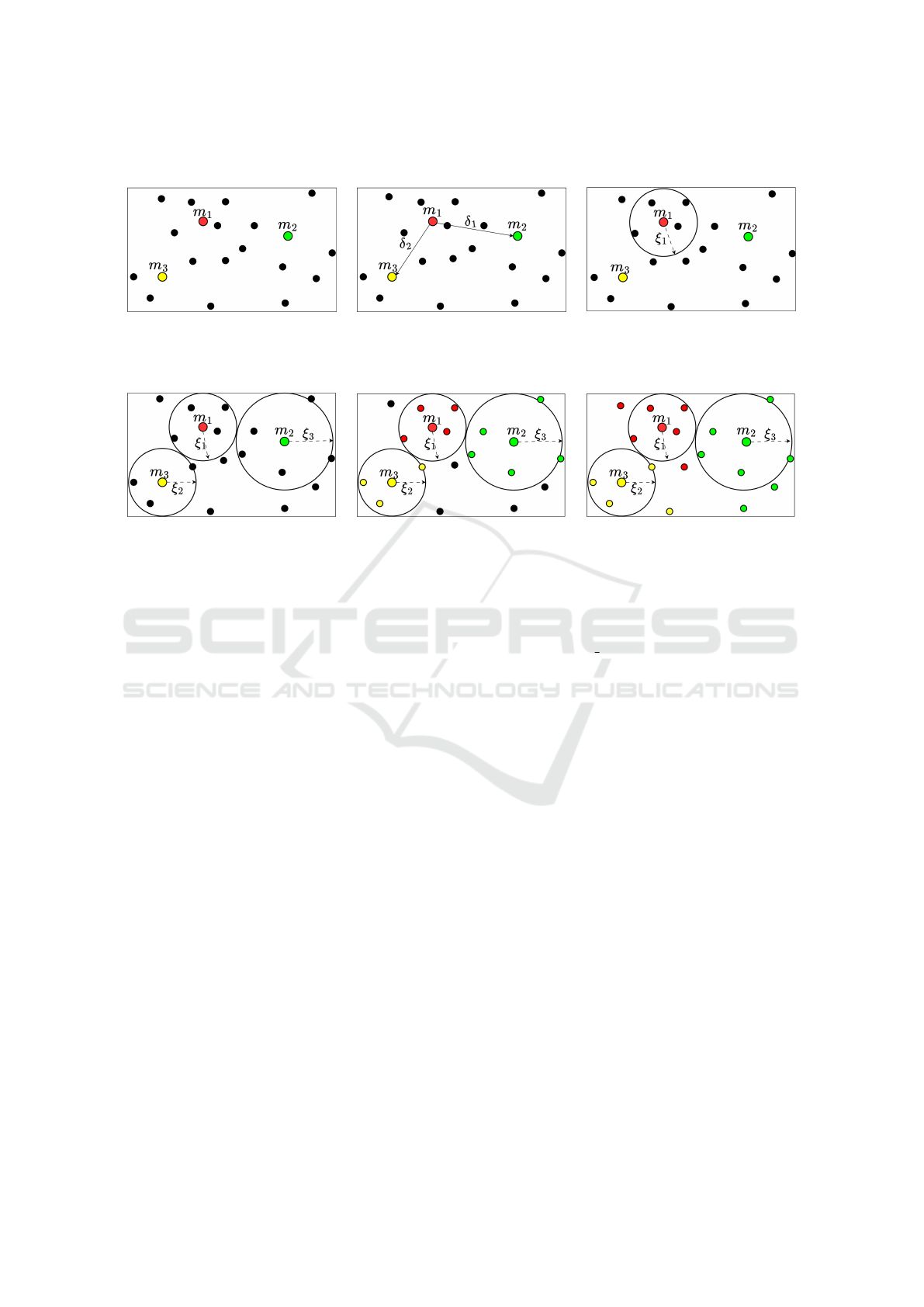
4.1.1 Definition of Coverage Radius
For each medoid m
i
∈ M, |M| = k, calculate the dis-
tance δ(m
i
,m
j
), where 1 ≤ j ≤ k and i ̸= j. The cover-
age radius ξ
i
for medoid m
i
is defined as
δ
min
/2, where
δ
min
represents the smallest distance obtained from
the distance of m
i
to all other medoids.
Figure 2 illustrates the process to define the cov-
erage radius ξ of the medoid m
1
. Considering k = 3
(a), the first step computes the distance (b) from the
medoid m
1
to m
2
and m
3
. Then, in (c) the coverage
radius ξ of m
1
is determined as
δ
min
/2. This process
repeats for the remaining medoids in (a).
4.1.2 Assignment of Nearest Objects
Figure 3(a) shows the coverage radius of each
medoid. For each medoid m
i
∈ M, a range query us-
ing MAM is conducted with the coverage radius ξ
i
,
as in (b). The objects covered by ξ
i
from medoid m
i
will form an initial cluster. Then, KluSIM employs
the instantiated MAM to prune the search space and
efficiently identify regions within the specified radius.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
78
(a) Initial medoids m
1
,m
2
e m
3
.
(b) Computing distance from m
1
to
m
2
and m
3
medoids.
(c) Choosing the coverage radius
δ
min
/2 for medoid m
1
.
Figure 2: Finding the coverage radius ξ of medoid m
1
, for k = 3.
(a) Coverage radius ξ of each medoid.
(b) Assigning objects to the group
covered by radius ξ of each medoid.
(c) Assignment remaining objects to
the nearest medoid.
Figure 3: Assigning each object to the nearest medoid, for k = 3.
4.1.3 Association of Remaining Objects
Objects outside the covering radius from any medoid
are treated as exceptions. In Figure 3(c), the algorithm
calculates the distance from each non-covered object
to all medoids. Then, each object is assigned to the
group of the nearest medoid.
4.2 Aspects and Advantages of KluSIM
KluSIM employs a clever heuristic that relies on prun-
ing the search space to significantly reduce the dis-
tance calculations. The swap step considers only a
subset of objects near the cluster centroid to be con-
sidered as better medoids during clustering iterations.
The swap step is the bottleneck of k-medoid-based
approaches, and KluSIM reduces the computational
cost of such approaches. The proposed approach ben-
efits clustering tasks over large datasets, since it ex-
changes the distance matrix of the objects with locally
based subsets of distances maintained by MAM. Fol-
lowing, we experimentally evaluate the improvements
obtained by KluSIM.
5 EXPERIMENTS
In this section, we evaluate the efficiency (execution
time and number of distance calculations) and effec-
tiveness (cluster quality) of the KluSIM.
5.1 Datasets and Setup
The experiments were conducted on several datasets:
• Synthetic Datasets: All synthetic datasets are
from the make blobs generator (Pedregosa et al.,
2011). A total of 48 datasets were created, with
dimensions (8, 64, and 128), number of samples
(5,000, 25,000, 50,000, and 100,000), and num-
bers of clusters k (5, 20, 50, and 100).
• Real Datasets: We selected the image datasets
ds-Mammoset (3457 tuples, 2 clusters) (Oliveira
et al., 2017), ds-DeepLesion (33334 tuples, 5 clus-
ters) (Yan et al., 2017) and ds-MNIST (70000 tu-
ples, 10 clusters)(Lecun et al., 1998). We em-
ployed the feature vectors provided by (Cazzolato
et al., 2022), generated with descriptors Texture
Spectrum (TS, 8 dimensions), Normalized Color
Histogram (NCH64, 64 dimensions), and Color
Structure (CS, 128 dimensions).
The choice of an optimal initialization method
is crucial. We evaluate two initialization strategies:
the build method and k-means++. The results re-
ported for each dataset are the average performance
across 10 iterations. In all experiments, our proposed
algorithm was compared against the state-of-the-art
FasterPAM algorithm, our baseline.
All experiments were performed on an Intel®
Core
TM
i5-7300U (2.60Ghz) with 16GB of RAM and
Ubuntu 20.04 LTS (64-Bit) GNU/ Linux OS. All al-
gorithms are implemented in Cython with the same
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method
79
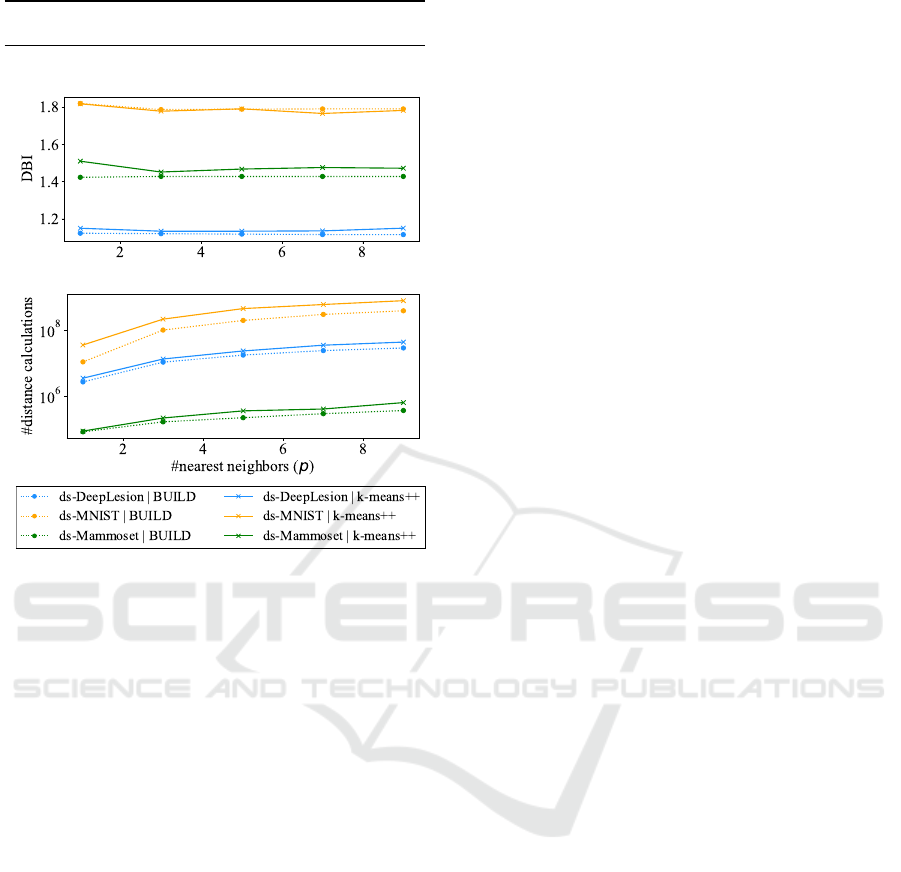
Real Datasets: Cluster quality and #distance
calculations (log scale)
(a) Cluster Quality by p value (DBI: the lower the
better)
(b) Number of distance calculations by p value
Figure 4: The plots show the cluster quality and number of
distance calculations varying p value. For p ≥ 3, the cluster
quality does not improve significantly, but the number of
distance calculation increases.
configuration, and none of them utilizes a distance
matrix in memory, ensuring a fair comparison.
Reproducibility. Our codes are available at https://
github.com/teixeiralari/KluSIM.
5.2 Estimating the Best Value for p
In step (v) (k-NN to find medoids candidates) of
KluSIM (see Figure 1), we perform a kNNq opera-
tion to find the p objects nearest to the centroid. We
evaluate the influence of p in the cluster quality. We
tested KluSIM with p values set to 1, 3, 5, 7, and 9,
analyzing the effect on both cluster quality and num-
ber of distance calculations. Figure 4 shows (a) the
cluster quality and (b) the number of distance calcu-
lations across different k values for real datasets. No
significant improvement occured in the cluster quality
for the number of nearest neighbors p ≥ 3, although
there is an almost linear increase in the number of dis-
tance calculations. Thus, the remaining experiments
were conducted using p = 3.
5.3 Evaluating the Execution Time
The efficiency of clustering algorithms is critical for
large-scale datasets. We evaluate execution time for
KluSIM against FasterPAM. The primary focus here
is to understand how the execution time varies regard-
ing the number of samples and clusters.
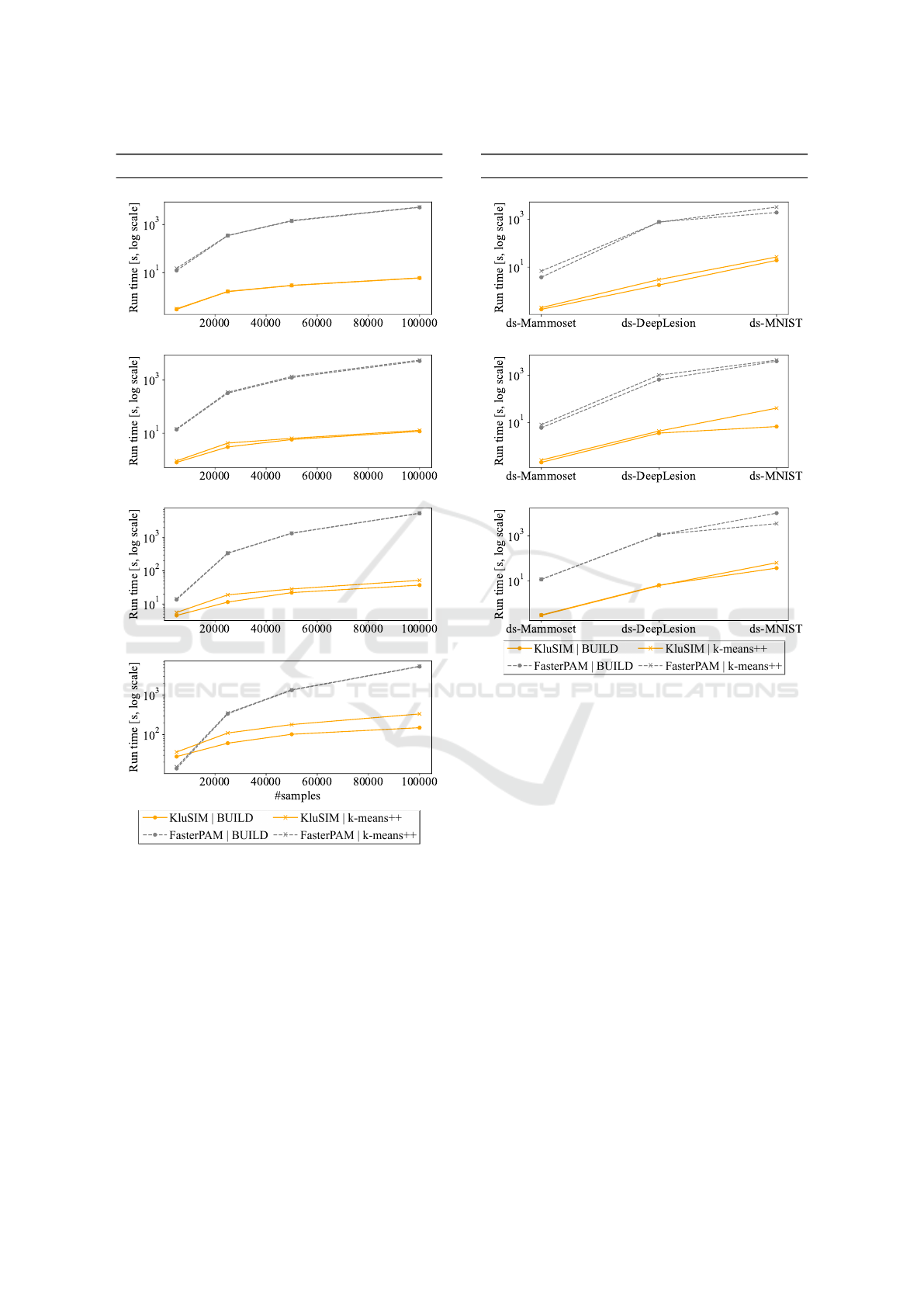
Figure 5 shows the experiments over synthetic
datasets with the number of samples from 5,000 to
100,000, and the average run time with build and
k-means++ initialization. All results are averaged
across each dimension (8, 64,128), for each dataset.
We compared the results of KluSIM and FasterPAM.
In (a), the scenario with a small number of clusters
(k = 5) and a large number of samples (S = 100, 000)
using build initialization, FasterPAM’s swap run time
was 5,144 seconds, and KluSIM took 5.96 sec-
onds, being approximately 863 times faster. With k-
means++ initialization, KluSIM outperformed Faster-
PAM by a speedup factor of 881 times. As the num-
ber of clusters increased to k = 100 with large sam-
ple size of S = 100,000 using build (Figure 5-d), a
speedup of about 35 times was observed. Similarly,
with k-means++, KluSIM exhibited still a substantial
speedup of about 16 times compared to FasterPAM.
Figure 6 shows the results for real datasets.
Specifically, for ds-Mammoset with a dimension of 8
(a), there is an approximate 21-fold acceleration in the
swap step with the build initialization and roughly 35
times using k-means++ when compared to FasterPAM
under the same conditions. On the other hand, for ds-
MNIST dataset, our algorithm achieved a speedup of
approximately 99 times with build initialization and
123 times with k-means++ in comparison to Faster-
PAM in the same scenarios. Furthermore, consider-
ing a higher dimension (c), the gain of our method is
nearly 40 times using build or k-means++ initializa-
tion for ds-Mammoset, and 275 times for ds-MNIST
dataset using build initialization.
5.4 Evaluating the Number of Distance
Calculations
We evaluated the number of distance calculations re-
quired in the clustering process for both KluSIM and
FasterPAM, varying the number of samples and clus-
ters. This analysis aims to understand the computa-
tional cost associated with KluSIM when compared
with the baseline FasterPAM approach. Given that
one of the goals of KluSIM is to provide efficiency
and scalability when clustering large datasets, as well
as reducing potential computational overhead in en-
vironments with limited memory, we conducted the
experiments, for both KluSIM and FasterPAM, with-
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
80
Synthetic Datasets - Execution time
(a) Number of clusters (k) = 5
(b) Number of clusters (k) = 20
(c) Number of clusters (k) = 50
(d) Number of clusters (k) = 100
Figure 5: KluSIM outperforms the main competitor by up to
881 times on synthetic datasets of different sizes. In nearly
all cases, KluSIM achieves faster times.
out storing a distance matrix in memory. All required
distances were calculated on the fly.
Figure 7 displays the total number of distance
calculations taken by the algorithms in the cluster-
ing process, for both build and k-means++ initializa-
tion. The results are averaged across all dimensions
(8,64,128) for each dataset. With a large number of
samples (S = 100, 000) and a small number of clusters
(k = 5) (a), our proposed algorithm KluSIM converges
with approximately 3,500 times fewer distance calcu-
lations than FasterPAM, using build or k-means++.
Real Datasets - Execution time
(a) 8 dimensions
(b) 64 dimensions
(c) 128 dimensions
Figure 6: KluSIM is consistently faster than the main com-
petitor for real datasets. The plots shows the run time
of KluSIM and FasterPAM on real datasets. In all cases,
KluSIM speeds up clustering.
As the number of clusters increases to k = 100 (d),
there is a notable reduction of roughly 39 times for
build initialization and 15 times for k-means++ ini-
tialization in the number of distance calculations in
comparison with the baseline FasterPAM.
Figure 8 presents the results for real datasets.
With 8 dimensions (a), our method exhibits a sig-
nificant reduction in the number of distance calcu-
lations. Specifically, there is a 184-fold reduction
when opting for the build initialization, compared to a
151-fold reduction with k-means++ initialization for
a smaller dataset (ds-Mammoset). In contrast, for
a larger dataset (ds-MNIST), there is a reduction of
59 times in the number of distance calculations with
build, and 56 times with k-means++. All these im-
provements are observed in comparison to FasterPAM
under same conditions. Considering a highest dimen-
sionality (c), KluSIM reduces by 62 times the num-
ber of distance calculations with build initialization,
and 54 times with k-means++ initialization for the ds-
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method
81
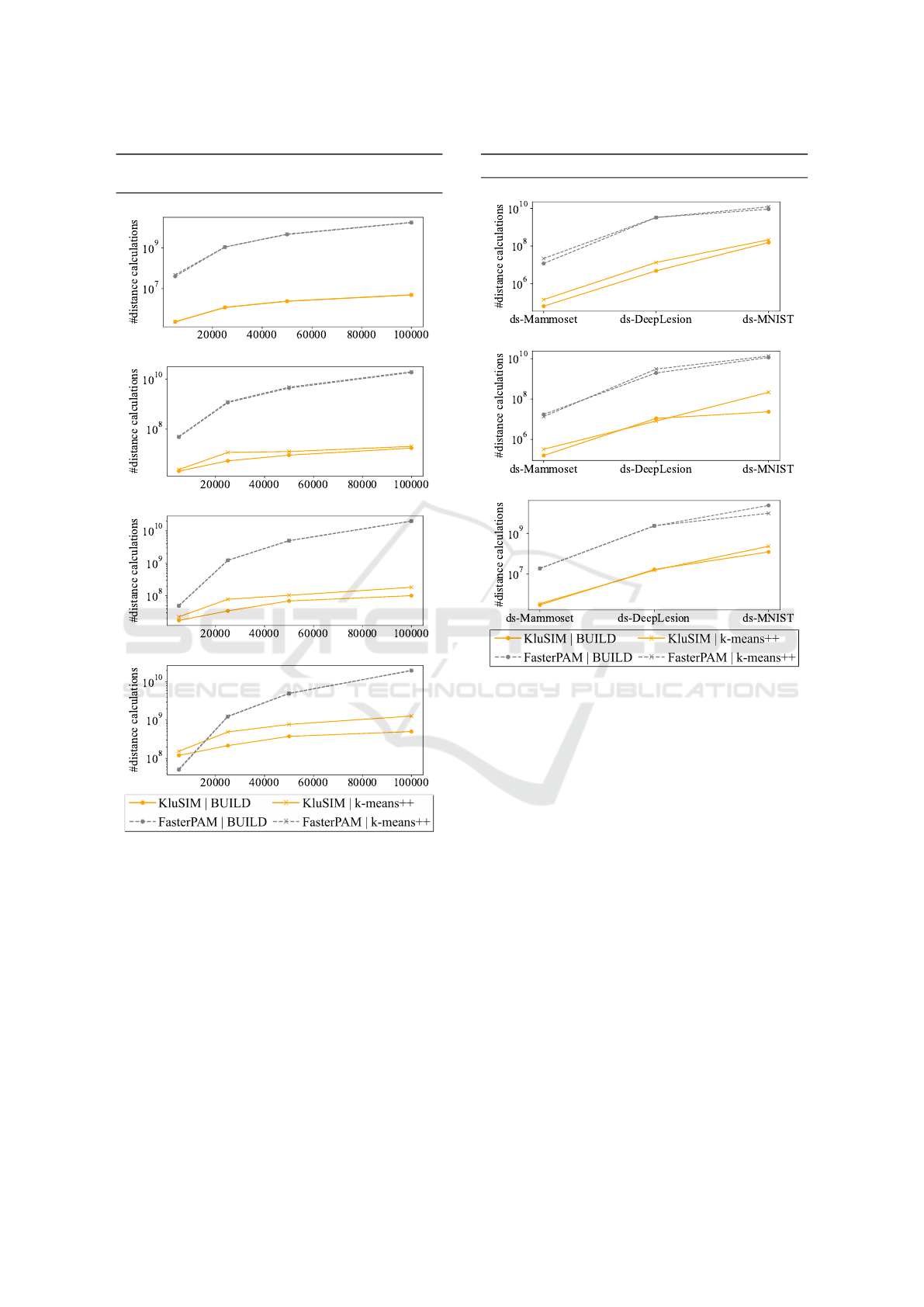
Synthetic Datasets - #distance calculations (log
scale)
(a) Number of clusters (k) = 5
(b) Number of clusters (k) = 20
(c) Number of clusters (k) = 50
(d) Number of clusters (k) = 100
Figure 7: KluSIM reduces the number of distance calcula-
tions by up to 3,500 times when compared to FasterPAM.
The plots show the number of distance calculations on syn-
thetic datasets for KluSIM and FasterPAM.
Mammoset. For the ds-MNIST dataset, KluSIM needs
193 times fewer distance calculations with build ini-
tialization when compared with FasterPAM.
5.5 Evaluating Cluster Quality
Finally, we evaluated the cluster quality. Enhancing
the speed of clustering algorithms is essential for han-
dling large datasets, while maintaining or even im-
proving the quality of the results. We investigate how
our proposal KluSIM strikes a balance between time
Real Datasets - #distance calculations (log scale)
(a) Dimension= 8
(b) Dimension= 64
(c) Dimension= 128
Figure 8: KluSIM shows a reduction in the number of dis-
tance calculations in the clustering process when compared
to FasterPAM. The plots shows the number of distance cal-
culations on real datasets for KluSIM and FasterPAM.
efficiency and cluster quality in comparison to Faster-
PAM employing synthetic and real datasets.
Figure 9 displays the average silhouette score (SS)
across all dimensions categorized by the number of
clusters and samples over synthetic datasets. The re-
sults show that KluSIM exhibits comparable cluster
quality to FasterPAM. Figure 10 illustrates the clus-
ter quality over real datasets. Similar to the synthetic
datasets, we observed an equivalent quality between
our method KluSIM and FasterPAM.
5.6 Discussion
The choice of the initialization method is a crucial as-
pect that influences our method KluSIM convergence
and overall cluster quality. Our experiments show the
build initialization method outperforming k-means++
in terms of quality, time efficiency, and number of dis-
tance calculations required for convergence. This su-
periority appears because build chooses each medoid
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
82
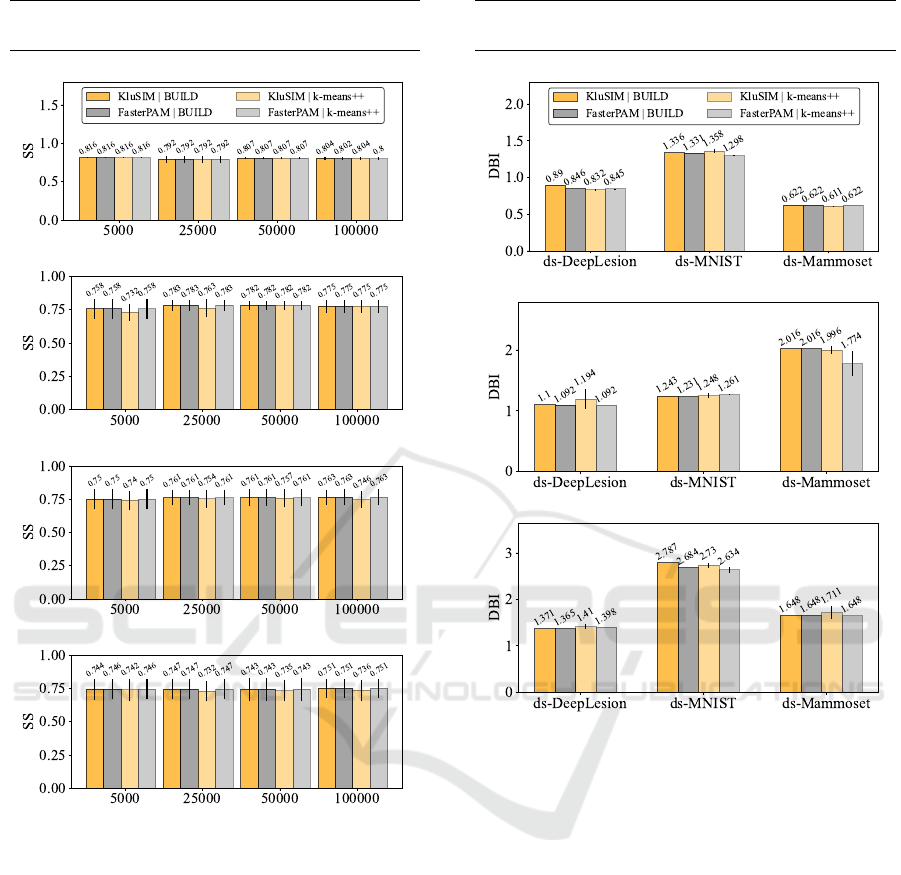
Synthetic Datasets - Cluster quality (SS: the
higher the better)
(a) Number of clusters (k) = 5
(b) Number of clusters (k) = 20
(c) Number of clusters (k) = 50
(d) Number of clusters (k) = 100
Figure 9: KluSIM quality of clustering is equivalent to the
quality of main competitor. Cluster quality over synthetic
datasets.
with an optimal quality (i.e. less TD). On the other
hand, k-means++ randomly selects the first and sub-
sequent objects that are likely to be far from each
other, without a focus on minimizing TD. Notably,
using build showcased a substantial speed-up of up
to 881 times. The improvement was particularly high
in scenarios where the number of clusters k was sub-
stantially smaller than the number of objects in the
dataset S (k ≪ n).
All in all, KluSIM has shown a significant reduc-
tion in the number of distance calculations, speeding
up existing approaches, and maintaining comparable
quality results.
Real Datasets - Cluster quality (DBI: the lower the
better)
(a) = 8 dimensions
(b) 64 dimensions
(c) 128 dimensions
Figure 10: KluSIM quality of clustering is equivalent to the
quality of the main competitor. Cluster quality over real
datasets.
6 CONCLUSION
This paper introduced the KluSIM approach, designed
to improve the efficiency of k-medoids algorithms by
incorporating a metric access methods to speed up
clustering. KluSIM focuses on the swap step pro-
cess, exhibiting significant improvements. Through
experimentation, KluSIM was compared to the base-
line FasterPAM, significantly reducing time in clus-
tering processing.
Our algorithm exhibited an average speedup of up
to 881 times when compared with the baseline Faster-
PAM, and a reduction of up to 3,500 times in distance
calculations, whereas maintaining a comparable clus-
tering quality. Unlike several methods in the litera-
ture, our algorithm does not store a distance matrix in
memory, eliminating a potential bottleneck faced by
other methods when clustering large datasets.
KluSIM: Speeding up K-Medoids Clustering over Dimensional Data with Metric Access Method
83

Our KluSIM proposal stands out as an efficient
and scalable solution for k-medoids clustering tasks.
The combination of metric access methods, opti-
mized initialization heuristics, and the elimination
of the need for a distance matrix in memory col-
lectively contribute to the outstanding performance
gains. Thus, KluSIM is a powerful tool for scalable
and high-performance clustering tasks, particularly
in scenarios with limited computational resources or
large datasets.
In future work, we intend to explore MAM-based
initialization heuristics to leverage the index structure
in the entire clustering process. We also want to eval-
uate KluSIM with other distance functions.
ACKNOWLEDGEMENT
This research was financed in part by the
Coordenac¸
˜
ao de Aperfeic¸oamento de Pessoal de
N
´
ıvel Superior - Brasil (CAPES) - Finance Code 001
and 12620352/M, by the S
˜
ao Paulo Research Founda-
tion (FAPESP, grants 2016/17078-0, 2020/11258-2),
the National Council for Scientific and Technological
Development (CNPq) and JIT Educac¸
˜
ao.
REFERENCES
Arthur, D. and Vassilvitskii, S. (2007). K-means++ the ad-
vantages of careful seeding. In Proceedings of the
eighteenth annual ACM-SIAM symposium on Discrete
algorithms, pages 1027–1035.
Barioni, M. C. N., Razente, H. L., Traina, A. J., and
Traina Jr, C. (2008). Accelerating k-medoid-based al-
gorithms through metric access methods. Journal of
Systems and Software, 81(3):343–355.
Bentley, J. L. (1975). Multidimensional binary search trees
used for associative searching. Communications of the
ACM, 18(9):509–517.
Cazzolato, M. T., Scabora, L. C., Zabot, G. F., Gutier-
rez, M. A., Traina Jr, C., and Traina, A. J. (2022).
Featset+: Visual features extracted from public image
datasets. Journal of Information and Data Manage-
ment, 13(1).
Davies, D. L. and Bouldin, D. W. (1979). A cluster separa-
tion measure. IEEE Transactions on Pattern Analysis
and Machine Intelligence, PAMI-1(2):224–227.
Han, J., Kamber, M., and Pei, J. (2011). Data Mining: Con-
cepts and Techniques, 3rd edition. Morgan Kaufmann.
Kaufman, L. (1990). Rousseeuw, pj: Finding groups in
data: An introduction to cluster analysis. Applied
Probability and Statistics, New York, Wiley Series in
Probability and Mathematical Statistics.
Kenger, O. N., Kenger, Z. D.,
¨
Ozceylan, E., and Mru-
galska, B. (2023). Clustering of cities based on
their smart performances: A comparative approach of
fuzzy c-means, k-means, and k-medoids. IEEE Ac-
cess, 11:134446–134459.
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
Lenssen, L. and Schubert, E. (2024). Medoid silhouette
clustering with automatic cluster number selection.
Information Systems, 120:102290.
Oliveira, P. H., Scabora, L. C., Cazzolato, M. T., Bedo,
M. V., Traina, A. J., and Traina-Jr, C. (2017). Mam-
moset: An enhanced dataset of mammograms. In
Satellite Events of the Brazilian Symp. on Databases.
SBC, pages 256–266.
Omohundro, S. M. (1989). Five balltree construction al-
gorithms. International Computer Science Institute
Berkeley.
Park, H.-S. and Jun, C.-H. (2009). A simple and fast algo-
rithm for k-medoids clustering. Expert systems with
applications, 36(2):3336–3341.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P.,
Weiss, R., Dubourg, V., et al. (2011). Scikit-learn:
Machine learning in python. the Journal of machine
Learning research, 12:2825–2830.
Qaddoura, R., Faris, H., and Aljarah, I. (2020). An efficient
clustering algorithm based on the k-nearest neighbors
with an indexing ratio. International Journal of Ma-
chine Learning and Cybernetics, 11(3):675–714.
Ran, X., Xi, Y., Lu, Y., Wang, X., and Lu, Z. (2023).
Comprehensive survey on hierarchical clustering al-
gorithms and the recent developments. Artificial In-
telligence Review, 56(8):8219–8264.
Schubert, E. and Rousseeuw, P. J. (2021). Fast and eager
k-medoids clustering: O (k) runtime improvement of
the pam, clara, and clarans algorithms. Information
Systems, 101:101804.
Tiwari, M., Zhang, M. J., Mayclin, J., Thrun, S., Piech, C.,
and Shomorony, I. (2020). Banditpam: Almost linear
time k-medoids clustering via multi-armed bandits. In
NeurIPS.
Traina, C., Traina, A., Faloutsos, C., and Seeger, B. (2002).
Fast indexing and visualization of metric data sets us-
ing slim-trees. IEEE Transactions on Knowledge and
Data Engineering, 14(2):244–260.
Vandanov, S., Plyasunov, A., and Ushakov, A. (2023). Par-
allel clustering algorithm for the k-medoids problem
in high-dimensional space for large-scale datasets.
In 2023 19th International Asian School-Seminar on
Optimization Problems of Complex Systems (OPCS),
pages 119–124.
Yan, K., Wang, X., Lu, L., and Summers, R. M. (2017).
Deeplesion: Automated deep mining, categorization
and detection of significant radiology image findings
using large-scale clinical lesion annotations. arXiv
preprint arXiv:1710.01766.
Yianilos, P. N. (1993). Data structures and algorithms for
nearest neighbor. In Proceedings of the fourth annual
ACM-SIAM Symposium on Discrete algorithms, vol-
ume 66, page 311. SIAM.
Zezula, P., Amato, G., Dohnal, V., and Batko, M. (2006).
Similarity search: the metric space approach, vol-
ume 32. Springer Science & Business Media.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
84
