
Applying Multiple Instance Learning for Breast Cancer Lesion Detection
in Mammography Images
Nedra Amara
1 a
and Said Gattoufi
2
1
INSA Centre Val de Loire, University of Orleans, LIFO EA 4022, F-45067, Orleans, France
2
SMART Laboratory, University of Tunis, Institut Sup
´
erieur de Gestion de Tunis, Tunisia
fi
Keywords:
Breast Cancer, Computer-Aided Detection, Multiple Instance Learning, Transfer Learning,
Mammography Images, Early Detection.
Abstract:
Breast cancer remains a major global health problem and early detection is essential to improve patient out-
comes. Current computer-aided detection (CAD) systems for breast cancer are often based on fully supervised
training, which requires careful manual annotation and accurate tumor segmentation. This paper presents a
novel approach based on multiple instance and transfer learning techniques. Our method uses an adapted
threshold segmentation technique to extract many small spots from mammography images. Instance features
are then extracted using a pre-trained model and grouped into a unified representation. A classifier trained on
these representations is used to classify the data. The proposed method eliminates the need for precise tumor
segmentation while demonstrating high accuracy in breast cancer detection.
1 INTRODUCTION
According to recent American Cancer Society statis-
tics, breast cancer will have the highest incidence and
mortality rate of any cancer type in 2020 Siegel et al.
(2023). The majority of breast cancers are detected by
abnormalities in breast tissue. It can take years for an
abnormality to develop into a malignant tumor. Early
detection can thus play an important role in breast
cancer prevention.
Currently, mammography is one of the most com-
mon methods of breast cancer screening. How-
ever, interpreting mammography results can be time-
consuming and inconsistent across radiologists, even
for the same patient. To address these limitations, a
variety of computer-aided diagnostic (CAD) systems
have been developed to detect abnormalities in mam-
mogram images.
Breast cancer decision support systems typically
include three major components: breast region seg-
mentation, feature extraction, and abnormality classi-
fication. Potential lesions are identified during breast
segmentation. For example, Khoulqi and Idrissi
(2019) used a mathematical morphology-based seg-
mentation algorithm to identify suspicious regions in
mammographic images Khoulqi and Idrissi (2019).
a
https://orcid.org/0000-0001-8794-7499
Gomez and his team used texture analysis to de-
tect the contours of breast lesions Gomez-Flores and
Ruiz-Ortega (2016). Reig and colleagues proposed
another method for segmenting suspicious tissue in
breast MRI images, which combines adaptive thresh-
olding techniques Reig et al. (2020)
Hirra et al. (2021) proposed a deep learning-based
method to improve lesion segmentation. Militello and
his team also used a semi-automatic segmentation ap-
proach, integrating clinical information to improve
tumor segmentation accuracy Militello et al. (2022).
These new methods emphasize the growing impor-
tance of advanced segmentation approaches for im-
proving lesion detection and characterization in breast
cancer.
Shape, size, texture, edge features, vasculariza-
tion, and kinetic features are distinguishing charac-
teristics of malignant tumours in breast cancer feature
extraction (Agner et al., 2011; Fusco et al., 2016). For
example, Hirra et al. (2021) investigated the use of
shape and texture features to characterize breast tu-
mors, extracting these distinguishing characteristics
using deep learning techniques. Similarly, sutton and
colleagues used morphology-based features to distin-
guish breast tumor types, incorporating multiparamet-
ric MRI data to improve accuracy Sutton et al. (2015).
In a similar manner Moura and Guevara L
´
opez
(2013) used texture and edge features to character-
Amara, N. and Gattoufi, S.
Applying Multiple Instance Learning for Breast Cancer Lesion Detection in Mammography Images.
DOI: 10.5220/0012689500003699
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2024), pages 93-97
ISBN: 978-989-758-700-9; ISSN: 2184-4984
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
93
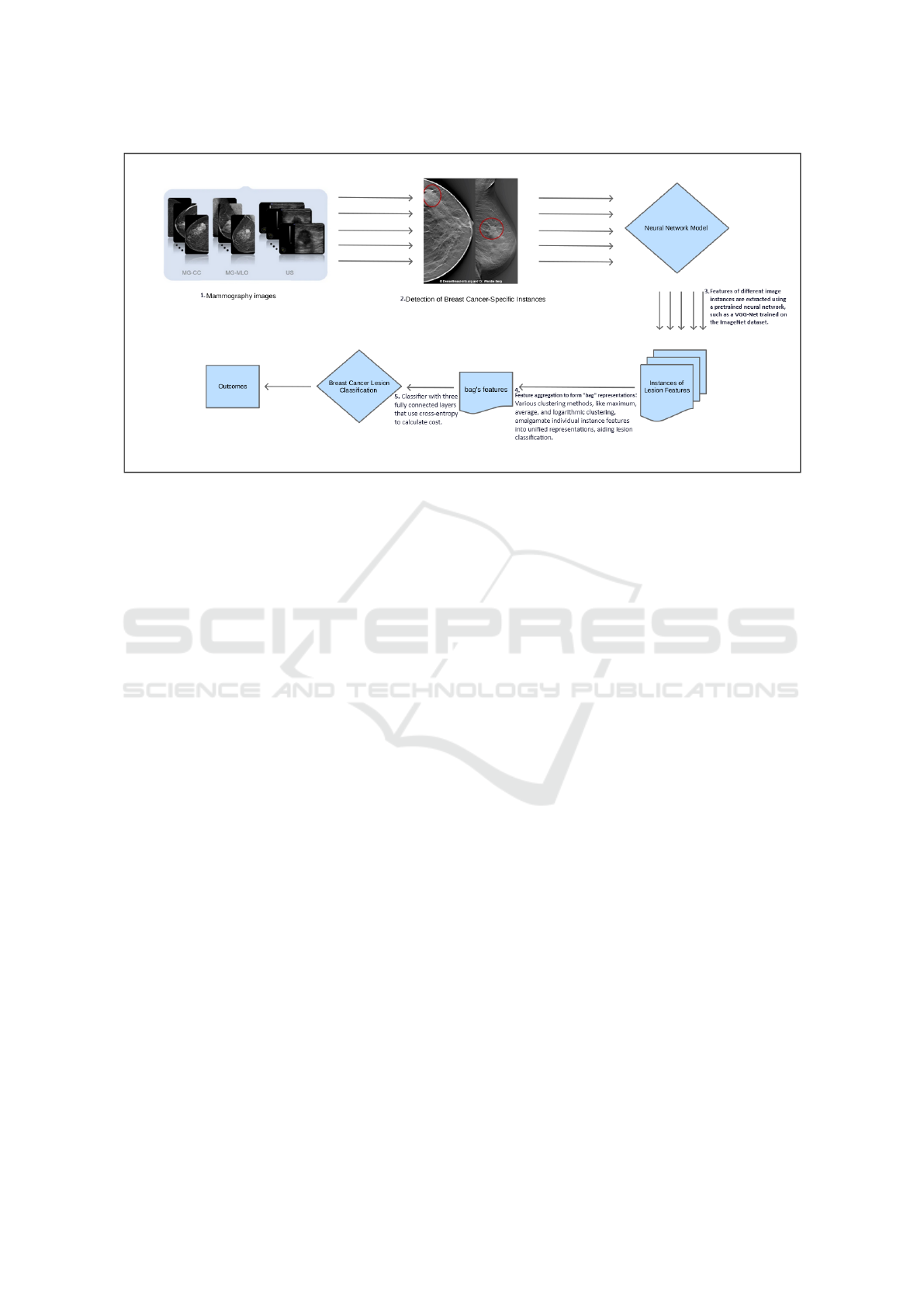
Figure 1: Proposed system for Lesion Detection and Classification in Breast Mammography Images.
ize breast tumors, employing geometric transforma-
tions to generate new discriminating features. Fur-
thermore, Agner et al. (2011) used dynamic kinetic
features extracted from dynamic imaging sequences
to determine the malignancy of breast tumors. These
approaches emphasize the importance of extracting
specific features from breast tumors and using a vari-
ety of techniques to better differentiate malignant tu-
mors.
The morphological, statistical, and textural fea-
tures of tumors in mammographic images are ex-
tracted and classified using various classification al-
gorithms. Most existing breast cancer decision sup-
port systems have three steps: identify tumor can-
didates in images, extract features from each tumor,
and classify each breast tumor as negative or posi-
tive. These methods rely on fully supervised learning,
which necessitates tedious manual annotation of ob-
ject locations in a training set. Furthermore, there are
no publicly accessible mammography datasets with
annotated tumors.
Because tumors are small in comparison to the im-
age size, and there are numerous artifacts in mammo-
graphic images, classification of the image set yields
poor results. To address these limitations, we pro-
posed a recent approaches, based on transfer learning
to improve mammography image classification.
The rest of this article is organized as follows:
Section II describes our method, while Section III
presents experiments and results. Section IV is de-
voted to discussions and conclusions.
2 BREAST CANCER LESION
DETECTION: MIL APPROACH
AND LEARNING TRANSFER
This section presents the Multiple Instance Learning
(MIL) formulation, defines learning transfer, and out-
lines the proposed system structure.
A) MIL.
MIL aims to learn f : X → Y using a training data set
D = (x
1
, y
1
), . . . , (x
m
, y
m
), where X
i
= x
i1
, . . . , x
im
. X
is referred to as a bag, while X
( j1,...,m
i
)
represents an
instance. The number of instances in X
i
is denoted by
m
i
, and y
i
∈ Y = {Y, N}. X
i
is a positive bag, which
means that y
i
= Y if there is a positive x
ip
, whereas
p ∈ {1, . . . , m
i
} are unknown. The goal is to predict
labels for unseen bags. In the case of breast cancer,
this method could be used to learn how to identify
and characterize lesion features from mammographic
image datasets. These lesions could be referred to as
”bags,” and the features to be extracted would be the
”instances” of these bags.
The basic idea behind MIL is to assign class labels
globally, rather than individually to each instance.
This implies that if a bag contains at least one posi-
tive instance (such as a region with a lesion), it is con-
sidered positive. When using MIL to classify breast
cancer lesions, bags may represent complete mam-
mographic images, while instances may represent re-
gions of these images that could contain lesions. The
features of these instances are then aggregated to cre-
ate bag representations, and the bag is classified ac-
cording to these aggregated representations.
ICT4AWE 2024 - 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health
94
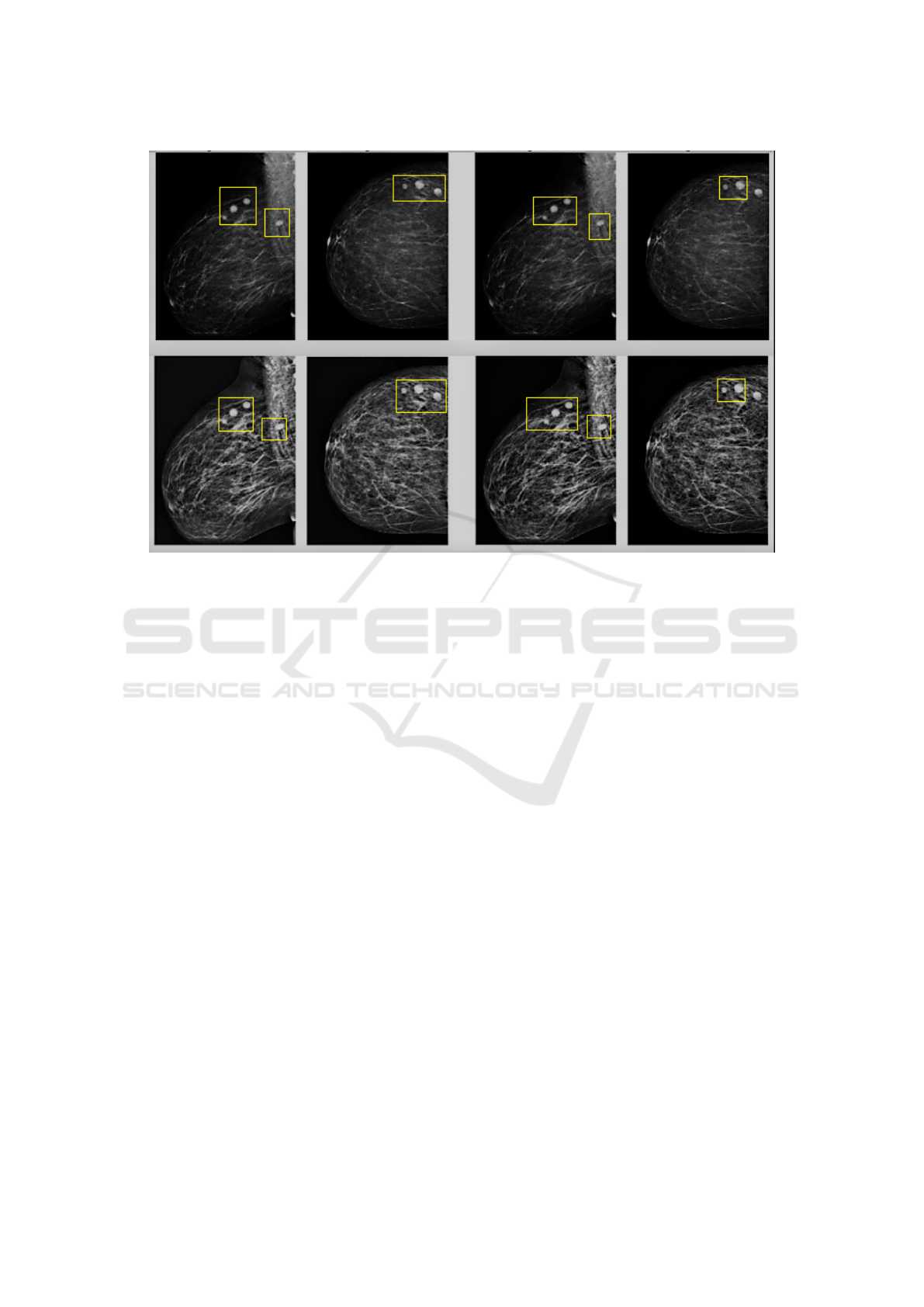
Figure 2: Breast region segmentation and instance identification.
B) Learning Transfer.
In recent years, deep convolutional neural networks
(DCNNs) have quickly become the preferred method-
ology for medical image analysis. However, robust
supervised training of a DCNN necessitates the use
of a large number of annotated images Papandreou
et al. (2015). Transfer learning entails using pre-
trained networks to avoid the need for large datasets in
deep network training Marcelino (2018); Baykal et al.
(2020). In medicine, two learning transfer strategies
have been used: the first uses a pre-trained network
as a feature extractor, and the second refines a pre-
trained network using training data. In the breast can-
cer context, transfer learning could be applied to pre-
train neural networks on large datasets of general im-
ages, and then adapt these models to analyze mammo-
graphic images more specifically. This would make it
possible to use and adjust features learned from big
data to enhance the detection and identification of le-
sions in breast cancer images.
C) Proposed System for Lesion Detection and
Classification.
Our proposed system, illustrated in Figure 1, outlines
the learning structure. First, mammographic images
are used for segmenting the breast region. The im-
age is then split into a number of smaller regions. In
our case, an image can be thought of as a bag, and
the regions extracted from it as instances. We then
use a pretrained network to learn these instance fea-
tures, and a clustering layer to aggregate these in-
stance scores into a score for the whole bag. Fi-
nally, we initialize the classification layer with ran-
dom weights and set it up for mammography image
classification.
This approach can be tailored to breast cancer by
segmenting relevant areas of breast images, extract-
ing features from regions of interest and using a pre-
trained neural network to classify and identify rele-
vant features of lesions or tumoral tissues.This would
result in an efficient system for automatically ana-
lyzing mammographic images in order to detect and
characterize abnormalities associated with breast can-
cer.
a) Breast Cancer Segmentation.
Firstly, threshold segmentation is employed to detect
the breast area, followed by morphological processing
to eliminate noise.
b) Instance Identification for Breast Cancer Lesion
Localization.
The mammographic images are divided into several
parts based on the segmentation results of the breast
region. Each part is treated as a bag, with each area
acting as an instance within the bag. Figure 2 il-
lustrates the breast region segmentation and instance
identification.
c) Feature Extraction for Lesion Detection.
To extract fixed features, we employ a VGG-Net that
has already been trained on the ImageNet dataset. We
Applying Multiple Instance Learning for Breast Cancer Lesion Detection in Mammography Images
95

Table 1: Comparison of Breast Cancer Detection Algorithms.
Approaches Accuracy Precision Recall AUC
ResNet 0.8323 0.7750 0.8611 0.8323
VGG 0.7688 0.7648 0.8056 0.7758
Mean MILIL 0.8472 0.8049 0.9277 0.8333
Max MILTL 0.9182 0.8260 0.9277 0.9277
Log MILIL 0.8790 0.8039 0.8789 0.8867
commence by extracting features from each instance
using the feature extractor, and then utilize a clus-
tering layer to aggregate these instance features into
a bag. The proposed system explores three cluster-
ing methods: maximum clustering, average cluster-
ing, and logarithmic clustering.
d) Classification of Cancer Lesions.
For classification, we construct a classifier with three
fully connected layers that utilize cross-entropy to
calculate cost. These steps are adapted to ana-
lyze mammographic breast cancer images specifically
by identifying relevant regions, extracting significant
features, and using a pre-trained network to classify
and select relevant lesion or tumor tissue features.
3 EXPERIMENTS AND RESULTS
IN BREAST CANCER
DETECTION
A) Materials.
The mammography data used in this study consist of
78 cases from The Cancer Imaging Archive (TCIA),
comprising 41 cases with lesions ranging in size from
5 to 9 mm and 37 cases with at least one lesion mea-
suring 10 mm or larger. Each patient case includes
two images, one in front and one in profile, totaling
two positive images (with a lesion). We also ran-
domly selected an equal number of negative images
from cases where no lesion was found.
Although our data case is limited, it still has rele-
vant features for our research on detecting breast can-
cer lesions. It’s curcial to consider that our dataset
may not be fully representative, and that the results
of our study may be influenced by its specific compo-
sition. As researchers, we have taken numbres steps
to minimize the potential biases associated with using
this dataset. For example, we use a ten-point cross-
validation method to assess classification results and
reduce the risk of assessment bias. In addition, 10 it-
erations were carried out to thoroughly evaluate the
statistical results of our study. All these steps allowed
to improve the consistency of our results.
B) Experimental and Evaluation Setup.
In this section, we conducted two comparative exper-
iments with the VGG-16 and ResNet50 pre-trained
networks, respectively. These models are available
through the TensorFlow model repository. We uti-
lized a ten-fold cross-validation method to evalu-
ate classification performance and mitigate evalua-
tion bias. Our study’s evaluation metrics include ac-
curacy, precision, recall, and AUC. Additionally, we
conducted 10 trials to assess the statistical results.
C) Results.
Table 1 indicates that the models we constructed out-
perform the existing VGG-16 and ResNet-50 pre-
trained networks. Furthermore, MILTL with a maxi-
mum clustering layer outperforms the other two meth-
ods, with an accuracy of 0.9182 and an AUC of
0.9277. These findings demonstrate the efficacy of
the developed methods, which were specifically tai-
lored for the analysis of mammographic images for
breast cancer. They emphasize the importance of us-
ing specialized methods to enhance classification per-
formance in this context.
4 CONCLUSION
Multiple Instance Learning (MIL) provides an ex-
cellent framework for classifying mammography im-
ages. In this work, we propose a new approach for
the automatic detection of breast lesions using mam-
mography. The method includes breast region seg-
mentation, instance identification, feature extraction,
and classification. Because of the nature of the MIL
method, breast region segmentation does not neces-
sitate precise segmentation results, which undeniably
simplifies and saves time for lesion detection. Our
method allows for improved classification accuracy.
In the future, we will focus on the probability rela-
tionship between the bag and the instances to ensure
instance labeling, especially for positive instances.
ACKNOWLEDGEMENTS
We sincerely thank the Regional Hospital Center of
Orleans, France, for collaborating with us and sup-
porting our research.
ICT4AWE 2024 - 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health
96

REFERENCES
Agner, S. C., Soman, S., Libfeld, E., McDonald, M.,
Thomas, K., Englander, S., Rosen, M. A., Chin, D.,
Nosher, J., and Madabhushi, A. (2011). Textural ki-
netics: a novel dynamic contrast-enhanced (dce)-mri
feature for breast lesion classification. Journal of dig-
ital imaging, 24:446–463.
Baykal, E., Dogan, H., Ercin, M. E., Ersoz, S., and Ek-
inci, M. (2020). Transfer learning with pre-trained
deep convolutional neural networks for serous cell
classification. Multimedia Tools and Applications,
79:15593–15611.
Fusco, R., Sansone, M., Filice, S., Carone, G., Amato,
D. M., Sansone, C., and Petrillo, A. (2016). Pattern
recognition approaches for breast cancer dce-mri clas-
sification: a systematic review. Journal of medical and
biological engineering, 36:449–459.
Gomez-Flores, W. and Ruiz-Ortega, B. A. (2016). New
fully automated method for segmentation of breast le-
sions on ultrasound based on texture analysis. Ultra-
sound in medicine & biology, 42(7):1637–1650.
Hirra, I., Ahmad, M., Hussain, A., Ashraf, M. U., Saeed,
I. A., Qadri, S. F., Alghamdi, A. M., and Alfa-
keeh, A. S. (2021). Breast cancer classification
from histopathological images using patch-based deep
learning modeling. IEEE Access, 9:24273–24287.
Khoulqi, I. and Idrissi, N. (2019). Breast cancer image seg-
mentation and classification. In Proceedings of the 4th
International Conference on Smart City Applications,
pages 1–9.
Marcelino, P. (2018). Transfer learning from pre-trained
models. Towards data science, 10:23.
Militello, C., Rundo, L., Dimarco, M., Orlando, A., Conti,
V., Woitek, R., D’Angelo, I., Bartolotta, T. V., and
Russo, G. (2022). Semi-automated and interactive
segmentation of contrast-enhancing masses on breast
dce-mri using spatial fuzzy clustering. Biomedical
Signal Processing and Control, 71:103113.
Moura, D. C. and Guevara L
´
opez, M. A. (2013). An evalua-
tion of image descriptors combined with clinical data
for breast cancer diagnosis. International journal of
computer assisted radiology and surgery, 8:561–574.
Papandreou, G., Chen, L.-C., Murphy, K. P., and Yuille,
A. L. (2015). Weakly-and semi-supervised learning of
a deep convolutional network for semantic image seg-
mentation. In Proceedings of the IEEE international
conference on computer vision, pages 1742–1750.
Reig, B., Heacock, L., Geras, K. J., and Moy, L. (2020).
Machine learning in breast mri. Journal of Magnetic
Resonance Imaging, 52(4):998–1018.
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A.
(2023). Cancer statistics, 2023. Ca Cancer J Clin,
73(1):17–48.
Sutton, E. J., Oh, J. H., Dashevsky, B. Z., Veeraraghavan,
H., Apte, A. P., Thakur, S. B., Deasy, J. O., and Mor-
ris, E. A. (2015). Breast cancer subtype intertumor
heterogeneity: Mri-based features predict results of a
genomic assay. Journal of Magnetic Resonance Imag-
ing, 42(5):1398–1406.
Applying Multiple Instance Learning for Breast Cancer Lesion Detection in Mammography Images
97
